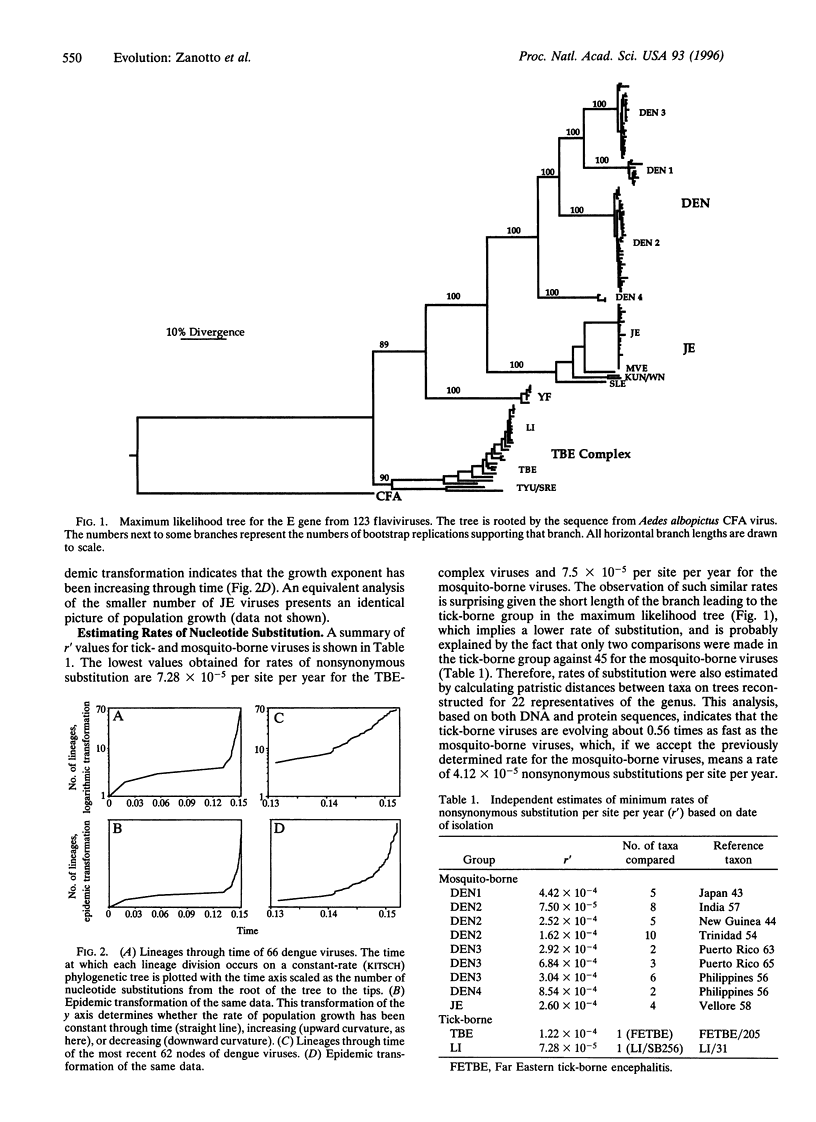
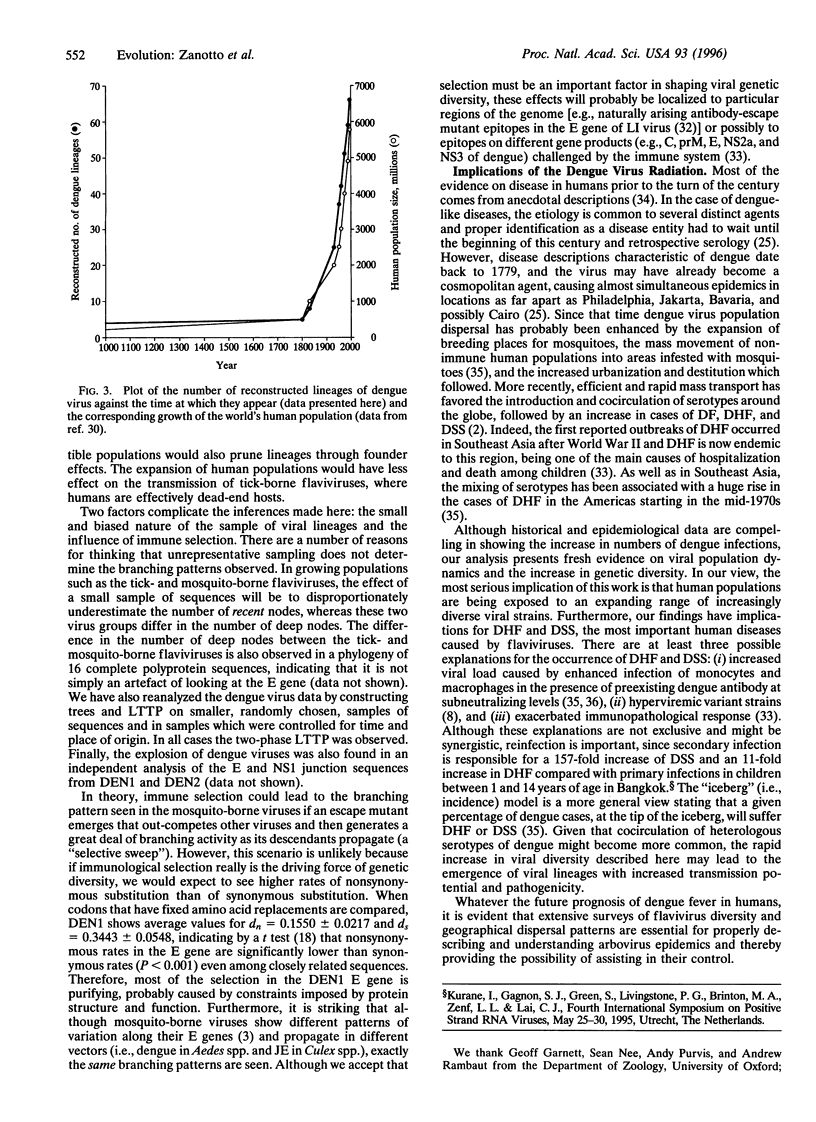
Abstract
The phylogeny of 123 complete envelope gene sequences was reconstructed in order to understand the evolution of tick- and mosquito-borne flaviviruses. An analysis of phylogenetic tree structure reveals a continual and asymmetric branching process in the tick-borne flaviviruses, compared with an explosive radiation in the last 200 years in viruses transmitted by mosquitoes. The distinction between these two viral groups probably reflects differences in modes of dispersal, propagation, and changes in the size of host populations. The most serious implication of this work is that growing human populations are being exposed to an expanding range of increasingly diverse viral strains.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blok J., McWilliam S. M., Butler H. C., Gibbs A. J., Weiller G., Herring B. L., Hemsley A. C., Aaskov J. G., Yoksan S., Bhamarapravati N. Comparison of a dengue-2 virus and its candidate vaccine derivative: sequence relationships with the flaviviruses and other viruses. Virology. 1992 Apr;187(2):573–590. doi: 10.1016/0042-6822(92)90460-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher C. H., Karabatsos N., Dalrymple J. M., Shope R. E., Porterfield J. S., Westaway E. G., Brandt W. E. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J Gen Virol. 1989 Jan;70(Pt 1):37–43. doi: 10.1099/0022-1317-70-1-37. [DOI] [PubMed] [Google Scholar]
- Cammisa-Parks H., Cisar L. A., Kane A., Stollar V. The complete nucleotide sequence of cell fusing agent (CFA): homology between the nonstructural proteins encoded by CFA and the nonstructural proteins encoded by arthropod-borne flaviviruses. Virology. 