Abstract
In most studied microbial rhodopsins two conserved carboxylic acid residues (the homologs of Asp-85 and Asp-212 in bacteriorhodopsin) and an arginine residue (the homolog of Arg-82) form a complex counterion to the protonated retinylidene Schiff base, and neutralization of the negatively charged carboxylates causes red shifts of the absorption maximum. In contrast, the corresponding neutralizing mutations in some relatively low-efficiency channelrhodopsins (ChRs) result in blue shifts. These ChRs do not contain a lysine residue in the second helix, conserved in higher efficiency ChRs (Lys-132 in the crystallized ChR chimera). By action spectroscopy of photoinduced channel currents in HEK293 cells and absorption spectroscopy of detergent-purified pigments, we found that in tested ChRs the Lys-132 homolog controls the direction of spectral shifts in the mutants of the photoactive site carboxylic acid residues. Analysis of double mutants shows that red spectral shifts occur when this Lys is present, whether naturally or by mutagenesis, and blue shifts occur when it is replaced with a neutral residue. A neutralizing mutation of the Lys-132 homolog alone caused a red spectral shift in high-efficiency ChRs, whereas its introduction into low-efficiency ChR1 from Chlamydomonas augustae (CaChR1) caused a blue shift. Taking into account that the effective charge of the carboxylic acid residues is a key factor in microbial rhodopsin spectral tuning, these findings suggest that the Lys-132 homolog modulates their pKa values. On the other hand, mutation of the Arg-82 homolog that fulfills this role in bacteriorhodopsin caused minimal spectral changes in the tested ChRs. Titration revealed that the pKa of the Asp-85 homolog in CaChR1 lies in the alkaline region unlike in most studied microbial rhodopsins, but is substantially decreased by introduction of a Lys-132 homolog or neutralizing mutation of the Asp-212 homolog. In the three ChRs tested the Lys-132 homolog also alters channel current kinetics.
Introduction
Phototaxis and the photophobic response in green flagellate algae, such as the model organism Chlamydomonas reinhardtii, are mediated by a specific subclass of sensory rhodopsins (1,2) known as channelrhodopsins (ChRs) for their ability to act as cation channels in heterologous systems (3,4). The use of these unique proteins for light control of membrane voltage and intracellular cation concentrations has revolutionized the field of optogenetics, and they are now considered for potential clinical applications; for review see (5–7). Fourteen native ChR sequences have been reported so far, but this number is only the tip of the iceberg, taking into account the thousands of known phototactic chlorophyte species.
All ChRs comprise a seven transmembrane-helix (7TM) scaffold homologous to other microbial rhodopsins and an additional large cytoplasmic domain. A recently obtained crystal structure of the 7TM domain of C1C2, a chimeric ChR (8), shows essential similarity with those of better characterized haloarchaeal rhodopsins, such as bacteriorhodopsin (BR) (9,10) and Natronomonas pharaonis sensory rhodopsin II (NpSRII) (11,12). Nevertheless, there are also clear structural differences between ChRs and other known microbial rhodopsins, especially in the first two helices. Identification of structural differences responsible for the unique channel function of ChRs is a goal of ongoing research.
As in all rhodopsins, the chromophore in ChRs is attached via a Schiff base linkage to a conserved lysine in the seventh helix. Retinal reconstitution studies in blind C. reinhardtii mutants identified all-trans-retinal as the functional isomer in vivo (13–15). However, HPLC analysis and resonance Raman spectroscopy also indicated a significant amount of 13-cis isomer (up to 30% of the total) in the unphotolyzed state of the 7TM domains of some ChRs (16,17). Two independent photocycles for all-trans, 15-anti, and 13-cis, 15-syn pigment forms have been proposed at least for the C128T mutant of channelrhodopsin 2 from C. reinhardtii (CrChR2) (18).
The photoactive sites of BR and NpSRII contain two carboxylates that form a complex counterion to the protonated Schiff base (Asp-85/75 and Asp-212/201), an arginine residue (Arg-82/72) and three water molecules. This overall arrangement was also found in the ChR chimera, although only one water molecule (W402) was resolved in the vicinity of the Schiff base (8). Computational analysis of homology models showed increased flexibility of W401 in the retinal binding pocket of Chlamydomonas-type ChRs (19), which may explain its absence from the crystal structure. The side chain of the Arg-82 homolog in the ChR chimera is found in a position similar to that in NpSRII rather than in BR, i.e., it faces away from the Schiff base.
The interaction between the positively charged Schiff base and its negatively charged counterion is the major factor that determines the absorption maximum of retinylidene proteins (20–22). Neutralization of the counterion carboxylates leads to reduction in the strength of this interaction and a bathochromic shift of the absorption maximum that has been observed in many prokaryotic rhodopsins (23–28). Similar bathochromic shifts were also reported for relatively high-efficiency ChRs: CrChR2 and PsChR, a channelrhodopsin from Platymonas subcordiformis (17,29).
Despite the overall similarity between the photoactive sites of C1C2 and BR, the distances between the protonated Schiff base and the photoactive site carboxylic acid residues are different in the two structures, as are their hydrogen bonding patterns (8). Empirical calculation yielded a lower pKa value for the Asp-212 homolog than for the Asp-85 homolog in C1C2. In CrChR2 protonation of the Asp-212 homolog (Asp-253) concomitant with light-induced Schiff base deprotonation was detected by time-resolved Fourier transform infrared spectroscopy (FTIR), whereas no such signal could be assigned to the Asp-85 homolog (Glu-123) (30).
