Abstract
The neck-linker is a structurally conserved region among most members of the kinesin superfamily of molecular motor proteins that is critical for kinesin’s processive transport of intracellular cargo along the microtubule surface. Variation in the neck-linker length has been shown to directly modulate processivity in different kinesin families; for example, kinesin-1, with a shorter neck-linker, is more processive than kinesin-2. Although small differences in processivity are likely obscured in vivo by the coupling of most cargo to multiple motors, longer and more flexible neck-linkers may allow different kinesins to navigate more efficiently around the many obstacles, including microtubule-associated proteins (MAPs), that are found on the microtubule surface within cells. We hypothesize that, due to its longer neck-linker, kinesin-2 can more easily navigate obstacles (e.g., MAPs) on the microtubule surface than kinesin-1. We used total internal reflection fluorescence microscopy to observe single-molecule motility from different kinesin-1 and kinesin-2 neck-linker chimeras stepping along microtubules in the absence or presence of two Tau isoforms, 3RS-Tau and 4RL-Tau, both of which are MAPs that are known to differentially affect kinesin-1 motility. Our results demonstrate that unlike kinesin-1, kinesin-2 is insensitive to the presence of either Tau isoform, and appears to have the ability to switch protofilaments while stepping along the microtubule when challenged by an obstacle, such as Tau. Thus, although kinesin-1 may be more processive, the longer neck-linker length of kinesin-2 allows it to be better optimized to navigate the complex microtubule landscape. These results provide new insight, to our knowledge, into how kinesin-1 and kinesin-2 may work together for the efficient delivery of cargo in cells.
Introduction
Intracellular transport is a highly regulated bidirectional process that is required for normal cellular function, particularly in neurons, where anterograde cargo is transported to specific locations throughout the cell periphery and retrograde cargo is transported in the opposite direction (1,2). Through specific adaptor proteins, cargos couple with an ensemble of molecular motors, including members of the kinesin, myosin, and dynein families. Different combinations of these motors, even from the same family, modulate cargo transport (3). For example, purified vesicles from adult mouse brains have been shown to contain dynein, kinesin-1, and kinesin-2, indicating an important regulatory role for that ensemble and other potential motor combinations in microtubule-based cargo transport (4). Similar observations have been made for intraflagellar transport (IFT) particles that contain two kinesin-2 family members in cilia/flagella (5) and ribonucleoprotein (RNP) complexes in Xenopus oocytes (6) that contain both kinesin-1 and kinesin-2. Although models of bidirectional transport have traditionally focused on two opposing motors, kinesin-1 and dynein, an unresolved question is why cargos need two directionally similar motors to drive anterograde transport. Presumably, different motor domains contribute to functionally distinct transport characteristics, but the significance of having at least two different plus-end-directed kinesin motors on the same cargo within the cell is presently unknown.
Kinesin-1 and kinesin-2 are two plus-end-directed microtubule-based motors that are colocalized on the same cargo in a number of different intracellular transport processes (4,7,8). Although it is known that kinesin-1 is more processive than kinesin-2 due to differences in the contour length of their neck-linker regions (9–11), the physiological relevance of this difference is unclear given that many cargos are bound to multiple motors, which minimizes any potential differences in processivity at the single-motor level (12). We hypothesize that, even more important than its effect on processivity, the neck-linker region contour length in kinesin-1 and kinesin-2 (Fig. 1, A and B) determines the efficiency in which specific kinesin family members can navigate obstacles on the microtubule surface in the crowded intracellular environment. Specifically, we expect kinesin-2’s longer neck-linker region (17 amino acids) to be more flexible than kinesin-1’s (14 amino acids), which would allow it to more easily navigate the complex microtubule landscape within the cell.
Figure 1.
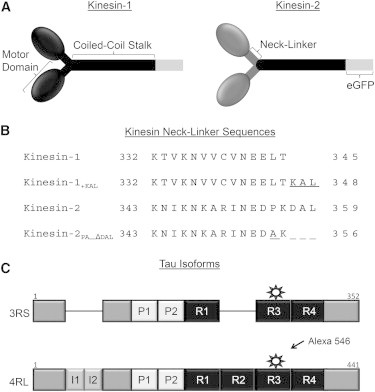
Experimental reagents. (A) Schematic of kinesin constructs illustrating the N-terminal globular motor domains, C-terminal coiled-coil stalk, and the random-coil neck-linker connecting the two motor domains. The C-terminal end of Drosophila kinesin-1 was truncated at 559 and fused with an eGFP. The kinesin-2 construct contained two mouse kif3A motor domains and their neck-linkers fused with the coiled-coil stalk of the kinesin-1 construct, which has been shown to be functionally equivalent to the wild-type kif3A/B heterodimer (9). (B) Primary amino acid sequence of the neck-linker regions of all four kinesin constructs used in the experiments. Kinesin-1’s 14 amino acid neck-linker was lengthened to 17 (kinesin-1+KAL), and kinesin-2’s 17 amino acid neck-linker was shortened to 14 (kinesin-2PA_ΔDAL) as described by Shastry and Hancock (10). (C) Linear schematic of 3RS- and 4RL-Tau isoforms containing an acidic N-terminal region, a central proline-rich region (P1 and P2), and a microtubule-binding region with three or four microtubule-binding repeats (R1–R4). 4RL-Tau contains two additional N-terminal acidic inserts (I1 and I2) and one additional C-terminal microtubule-binding repeat (R2). Tau isoforms were labeled with Alexa 546 at a single cysteine residue in R3.
This landscape contains a number of different microtubule-associated proteins (MAPs) that kinesin-1 and kinesin-2 must navigate to deliver the cargos to their target destinations. Many cell types, including neurons, contain microtubules decorated with Tau/MAP2/MAP4 family members, which have similar C-terminal microtubule-binding repeats and have been shown to impede kinesin-1 motility in vitro and in vivo (13,14). Tau is a highly expressed MAP in the axon of nerve cells that decorates the microtubule surface (15). There are six human isoforms of Tau (16) and they have been shown to aid in polymerization and stabilization of microtubules in vitro (17). 3RS-Tau is the shortest isoform, containing three C-terminal microtubule-binding repeats and no N-terminal acidic inserts, whereas the longest 4RL isoform contains four C-terminal microtubule-binding repeats and two N-terminal acidic inserts (Fig. 1 C). Both isoforms were shown to impede kinesin-1 motility in vitro, with 3RS-Tau being more inhibitory than 4RL-Tau (18–20), but the effect of Tau on kinesin-2’s motility is unknown. Intriguingly, nonmammalian kinesin-2 motor proteins have been observed in vitro to be capable of sidestepping to adjacent protofilaments on the microtubule surface, unlike kinesin-1 and mammalian kinesin-2, which have been shown to track a single protofilament in their processive walk along the microtubule surface (21). We hypothesize that, at the expense of its processivity, kinesin-2’s longer neck-linker region allows this motor (even mammalian kinesin-2) to navigate around obstacles such as Tau.