1992 Aug;189(2):511–524. doi: 10.1016/0042-6822(92)90575-a. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988 Nov 25;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G. F., Hussain M. H., Reid H. W., Gould E. A. Identification of naturally occurring monoclonal antibody escape variants of louping ill virus. J Gen Virol. 1994 Mar;75(Pt 3):609–614. doi: 10.1099/0022-1317-75-3-609. [DOI] [PubMed] [Google Scholar]
- Gritsun T. S., Holmes E. C., Gould E. A. Analysis of flavivirus envelope proteins reveals variable domains that reflect their antigenicity and may determine their pathogenesis. Virus Res. 1995 Mar;35(3):307–321. doi: 10.1016/0168-1702(94)00090-y. [DOI] [PubMed] [Google Scholar]
- Gubler D. J., Clark G. G. Dengue/dengue hemorrhagic fever: the emergence of a global health problem. Emerg Infect Dis. 1995 Apr-Jun;1(2):55–57. doi: 10.3201/eid0102.952004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuda M., Nuttall P. A., Kozuch O., Elecková E., Williams T., Zuffová E., Sabó A. Non-viraemic transmission of tick-borne encephalitis virus: a mechanism for arbovirus survival in nature. Experientia. 1993 Sep 15;49(9):802–805. doi: 10.1007/BF01923553. [DOI] [PubMed] [Google Scholar]
- Lanciotti R. S., Lewis J. G., Gubler D. J., Trent D. W. Molecular evolution and epidemiology of dengue-3 viruses. J Gen Virol. 1994 Jan;75(Pt 1):65–75. doi: 10.1099/0022-1317-75-1-65. [DOI] [PubMed] [Google Scholar]
- Lewis J. A., Chang G. J., Lanciotti R. S., Kinney R. M., Mayer L. W., Trent D. W. Phylogenetic relationships of dengue-2 viruses. Virology. 1993 Nov;197(1):216–224. doi: 10.1006/viro.1993.1582. [DOI] [PubMed] [Google Scholar]