The purpose of this work was to determine the role of the neck-linker composition in kinesin’s ability to step along microtubules in the presence or absence of MAPs known to impede kinesin-1 motility (3RS-Tau and 4RL-Tau). We compare and contrast the characteristic run lengths, average velocities, and pause behavior for kinesin-1 and kinesin-2 constructs on different microtubule complexes in vitro, including chimeras in which the kinesin-1 and kinesin-2 neck-linker regions are effectively switched. Our results demonstrate the importance of kinesin’s neck-linker in its ability to navigate around obstacles such as Tau on the microtubule surface, and provide important new insight (to our knowledge) into the role of multiple kinesin family members in transporting single cargos through the complex intracellular environment.
Materials and Methods
Protein expression and purification
All kinesin constructs contained Drosophila kinesin-1 neck and stalk domains (residues 346–559) fused with C-terminal eGFP and hexahistidine tags, and were expressed and purified as previously described (10). Kinesin-1 constructs contained the N-terminal motor domain and neck-linker from Drosophila KIF5 (residues 1–345), and kinesin-2 constructs contained the motor domain and neck-linker from mouse kif3A (residues 1–359). The kinesin-2pa_ΔDAL construct also included the deletion of the last three amino acids (D, A, and L) and a single amino acid substitution (P355A) in the kif3A neck-linker. The kinesin-1+KAL construct included the addition of three amino acids (K, A, and L) between T344 and A345 of the Drosophila KIF5 neck-linker.
Tau was expressed from 3RS- and 4RL-Tau plasmids (a generous gift from Dr. Stephen King) in BL21-CodonPlus(DE3)-RP E. coli cells (Stratagene, La Jolla, CA) using the isopropyl 1-thio-β-D-galactopyranoside-inducible pET vector system (Novagen, Madison, WI) and purified as previously described (20,22). The 4RL-Tau construct contained a single amino acid substitution, C291I, to limit the labeling to one binding site, and was created using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). Tau concentration was determined using the Bicinchoninic Acid Protein Assay (Pierce, Rockford, IL) using desalted, lyophilized 3RS- or 4RL-Tau as standards. Samples were dialyzed against BRB80 (80 mM PIPES, pH 6.9 at room temperature, 1 mM EGTA, and 1 mM MgCl2). Tubulin was isolated from bovine brain (obtained from Vermont Livestock & Slaughter, Ferrisburgh, VT) using high-molarity PIPES buffer (1 M PIPES, pH 6.9 at room temperature, 10 mM MgCl2, and 20 mM EGTA) as previously described (23). Tubulin concentration was determined using the Bradford Assay (Sigma-Aldrich, St. Louis, MO).
Fluorescent labeling of Tau
Tau protein was incubated with a 10-fold molar excess of dithiothreitol (DTT) for 2 h at room temperature, and the DTT was removed using a 2 mL 7K MWCO Zeba spin desalting column (Pierce, Rockford, IL). Tau was then incubated in a 10-fold molar excess of Alexa Fluor 546-C5 maleimide (Invitrogen, Grand Island, NY) for an additional 2 h at room temperature, and excess fluorophore was removed using a second desalting column. The labeling efficiency of Tau was determined by comparing the concentration of fluorophore with that of protein. Alexa Fluor 546 concentration was determined using an extinction coefficient of 93,000 cm−1 M−1 at 554 nm in a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Rockford, IL). The 3RS-isoform was labeled at C233 and the 4RL-isoform was labeled at C322. Labeling efficiency was determined to be 79–85% for both Tau isoforms.
Microtubule preparation and labeling
Tubulin was thawed on ice and centrifuged at 350,000 × g for 20 min at 4°C before polymerization. The tubulin was then supplemented with either 1 mM GTP (Sigma-Aldrich) or guanosine-5′-[(α,β)-methyleno]triphosphate sodium salt (GMPCPP; Jena Bioscience, Jena, Germany), and unlabeled tubulin was mixed with rhodamine-labeled tubulin (Cytoskeleton, Denver, CO) at a 1:10 labeled/unlabeled ratio. For paclitaxel microtubules, 100 μL of 39 μM tubulin was incubated in BRB80 at 37°C for 20 min, followed by addition of paclitaxel (Sigma-Aldrich) to a final concentration of 20 μM. For GMPCPP microtubules, 20 μL of 39 μM GMPCPP-tubulin was incubated in BRB80 at 37°C for 20 min, followed by four additions of 20 μL of 39 μM GMPCPP-tubulin, each followed with a 20 min incubation before the next addition (20). This process ensured sufficiently long microtubules suitable for use in the single-molecule imaging experiments.
For experiments performed in the presence of Tau, tubulin polymerization was performed as described above except that labeled tubulin was excluded. Instead, stabilized microtubules were incubated with either Alexa 546 3RS- or 4RL-Tau at a 1:5 Tau/tubulin ratio at 37°C for an additional 20 min. The samples were centrifuged at room temperature for 30 min at 16,000 × g and the pellet was resuspended at 37°C in motility assay buffer (MAB) (10 mM PIPES, pH 7.4 at room temperature, 50 mM potassium acetate, 4 mM magnesium acetate, 1 mM EGTA), 10 mM DTT, and an oxygen scavenger system (5.8 mg/mL glucose, 0.045 mg/mL catalase, and 0.067 mg/mL glucose oxidase; Sigma-Aldrich). All solutions containing paclitaxel microtubules were supplemented with 20 μM paclitaxel.
Single-molecule total internal reflection fluorescence assay
Flow chambers were prepared by adhering ARTUS shims (ARTUS, Englewood, NJ) with Norland optical adhesive (Norland Products, Cranbury, NJ) to siliconized glass coverslips. Samples were prepared by incubating the flow chamber with monoclonal anti-β III (neuronal) antibodies (Sigma-Aldrich) at 33 μg/mL in MAB for 5 min. The chambers were washed and blocked with 0.5 mg/mL of bovine serum albumin (BSA) in MAB for an additional 2 min before addition of 1 μM of the desired microtubule preparation, followed by incubation for another 12 min. The chambers were washed with MAB, and 1 nM of the desired kinesin construct along with 1 mM ATP was added just before image acquisition for all experimental conditions examined. It should be noted that due to differences in buffer conditions, the kinesin run-length data we obtained in MAB differ from values previously measured in BRB80 with the same constructs (10).
We performed total internal reflection fluorescence (TIRF) microscopy at room temperature using an inverted microscope (Eclipse Ti-U; Nikon, Melville, NY) equipped with a 100× plan apochromatic objective lens (1.49 NA) and auxiliary 1.5× magnification. Kinesin-eGFP constructs were excited with a 473 nm argon laser and imaged through an emission 525/50 band-pass filter. Alexa 546-labeled 3RS-Tau or 4RL-C291I Tau and rhodamine-labeled tubulin were excited with a 532 nm argon laser and imaged through an emission 605/70 band-pass filter. We obtained images using an XR/Turbo-Z camera (Stanford Photonics, Palo Alto, CA) running Piper Control software (v2.3.39). The pixel resolution was 95.0 nm and all movies were acquired at five frames per second, with the exception of kinesin-2 GMPCPP data, which were acquired at 3.33 frames per second. Representative movies of the single-molecule TIRF assay are provided in the Supporting Material (Movies S1, S2, S3, and S4). Kinesin photobleaching rate was determined to be 7.8 s (Fig S1).
Data analysis
Motility was measured using the MTrackJ plugin for ImageJ software, version 1.46r (National Institutes of Health, Bethesda, MD), and track lengths were measured using the segmented line tool in ImageJ. Average velocity values from events were plotted in a histogram and fit to a Gaussian distribution, and the mean and standard deviation (SD) were reported. Characteristic run-length measurements were calculated as described by Thompson et al. (24). In brief, run-length events were fit to a cumulative frequency plot to determine the characteristic run length. A calculated characteristic run length, Xexpected, was determined to minimize any track distribution bias effects (error was the 99% confidence level of the resampled data set repeated 1000 times). All reported characteristic run lengths are reported as Xexpected. Lastly, a permutation resampling algorithm was used to determine significance difference between two data sets (24).
Pause events were scored from kymographs generated from kinesin-1 and kinesin-2 motility in the presence or absence of 3RS-Tau on paclitaxel microtubules using the multiple-kymograph plugin with ImageJ (see Fig. 4). Pauses were further categorized as stepping after a pause (pause-step) or terminating after a pause (pause-terminate). A pause was defined as movement ≤5 pixels in the spatial direction and ≥0.4 s in the temporal direction. (The estimated Rayleigh diffraction-limited spot for an eGFP fluorophore is 274 nm. The camera resolution was 95 nm/pixel and the spot size was 2.9 pixels, but to be conservative the spot was widened to 5 pixels or 475 nm.) A Z test was used to measure significance of proportions for all pause data, and a Mann-Whitney U test was used to determine significance for all pause dwell times.
Figure 4.
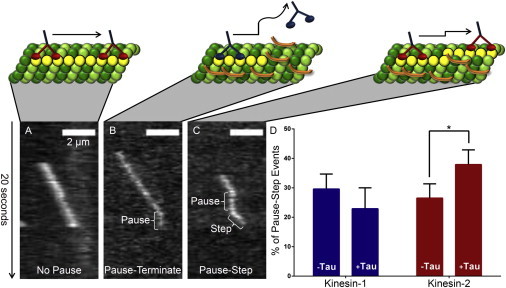
Representative kymograph images of kinesin-2 pausing events observed during processive movement along paclitaxel microtubules. (A) Uninterrupted processive movement (nonpause event) in the absence of Tau. (B) Pause-termination event in the presence of 3RS-Tau. (C) Pause-step event in the presence of 3RS-Tau. Scale bars represent 2 μm. Animations in panels A–C are not drawn to scale and are for visual effect. (D) Kinesin-1 and kinesin-2’s percentage of pause-step events in the absence and presence of 3RS-Tau. Kinesin-2, in the presence of 3RS-Tau, is more likely to step after a pause relative to kinesin-1. Error bars represent SE. ∗Represents significant difference between the absence and presence of 3RS-Tau, p = 0.05.
Results
Kinesin-2’s characteristic run length is not affected by Tau
The single-molecule motility of both kinesin-1 and kinesin-2 motors was observed by TIRF microscopy in the absence and presence of the 3RS and 4RL isoforms of Tau on either paclitaxel microtubules, representing a GDP nucleotide state, or GMPCPP microtubules, mimicking a GTP nucleotide state of the microtubule lattice. Although the presence of 3RS- and 4RL-Tau or the GMPCPP nucleotide state of the microtubule lattice was previously shown to reduce the in vitro motility of kinesin-1 (18–20), its effects on kinesin-2 motility are currently unknown.
In the absence of Tau, kinesin-1’s characteristic run length was observed to be 33% higher on paclitaxel microtubules than on GMPCPP microtubules (1.53 ± 0.27 μm vs. 1.15 ± 0.30 μm, p = 3 × 10−4; Figs. 2 A and 3 A; Tables 1 and 2), in agreement with previous results (20). Kinesin-2’s characteristic run length was also increased by 26% on paclitaxel microtubules relative to that observed on GMPCPP microtubules (1.03 ± 0.24 μm vs. 0.81 ± 0.16 μm, p = 1 × 10−3; Figs. 2 D and 3 D; Tables 1 and 2).
Figure 2.
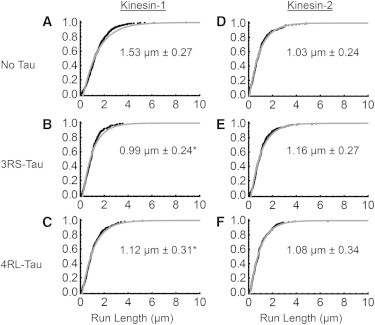
Characteristic run length comparison between kinesin-1 and kinesin-2 on paclitaxel microtubules. (A–C) Cumulative frequency plots of kinesin-1 in the absence or presence of 3RS- or 4RL-Tau. (D and E) Cumulative frequency plots of kinesin-2 in the absence or presence of 3RS- or 4RL-Tau. Black dots represent the raw run-length data and the gray curve is the observed cumulative frequency. The expected characteristic run length, derived from the microtubule length distribution, is shown within each graph. The error represents the 99% confidence interval and a p-value of less than 0.01 was considered significant. ∗Represents a statistically significant difference from the characteristic run length observed in the absence of Tau.
Figure 3.
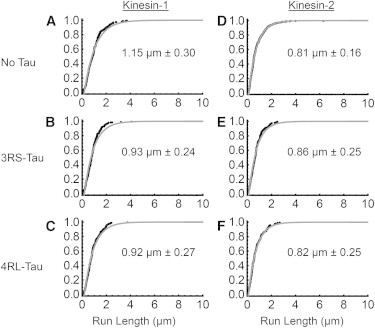
Characteristic run length comparison between kinesin-1 and kinesin-2 on GMPCPP microtubules. (A–C) Cumulative frequency plots of kinesin-1 in the absence or presence of 3RS- or 4RL-Tau. (D and E) Cumulative frequency plots of kinesin-2 in the absence or presence of 3RS- or 4RL-Tau. Black dots represent the raw run-length data and the gray curve is the observed cumulative frequency. The expected characteristic run length, derived from the microtubule length distribution, is shown within each graph. The error represents the 99% confidence interval, and a p-value of less than 0.01 was considered significant.
Table 1.