- Marin M. S., Zanotto P. M., Gritsun T. S., Gould E. A. Phylogeny of TYU, SRE, and CFA virus: different evolutionary rates in the genus Flavivirus. Virology. 1995 Feb 1;206(2):1133–1139. doi: 10.1006/viro.1995.1038. [DOI] [PubMed] [Google Scholar]
- Monath T. P. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nee S., Holmes E. C., Rambaut A., Harvey P. H. Inferring population history from molecular phylogenies. Philos Trans R Soc Lond B Biol Sci. 1995 Jul 29;349(1327):25–31. doi: 10.1098/rstb.1995.0087. [DOI] [PubMed] [Google Scholar]
- Nee S., May R. M., Harvey P. H. The reconstructed evolutionary process. Philos Trans R Soc Lond B Biol Sci. 1994 May 28;344(1309):305–311. doi: 10.1098/rstb.1994.0068. [DOI] [PubMed] [Google Scholar]
- Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986 Sep;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Matsuda H., Hagstrom R., Overbeek R. fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994 Feb;10(1):41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990 Feb;174(2):479–493. doi: 10.1016/0042-6822(90)90102-w. [DOI] [PubMed] [Google Scholar]
- Shiu S. Y., Jiang W. R., Porterfield J. S., Gould E. A. Envelope protein sequences of dengue virus isolates TH-36 and TH-Sman, and identification of a type-specific genetic marker for dengue and tick-borne flaviviruses. J Gen Virol. 1992 Jan;73(Pt 1):207–212. doi: 10.1099/0022-1317-73-1-207. [DOI] [PubMed] [Google Scholar]
- Taylor W. R., Jones D. T. Deriving an amino acid distance matrix. J Theor Biol. 1993 Sep 7;164(1):65–83. doi: 10.1006/jtbi.1993.1140. [DOI] [PubMed] [Google Scholar]
- Zanotto P. M., Gao G. F., Gritsun T., Marin M. S., Jiang W. R., Venugopal K., Reid H. W., Gould E. A. An arbovirus cline across the northern hemisphere. Virology. 1995 Jun 20;210(1):152–159. doi: 10.1006/viro.1995.1326. [DOI] [PubMed] [Google Scholar]