Summary of kinesin motility on paclitaxel microtubules in the absence and presence of Tau
| Tau isoform | Xobserved (μm) | N | Lobserved (μm) | N | Xexpected (μm) | p-Value | Velocity (μm/s) | |
|---|---|---|---|---|---|---|---|---|
| Kinesin-1 | – | 1.29 ± 0.11 | 591 | 4.63 ± 0.67 | 243 | 1.53 ± 0.27 | 0.74 ± 0.01 | |
| 3RS | 0.89 ± 0.12 | 243 | 4.57 ± 1.20 | 113 | 0.99 ± 0.24 | 2 × 10−6 | 0.65 ± 0.02 | |
| 4RL | 0.99 ± 0.17 | 200 | 4.55 ± 0.84 | 185 | 1.12 ± 0.31 | 5 × 10−4 | 0.58 ± 0.02 | |
| Kinesin-1+KAL | – | 0.79 ± 0.14 | 182 | 5.74 ± 0.63 | 387 | 0.86 ± 0.25 | 0.64 ± 0.03 | |
| 3RS | 0.65 ± 0.12 | 165 | 5.97 ± 0.81 | 298 | 0.70 ± 0.21 | 0.06 | 0.55 ± 0.03 | |
| 4RL | 0.64 ± 0.11 | 191 | 4.66 ± 0.90 | 204 | 0.69 ± 0.20 | 0.05 | 0.55 ± 0.03 | |
| Kinesin-2 | – | 0.92 ± 0.13 | 300 | 5.02 ± 0.75 | 268 | 1.03 ± 0.24 | 0.31 ± 0.01 | |
| 3RS | 0.92 ± 0.12 | 325 | 2.57 ± 0.21 | 1029 | 1.16 ± 0.27 | 0.03 | 0.32 ± 0.01 | |
| 4RL | 0.86 ± 0.16 | 210 | 2.50 ± 0.34 | 222 | 1.08 ± 0.34 | 0.03 | 0.32 ± 0.01 | |
| Kinesin-2PA_ΔDAL | – | 1.42 ± 0.27 | 182 | 4.05 ± 0.59 | 315 | 1.78 ± 0.59 | 0.40 ± 0.01 | |
| 3RS | 0.87 ± 0.14 | 215 | 3.92 ± 0.70 | 187 | 0.99 ± 0.27 | 1 × 10−7 | 0.35 ± 0.01 | |
| 4RL | 1.18 ± 0.17 | 280 | 3.46 ± 0.44 | 314 | 1.47 ± 0.37 | 0.04 | 0.36 ± 0.01 |
Xobserved is the measured characteristic run length, Lobserved is the characteristic microtubule track length, and Xexpected is the expected characteristic run length to adjust for differences in the microtubule track length distribution. The error represents the 99% confidence interval and p-values were calculated for the expected characteristic run length in the presence of Tau relative to that in the absence of Tau, where p < 0.01 was considered significant.
Table 2.
Summary of kinesin motility on GMPCPP microtubules in the absence and presence of Tau
| Tau isoform | Xobserved (μm) | N | Lobserved (μm) | N | Xexpected (μm) | p-Value | Velocity (μm/s) | |
|---|---|---|---|---|---|---|---|---|
| Kinesin-1 | – | 0.90 ± 0.13 | 255 | 2.49 ± 0.55 | 117 | 1.15 ± 0.30 | 3 × 10−4a | 0.68 ± 0.02 |
| 3RS | 0.78 ± 0.11 | 228 | 2.83 ± 0.76 | 104 | 0.93 ± 0.24 | 0.04 | 0.64 ± 0.02 | |
| 4RL | 0.77 ± 0.13 | 195 | 2.63 ± 0.53 | 138 | 0.92 ± 0.27 | 0.04 | 0.66 ± 0.02 | |
| Kinesin-2 | – | 0.67 ± 0.08 | 567 | 2.13 ± 0.25 | 355 | 0.81 ± 0.16 | 1 × 10−3a | 0.36 ± 0.01 |
| 3RS | 0.67 ± 0.11 | 196 | 1.80 ± 0.17 | 477 | 0.86 ± 0.25 | 0.28 | 0.33 ± 0.01 | |
| 4RL | 0.66 ± 0.12 | 206 | 1.96 ± 0.21 | 382 | 0.82 ± 0.25 | 0.35 | 0.31 ± 0.01 |
Xobserved is the measured characteristic run length, Lobserved is the characteristic track length, Xexpected is the expected characteristic run length to adjust for differences in the microtubule track distribution.
Represents a statistically significant difference from that observed on paclitaxel microtubules. The error represents the 99% confidence interval and a p-value of less than 0.01 was considered significant.
In both cases, the processivity was reduced going from a GDP to a GTP-like microtubule state, demonstrating that kinesin-2, like kinesin-1 (20), is sensitive to differences in the nucleotide state of the microtubule lattice. This nucleotide sensitivity may result from a structural change in the microtubule’s motor-binding site, since it was recently shown kinesin-1 preferentially binds to GMPCPP microtubules (25). This preference is thought to be due to β-tubulin’s C-terminal half of helix H4 being pushed toward kinesin, as well as to helix H4 making longitudinal contact with α-tubulin’s helix H11, which better positions the canonical kinesin-binding site to interact with kinesin-1’s loop L11 (26). Repositioning of kinesin’s binding site on the microtubule could change kinesin’s kinetics in the weak binding (ADP) state. As processivity is controlled by the race between front-head binding and rear-head detachment from the microtubule, an accelerated detachment on GMPCPP microtubules in the weak binding (ADP) state should decrease kinesin’s run length, consistent with our observations for both kinesin-1 and kinesin-2.
In the presence of Tau, both the 3RS- and 4RL-isoforms reduced kinesin-1’s motility on paclitaxel microtubules, as expected, with the 3RS isoform having a greater effect than the 4RL isoform (0.99 ± 0.24 μm, p = 2 × 10−6 vs. 1.12 ± 0.31 μm, p = 5 × 10−4, respectively; Fig. 2, B and C; Table 1) compared with bare microtubules (1.53 ± 0.27 μm; Fig. 2 A). Interestingly, and contrary to kinesin-1, neither Tau isoform impeded kinesin-2’s characteristic run length (3RS-Tau: 1.16 ± 0.27 μm, p = 0.03; 4RL-Tau: 1.08 ± 0.34 μm, p = 0.03; Fig. 2, E and F; Table 1) on paclitaxel microtubules compared with bare microtubules (1.03 ± 0.24 μm; Fig. 2 D), indicating that unlike kinesin-1, kinesin-2 is not sensitive to the presence of either isoform of Tau on the microtubule surface.
On GMPCPP microtubules, kinesin-1 was not impeded by either isoform of Tau (3RS: 0.93 ± 0.24 μm, 4RL: 0.92 ± 0.27 μm; Fig. 3, B and C; Table 2), as their characteristic run lengths were similar to that observed on bare microtubules (1.15 ± 0.30 μm; Fig. 3 A; Table 2). Kinesin-2, like kinesin-1, also was not impeded by either isoform of Tau (3RS: 0.86 ± 0.25 μm, 4RL: 0.82 ± 0.25 μm; Fig. 3, E and F; Table 2), as their characteristic run lengths were similar to that observed on bare microtubules (0.81 ± 0.16 μm; Fig. 3 D).
Kinesin-2 steps more frequently after a pause than kinesin-1
To further explore kinesin-2’s uninterrupted characteristic run length on Tau-decorated paclitaxel microtubules, we counted the number of pauses for kinesin-1 and kinesin-2 in the absence and presence of 3RS-Tau. Kinesin-1 is known to pause in its processive walk along the microtubule (27,28), and such events are even more likely to occur upon encountering an obstacle, such as Tau. Kinesin’s response after a pause will be either to keep stepping or to dissociate from the microtubule track, terminating the processive run. Thus, we predicted that both kinesin-1 and kinesin-2 would be more likely to pause during their processive runs along the microtubule in the presence of Tau than in its absence. Furthermore, if kinesin-2 can navigate Tau obstacles on the microtubule track but kinesin-1 cannot, we expected that kinesin-2 would be more likely to continue stepping after a pause, whereas kinesin-1 would be more likely to terminate its processive run. Observed pausing events were categorized as either terminating after a pause (pause-terminate) or stepping after a pause (pause-step; Fig. 4). In the absence of Tau, kinesin-2 is 7% more likely to pause than kinesin-1, but in the presence of 3RS-Tau, it is 18% (i.e., more than twice as likely to pause than kinesin-1) (Table 3). Interestingly, the pause-step percentages of kinesin-2 and kinesin-1 are similar in the absence of Tau; however, in the presence of 3RS-Tau, kinesin-2 is 15% more likely to step after a pause compared with kinesin-1, which is significant (p = 0.05; Table 3). Thus, kinesin-2 prefers to step rather than terminate after a pause, in contrast to kinesin-1, which is more likely to terminate its processive run after a pause, in the presence of 3RS-Tau (Fig. 4). Kinesin-2’s ability to step more efficiently after a pause is consistent with its unchanged characteristic run length in the presence of either isoform of Tau. Similarly, kinesin-1’s characteristic run length decreases in the presence of Tau, which is consistent with its decrease in the number of steps after a pause event.
Table 3.
Summary of kinesin-1 and kinesin-2 pausing behavior on paclitaxel microtubules in the absence and presence of 3RS-Tau
| Kinesin-1 |
Kinesin-2 |
|||
|---|---|---|---|---|
| No Tau (N = 392) | 3RS-Tau (N = 245) | No Tau (N = 305) | 3RS-Tau (N = 295) | |
| Number of pauses | 81 | 35 | 83 | 95 |
| % of total events | 20.6 | 14.3a | 27.2b | 32.5b |
| Dwell time (s) | 2.79 ± 0.45 | 3.84 ± 0.60a | 3.13 ± 0.31b | 2.38 ± 0.31a,b |
| Number of steps after a pause | 24 | 8 | 22 | 36 |
| % of pause events | 29.6 | 22.9 | 26.5 | 37.9a,b |
| Dwell time (s) | 2.41 ± 0.51 | 3.03 ± 0.87 | 3.01 ± 0.41 | 1.88 ± 0.18a |
| Number of terminations after a pause | 57 | 27 | 61 | 59 |
| % of pause events | 70.4 | 77.1 | 73.5 | 62.1a,b |
| Dwell time (s) | 2.94 ± 0.60 | 4.09 ± 0.73a | 3.17 ± 0.40b | 2.69 ± 0.49a,b |
Significant differences in dwell times were calculated with a Mann-Whitney U test. Histograms of dwell-time data found in the Supporting Material, Fig S2.
Represents significant difference (p ≤ 0.05) from the no-Tau condition.
Represents significant difference (p ≤ 0.05) from kinesin-1 under the same Tau condition. Dwell-time errors represent 95% confidence intervals.
Truncation of kinesin-2’s neck-linker confers susceptibility to inhibition by Tau
Kinesin-2’s insensitivity to Tau on paclitaxel microtubules could be due to changes in the biochemistry or mechanics of the motor domain, or to differences in the neck-linker length. We hypothesize that this difference in behavior between kinesin-1 and kinesin-2 originates in the neck-linker composition of the two motors. To directly test this hypothesis, we used a truncated neck-linker chimera that was previously shown to mimic kinesin-1’s processivity on paclitaxel microtubules (10), and sought to determine whether these changes in the neck-linker length also dictated sensitivity to the presence of Tau on the microtubule surface. The chimera, kinesin-2PA_ΔDAL, contained a proline-to-alanine switch (PA) to remove the kink and a deletion of the C-terminal end of the neck-linker (ΔDAL), mimicking the same number of amino acids as kinesin-1’s neck-linker (Fig. 1 B). On bare paclitaxel microtubules, the characteristic run length of the kinesin-2 neck-linker chimera, kinesin-2PA_ΔDAL, increased by 75% compared with our kinesin-2 construct (1.78 ± 0.59 μm vs. 1.03 ± 0.24 μm, p = 5 × 10−8; Fig. 5 D; Table 1), which indicates an increase in processivity, consistent with previous results (10). Interestingly, kinesin-2PA_ΔDAL also demonstrated an isoform-specific sensitivity to the presence of Tau similar to that observed for kinesin-1. In the presence of 3RS-Tau, kinesin-2PA_ΔDAL’s characteristic run length fell by 44% (0.99 ± 0.27 μm, p = 1 × 10−7), which was statistically significant compared with kinesin-2PA_ΔDAL’s motility on undecorated microtubules (Fig. 5 E; Table 1). In the presence of 4RL-Tau, the characteristic run length fell by 17%, which was not statistically significant compared with undecorated microtubules (1.47 ± 0.37 μm, p = 0.04; Fig. 5 F; Table 1).
Figure 5.
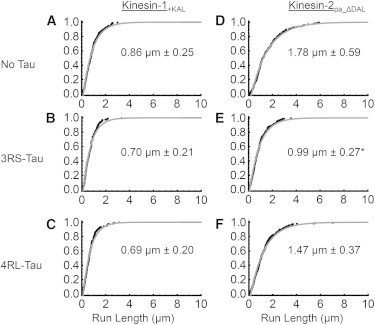
Characteristic run length comparison between kinesin-1+KAL and kinesin-2pa_ΔDAL on paclitaxel microtubules. (A–C) Cumulative frequency plots of kinesin-1+KAL in the absence or presence of 3RS- or 4RL-Tau. (D and E) Cumulative frequency plots of kinesin-2pa_ΔDAL in the absence or presence of 3RS- or 4RL Tau. Black dots represent the raw run length data and the gray curve is the observed cumulative frequency. The expected characteristic run length, derived from the microtubule length distribution, is shown within each graph. The error represents the 99% confidence interval and a p-value of less than 0.01 was considered significant. ∗Represents a statistically significant difference from the characteristic run length observed in the absence of Tau.
Lengthening kinesin-1’s neck-linker abolishes its sensitivity to the presence of Tau
Because the deletion of three amino acids in kinesin-2’s neck-linker increased the chimera’s sensitivity to Tau, we tested the corollary by lengthening kinesin-1’s neck-linker by three amino acids to see if its sensitivity to Tau was abolished. The chimera, kinesin-1KAL, contained a three amino acid (KAL) insert in the C-terminal end of its neck-linker region (Fig. 1 B), and has been shown to have a similar characteristic run length compared with wild-type kinesin-2 (10). On bare microtubules, kinesin-1KAL’s characteristic run length was 0.86 ± 0.25 μm (Fig. 5 A; Table 1) or 44% lower than kinesin-1 (1.53 ± 0.27 μm; Fig. 2 A), as expected from previous results (10). However, the kinesin-1KAL chimera lost the sensitivity to Tau displayed by wild-type kinesin-1, as the characteristic run length in the presence of either 3RS-Tau (0.70 ± 0.21 μm, p = 0.06; Fig. 5 B; Table 1) or 4RL-Tau (0.69 ± 0.20 μm, p = 0.05; Fig. 5 C; Table 1) did not change appreciably from that observed in the absence of Tau.
Discussion
This study demonstrates that kinesin-1 and kinesin-2 are sensitive to the microtubule nucleotide state, as both constructs have reduced processivity on GMPCPP microtubules compared with paclitaxel microtubules (Figs. 2 and 3; Tables 1 and 2). In addition, Tau does not impede the motility of kinesin-2 on paclitaxel microtubules, in contrast to kinesin-1, which is impeded by Tau in an isoform-specific manner (Fig. 2) (18–20). Additionally, we can rule out the possibility that these results are not dependent on the Tau isoforms binding differently from each other, or on different nucleotide states of the microtubule lattice (20). This is corroborated by the pausing data, which show that kinesin-2 is 15% more likely than kinesin-1 to step after a pausing event in the presence of 3RS-Tau on paclitaxel microtubules (Fig. 4; Table 3). Furthermore, we have shown the family-specific differences in kinesin’s neck-linker length play a critical role in kinesin’s ability to bypass Tau obstacles on the microtubule surface. Kinesin-2’s neck-linker length is sufficient to allow for uninterrupted run lengths on paclitaxel microtubules in the presence of either 3RS- or 4RL-Tau, whereas the shorter neck-linker of kinesin-1 confers inhibition by Tau.
The loss of kinesin-2’s sensitivity to Tau on paclitaxel microtubules, contrary to that of kinesin-1, directly supports our hypothesis that the flexibility of kinesin’s neck-linker region determines its ability to navigate obstacles, such as Tau, on the microtubule surface. There are multiple molecular explanations for kinesin-2’s ability to bypass Tau obstacles, which may not be mutually exclusive: 1), kinesin-2, due to its shorter characteristic run length, does not encounter Tau as frequently as kinesin-1; 2), kinesin-2, due to its longer neck-linker length, may step over or around Tau; and 3), kinesin-2, due to its slower velocity, may be able to wait for Tau to move out of its way during its processive walk along the microtubule surface.
To ensure that kinesin-2’s loss of sensitivity to Tau was not an artifact of its encountering fewer Tau molecules during its processive walk along the microtubule surface due to its shorter characteristic run length relative to kinesin-1, we measured the average density of Tau on the microtubule surface. We then determined the expected Tau encounter frequency for each motor and the expected effect on the observed run length in both cases (see Supporting Material). Under our conditions, kinesin-1 and kinesin-2 will, on average, take 49 8-nm steps before encountering a Tau molecule along a single protofilament, which is a distance of 0.39 μm. Given kinesin-2’s observed characteristic run length in the absence of Tau, 0.92 μm, it should encounter a Tau molecule 2.3 times per processive run (assuming it tracks along a single protofilament). We believe this to be an underestimate, as many of the shorter-lived Tau events (≤0.4 s) were hard to resolve in our kymographs. To further validate kinesin-2’s loss of sensitivity to Tau on the microtubule surface, we simulated the degree of 3RS-Tau inhibition on kinesin-1 as a 1:1 steric blocker and applied this to kinesin-2’s motility, assuming that it too was sensitive to 3RS-Tau (see Supporting Material). From the simulation, we calculated kinesin-2’s expected characteristic run length, Xexpected, in the presence of 3RS-Tau and compared it with the observed characteristic run length in the presence of Tau to evaluate whether there was a significant deviation between these two groups. The simulation of Xexpected for kinesin-2, unlike that for kinesin-1, using the simulated theoretical curve derived from a data set of identical size, indicated a significant deviation (p = 5 × 10−3) from the predicted, Xexpected, behavior of 3RS-Tau (Fig. S3, A and B, and Fig. S4). This suggests that kinesin-2 has the ability to bypass obstacles such as 3RS-Tau, and that this ability is not merely a feature of it encountering less 3RS-Tau due to its lower processivity.
We next considered how kinesin-2’s neck-linker contributes to its ability to step over or around Tau. There is electron microscopy evidence for Tau binding both across (29) and along (29,30) protofilaments, but ultimately it is not clear how Tau lies along the microtubule lattice. Kinesin-2’s neck-linker is three amino acids longer than kinesin-1’s (17 vs. 14 amino acids, respectively; Figs. 1 B and 6 A). The longer length of kinesin-2’s neck-linker may allow it to step over Tau if the latter is lying across multiple protofilaments. Tau binding has been shown to be centered on α-tubulin (29), which may limit interference with kinesin’s binding site on β-tubulin and allow kinesin to step over Tau given a sufficiently flexible neck-linker region. If Tau lies along a protofilament and blocks kinesin’s forward binding site, then kinesin is more likely to sidestep to an adjacent protofilament than to step over Tau. Nonmammalian kinesin-2 family members have been shown to be capable of switching protofilaments in the absence of MAPs such as Tau (21). However, Brunnbauer et al. (21) also observed that mammalian kinesin-2, Mmkif3A/B, predominantly tracks along a single protofilament like kinesin-1. Mmkif3A/B is similar to the construct we used in this study (Fig. 1 A). Our construct contains two kif3A motor domains (kif3A/A) and neck-linkers fused to a Drosophila kinesin-1 coiled-coil (see Materials and Methods), which is functionally equivalent to the kif3A/B heterodimer as demonstrated by Muthukrishnan et al. (9). Our results suggest that, given the opportunity, even mammalian kinesin-2 can sidestep to an adjacent protofilament to navigate around an obstacle (e.g., Tau) on the microtubule surface. Additionally, Bormuth et al. (31) demonstrated that kinesin-8, which like kinesin-2 has a 17 amino acid neck-linker, is capable of sidestepping to an adjacent protofilament. To further explore this possibility, we modeled the probability of our kinesin-1 and kinesin-2 constructs sidestepping to an adjacent protofilament as a function of neck-linker contour length (see Supporting Material). The probability for kinesin-1’s front head to bind to the next available binding site along the protofilament is 99.6%, which is as expected because kinesin-1 is known to track along a single protofilament (Fig. 6 B; Supporting Material, Table S1) (32), whereas the probability to sidestep is 0.41%, counterclockwise. For wild-type kif3A/B, the probability of the front head stepping forward along the protofilament is 98.9% and the probability of sidestepping is 1.1%, counterclockwise (see Supporting Material). For our construct, kif3A/A, the probability of forward stepping along the protofilament is 97.7% and the probability of sidestepping is 2.1%, counterclockwise (Fig. 6 B). The modeling suggests that the wild-type may not be able to sidestep as frequently as our construct; however, kif3A/B is still twice as likely to sidestep compared with kinesin-1. Thus, for an undecorated 13-protofilament microtubule, our kinesin-2 construct is likely to take a sidestep a little over 2% of the time. This small but significant probability provides kinesin-2 the opportunity to sidestep to an adjacent protofilament when challenged by an obstacle (e.g., Tau) during its processive walk along the microtubule.
Figure 6.
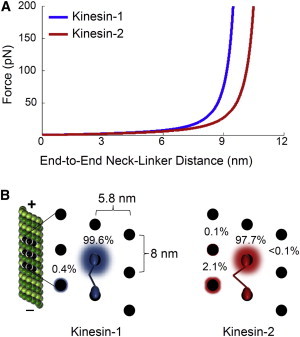
Probability of kinesin sidestepping as a function of neck-linker contour length. (A) Force-extension curves of kinesin-1 and kinesin-2 (kif3A/A) neck-linker regions determined assuming a worm-like chain model (41–43). Kinesin-2’s longer neck-linker allows for a longer reach at the same force and thus an increased probability of stepping to an off-protofilament binding site. (B) Probability of kinesin stepping to nearby binding sites. Black dots represent binding sites for a 13-protofilament microtubule. Kinesin-1 only steps along a single protofilament, whereas kinesin-2 is predicted to sidestep left to the adjacent protofilament 2.1% of the time. The animation is not drawn to scale and is for visual effect; see Supporting Material for further details about the modeling.
We also considered how kinesin-2, with its slower velocity, might be able to wait for Tau to move out of the way before continuing its processive march along the microtubule. The velocity of kinesin-2 is about half that of kinesin-1 (Table 1), which potentially translates into the rear head spending twice as much time bound to the microtubule. This would give the front head more time to diffusively search for an available off-protofilament binding site when there is an obstacle in front of it. Alternatively, kinesin-2 may be able to pause long enough before taking its next step to allow a Tau obstacle on the microtubule surface to dissociate or diffuse away, as suggested by Xu et al. (33), rather than stepping around or over it. Although we cannot completely rule out this possibility, the average time between steps for kinesin-2 (0.008 μm/step / 0.31 μm/s = 0.026 s/step) is significantly shorter than the mean dwell time for 3RS-Tau on the microtubule surface (21.6 s) (34). Another consideration in this analysis is Tau’s multivalent interaction with the microtubule lattice. Tau contains either three or four microtubule-binding repeats (Fig. 1 C) depending on the Tau isoform. Individually, the binding repeats interact weakly with the microtubule lattice, but when combined they have a much stronger affinity to the microtubule (35). These binding repeats interact transiently with the lattice on the scale of milliseconds, which may rapidly sample the lattice in different orientations (35). It is plausible that during these transient interactions the binding repeats, and thus Tau can move out of kinesin-2’s path. Currently, this is untestable because we lack a more detailed understanding of Tau’s interaction with the microtubule lattice. Finally, it is possible that kinesin-2 undergoes extended pausing at obstacles such as Tau compared with kinesin-1. However, the average pause duration for kinesin-2 is slightly shorter than in the absence of Tau or for kinesin-1 in the presence of 3RS-Tau (Table 3). Interestingly, kinesin-1’s dwell time increases in the presence of 3RS-Tau, which suggests there might be an interaction between kinesin-1 and 3RS-Tau, or that it simply waits longer before detaching because its forward binding site is blocked. Conversely, kinesin-2’s dwell time decreases in the presence of 3RS-Tau, indicating that 3RS-Tau may assist kinesin-2 in bypassing Tau or restricts kinesin-2’s access to the next forward binding site, thereby directing it to sidestep to an adjacent protofilament.
Conclusions
The ability of kinesin-2 to efficiently navigate obstacles such as Tau on the microtubule surface is likely to be important for its role as a molecular motor in a number of different intracellular transport processes. The results from this work are most directly relevant to fast axonal transport in neurons, in which microtubules are known to be heavily decorated with Tau (15). The extent to which Tau inhibits kinesin motility in the axon is dependent on a number of factors, including the isoform of Tau involved (18–20), the structural state of the microtubule lattice (20), and, as shown here, the kinesin family member involved. Evidence from in vivo studies is mixed as to whether Tau is inhibitory to axonal transport (36) or not (37,38). Both kinesin-1 and kinesin-2 are involved in axonal transport, and kinesin-2, through its extended neck-linker region, may be optimized to transport cargos around obstacles such as Tau in the crowded axonal landscape at the expense of its processivity relative to kinesin-1. Indeed, the ability to navigate obstacles on the microtubule surface may be of more value than single-molecule processivity in a crowded intracellular environment in which most cargos are bound to multiple anterograde motors anyway. Furthermore, although many axonal cargos are specifically transported by a particular motor protein, it is interesting to note that many cargo complexes are bound to multiple molecular motors, including kinein-1, kinesin-2, and cytoplasmic dynein (4,7,8). Although the benefit of having both a plus-end-directed (e.g., kinesin-1) and a minus-end-directed (e.g., cytoplasmic dynein) motor on the same cargo for bidirectional transport is obvious, the reason for having two plus-end-directed motors on the same cargo is less clear. Kinesin-1 is known to preferentially target cargos to specific microtubule tracks within the axon (39,40), but once in the axon, kinesin-2 bound to the same cargo could help kinesin-1 navigate the microtubule surface more efficiently in the presence of many potential obstacles, such as Tau. The ability of kinesin-2 to coordinate with another, more processive plus-end-directed motor is not limited to the neuronal axon, as RNPs in Xenopus oocytes are known to be complexed with both kinesin-1 and kinesin-2 (6), and IFT particles in the cilia of C. elegans sensory cells are known to contain both kinesin-2 and OSM-3, a faster, more processive member of the kinesin-2 family. The role of kinesin-2 in intracellular transport and its coordination with other kinesin family members on the same cargo require further understanding, but to our knowledge, the results presented in this work shed important new light on novel functions of the kinesin neck-linker in navigating around obstacles on the microtubule surface.
Acknowledgments
The authors thank Dr. David Warshaw and Guy Kennedy for equipment and operational support, and Dr. Jason Stumpff for helpful discussions during preparation of the manuscript.
This work was supported by the National Institutes of Health (R01-GM-101066 to C.L.B. and R01-GM-076476 to W.O.H.).
Supporting Material
References
- 1.Goldstein L.S., Yang Z. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu. Rev. Neurosci. 2000;23:39–71. doi: 10.1146/annurev.neuro.23.1.39. [DOI] [PubMed] [Google Scholar]
- 2.Chevalier-Larsen E., Holzbaur E.L. Axonal transport and neurodegenerative disease. Biochim. Biophys. Acta. 2006;1762:1094–1108. doi: 10.1016/j.bbadis.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Holzbaur E.L., Goldman Y.E. Coordination of molecular motors: from in vitro assays to intracellular dynamics. Curr. Opin. Cell Biol. 2010;22:4–13. doi: 10.1016/j.ceb.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendricks A.G., Perlson E., Ross J.L., Schroeder H.W., 3rd, Tokito M., Holzbaur E.L. Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol. 2010;20:697–702. doi: 10.1016/j.cub.2010.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snow J.J., Ou G., Scholey J.M. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat. Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- 6.Messitt T.J., Gagnon J.A., Mowry K.L. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev. Cell. 2008;15:426–436. doi: 10.1016/j.devcel.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maday S., Wallace K.E., Holzbaur E.L. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 2012;196:407–417. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Twelvetrees A., Hendricks A.G., Holzbaur E.L. SnapShot: axonal transport. Cell. 2012;149 doi: 10.1016/j.cell.2012.05.001. 950–950 e951. [DOI] [PubMed] [Google Scholar]
- 9.Muthukrishnan G., Zhang Y., Shastry S., Hancock W.O. The processivity of kinesin-2 motors suggests diminished front-head gating. Current biology: CB. 2009;19:442–447. doi: 10.1016/j.cub.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shastry S., Hancock W.O. Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Current biology: CB. 2010;20:939–943. doi: 10.1016/j.cub.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shastry S., Hancock W.O. Interhead tension determines processivity across diverse N-terminal kinesins. Proc. Natl. Acad. Sci. USA. 2011;108:16253–16258. doi: 10.1073/pnas.1102628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Block S.M., Goldstein L.S., Schnapp B.J. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 13.Al-Bassam J., Roger B., Milligan R.A. Analysis of the weak interactions of ADP-Unc104 and ADP-kinesin with microtubules and their inhibition by MAP2c. Cell Motil. Cytoskeleton. 2007;64:377–389. doi: 10.1002/cm.20190. [DOI] [PubMed] [Google Scholar]
- 14.Dehmelt L., Halpain S. The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 2005;6:204. doi: 10.1186/gb-2004-6-1-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binder L.I., Frankfurter A., Rebhun L.I. The distribution of tau in the mammalian central nervous system. J. Cell Biol. 1985;101:1371–1378. doi: 10.1083/jcb.101.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goedert M., Spillantini M.G., Crowther R.A. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron. 1989;3:519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 17.Weingarten M.D., Lockwood A.H., Kirschner M.W. A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. USA. 1975;72:1858–1862. doi: 10.1073/pnas.72.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vershinin M., Carter B.C., Gross S.P. Multiple-motor based transport and its regulation by Tau. Proc. Natl. Acad. Sci. USA. 2007;104:87–92. doi: 10.1073/pnas.0607919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixit R., Ross J.L., Holzbaur E.L. Differential regulation of dynein and kinesin motor proteins by tau. Science. 2008;319:1086–1089. doi: 10.1126/science.1152993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McVicker D.P., Chrin L.R., Berger C.L. The nucleotide-binding state of microtubules modulates kinesin processivity and the ability of Tau to inhibit kinesin-mediated transport. J. Biol. Chem. 2011;286:42873–42880. doi: 10.1074/jbc.M111.292987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunnbauer M., Dombi R., Ökten Z. Torque generation of kinesin motors is governed by the stability of the neck domain. Mol. Cell. 2012;46:147–158. doi: 10.1016/j.molcel.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Kar S., Fan J., Amos L.A. Repeat motifs of tau bind to the insides of microtubules in the absence of taxol. EMBO J. 2003;22:70–77. doi: 10.1093/emboj/cdg001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castoldi M., Popov A.V. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr. Purif. 2003;32:83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- 24.Thompson A.R., Hoeprich G.J., Berger C.L. Single-molecule motility: statistical analysis and the effects of track length on quantification of processive motion. Biophys. J. 2013;104:2651–2661. doi: 10.1016/j.bpj.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakata T., Niwa S., Hirokawa N. Preferential binding of a kinesin-1 motor to GTP-tubulin-rich microtubules underlies polarized vesicle transport. J. Cell Biol. 2011;194:245–255. doi: 10.1083/jcb.201104034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yajima H., Ogura T., Hirokawa N. Conformational changes in tubulin in GMPCPP and GDP-taxol microtubules observed by cryoelectron microscopy. J. Cell Biol. 2012;198:315–322. doi: 10.1083/jcb.201201161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asbury C.L., Fehr A.N., Block S.M. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guydosh N.R., Block S.M. Backsteps induced by nucleotide analogs suggest the front head of kinesin is gated by strain. Proc. Natl. Acad. Sci. USA. 2006;103:8054–8059. doi: 10.1073/pnas.0600931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santarella R.A., Skiniotis G., Hoenger A. Surface-decoration of microtubules by human tau. J. Mol. Biol. 2004;339:539–553. doi: 10.1016/j.jmb.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Al-Bassam J., Ozer R.S., Milligan R.A. MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J. Cell Biol. 2002;157:1187–1196. doi: 10.1083/jcb.200201048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bormuth V., Nitzsche B., Diez S. The highly processive kinesin-8, Kip3, switches microtubule protofilaments with a bias toward the left. Biophys. J. 2012;103:L4–L6. doi: 10.1016/j.bpj.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray S., Meyhöfer E., Howard J. Kinesin follows the microtubule’s protofilament axis. J. Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu J., King S.J., Nemec B. Interplay between velocity and travel distance of Kinesin-based transport in the presence of Tau. Biophys. J. 2013;105:L23–L25. doi: 10.1016/j.bpj.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McVicker D.P., Hoeprich G.J., Berger C.L. Tau interconverts between diffusive and stable populations on the microtubule surface in an isoform and lattice specific manner. Cytoskeleton. 2014 doi: 10.1002/cm.21163. February 24, 2014. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butner K.A., Kirschner M.W. Tau protein binds to microtubules through a flexible array of distributed weak sites. J. Cell Biol. 1991;115:717–730. doi: 10.1083/jcb.115.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoothoff W., Jones P.B., Hyman B.T. Differential effect of three-repeat and four-repeat tau on mitochondrial axonal transport. J. Neurochem. 2009;111:417–427. doi: 10.1111/j.1471-4159.2009.06316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaPointe N.E., Morfini G., Brady S.T. The amino terminus of tau inhibits kinesin-dependent axonal transport: implications for filament toxicity. J. Neurosci. Res. 2009;87:440–451. doi: 10.1002/jnr.21850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morfini G.A., Burns M., Brady S.T. Axonal transport defects in neurodegenerative diseases. J. Neurosci. 2009;29:12776–12786. doi: 10.1523/JNEUROSCI.3463-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konishi Y., Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat. Neurosci. 2009;12:559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- 40.Nakata T., Hirokawa N. Microtubules provide directional cues for polarized axonal transport through interaction with kinesin motor head. J. Cell Biol. 2003;162:1045–1055. doi: 10.1083/jcb.200302175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sahoo H., Roccatano D., Nau W.M. Distance distributions of short polypeptides recovered by fluorescence resonance energy transfer in the 10 A domain. J. Am. Chem. Soc. 2006;128:8118–8119. doi: 10.1021/ja062293n. [DOI] [PubMed] [Google Scholar]
- 42.Kutys M.L., Fricks J., Hancock W.O. Monte Carlo analysis of neck linker extension in kinesin molecular motors. PLOS Comput. Biol. 2010;6:e1000980. doi: 10.1371/journal.pcbi.1000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bustamante C., Marko J.F., Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 44.Dietz H., Rief M. Protein structure by mechanical triangulation. Proc. Natl. Acad. Sci. USA. 2006;103:1244–1247. doi: 10.1073/pnas.0509217103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chretien D., Wade R.H. New data on the microtubule surface lattice. Biol. Cell. 1991;71:161–174. doi: 10.1016/0248-4900(91)90062-r. [DOI] [PubMed] [Google Scholar]
- 46.Hinrichs M.H., Jalal A., Scholz T. Tau protein diffuses along the microtubule lattice. J. Biol. Chem. 2012;287:38559–38568. doi: 10.1074/jbc.M112.369785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