Previously, we probed intramolecular transfers of the Schiff base proton in ChRs directly by photoelectric measurements upon laser flash excitation (31). Fast outward proton transfer currents were detected in relatively low-efficiency ChRs, such as Chlamydomonas augustae channelrhodopsin 1 (CaChR1), but not in higher efficiency ChRs. An anticorrelation between the ability to generate detectable outward proton transfer current and channel activity was observed. We found that two residues can act as acceptors of the Schiff base proton in CaChR1. Both outward proton transfer current and formation of the M intermediate of the photocycle are slowed upon neutralizing mutation of the Asp-85 homolog (E169Q) and accelerated upon mutation of the Asp-212 homolog (D299N). These findings indicate that in the absence of the second photoactive site carboxylate, Glu-169 acts as a more efficient (i.e., primary) proton acceptor than Asp-299 in the absence of Glu-169.
In this study, we extend our analysis to pH dependence of the absorption spectrum in wild-type (WT) CaChR1 and its mutants. The results show that in the WT the Asp-85 homolog (Glu-169) has a pKa∼9, but this value is substantially decreased upon neutralization of the second photoactive site carboxylate (Asp-299). An even greater decrease in pKa was observed upon introducing a Lys residue in the position corresponding to Lys-132 of C1C2, which in WT CaChR1 is occupied by a Phe residue. Furthermore, we have found that the presence of the Lys-132 homolog determines the direction of the spectral shifts measured upon replacement of the photoactive site carboxylic acid residues with nonionizable residues in tested ChRs at neutral pH. Red spectral shifts were detected when this Lys was present, whether naturally or by mutagenesis, and blue shifts were measured when it was replaced with a neutral residue. These results indicate a role of the Lys-132 homolog in modulation of the effective charge of the photoactive site carboxylic acid residues in tested ChRs, whereas the influence of the Arg-82 homolog (BR numbering) on color tuning is minimal.
Materials and Methods
Whole-cell patch clamp measurements in HEK293 cells
The mammalian expression constructs that contained cDNAs encoding the 7TM domains of CaChR1 (amino acid residues 1–352), DsChR1 (residues 1–365), and PsChR (residues 1–326) in frame with an EYFP tag were as described previously (31). The construct for C1C2 (residues 1–356) was made as in (8). Single and double mutations were introduced using a QuikChange XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) and verified by DNA sequencing. HEK293 (human embryonic kidney) cells were transfected as described (31). Fabrication of patch pipettes and contents of pipette and bath solutions were as before (31). The pH of the bath solution was 7.4 unless otherwise stated.
Continuous illumination was provided by a Polychrome IV light source (T.I.L.L. Photonics GMBH, Grafelfing, Germany) pulsed with a mechanical shutter (Uniblitz model LS6, Vincent Associates, Rochester, NY; half-opening time 0.5 ms, maximal quantum density at the focal plane of the 40× objective lens ∼2 × 1022 photons × m−2 × s−1). Laser excitation was provided by Minilite Nd:YAG laser (532 nm, pulsewidth 6 ns, energy 12 mJ; Continuum, Santa Clara, CA). The signals were recorded with an Axopatch 200B amplifier and digitized with a Digidata 1440A using the pClamp 10.2 software (all from Molecular Devices, Union City, CA) at the sampling rate 4 and 200 μs/point, respectively, for laser and Polychrome excitation.
To determine the action spectra, photocurrents were recorded in response to 50-ms light pulses in the linear range of the stimulus-response curve (usually 5% of the maximal intensity) with a 30-s dark interval between consecutive pulses. The wavelength was increased in 10-nm steps and then decreased in a mirror sequence, and the signals from the forward and reverse series were averaged to eliminate possible changes in sensitivity of the response during experiments. The initial slopes of the photocurrent were corrected for the quantum output of the light source at a given wavelength. The precise positions of the spectral maxima were determined by Gaussian fit of 4–5 data points around the main peak with the baseline fixed at zero.
Expression and purification of pigments from Pichia pastoris
Expression constructs contained the 7TM domains of ChRs with a TEV protease site at the N-terminus and a nine-His tag at the C-terminus in the pPIC9K vector backbone (Invitrogen, Carlsbad, CA). Geneticin-resistant P. pastoris clones were selected according to the manufacturer’s instructions. Protein expression and purification on a Ni-NTA column (Qiagen, Hilden, Germany) were carried out as described (32). For absorption measurements the protein samples were concentrated in 100 mM NaCl, 0.01% DDM, 20 mM HEPES (pH 7.4, unless otherwise stated).
Absorption spectroscopy and flash photolysis
Absorption spectra of partially purified ChRs in the UV-Visible range were recorded on a Cary 4000 spectrophotometer (Varian, Palo Alto, CA). pH titration was carried out by the addition of small volumes of 1 M NaOH, 0.5 M Tris (pH 10), 0.5 M citric acid, or 1 M HCl. Light-induced absorption changes of Pichia-expressed pigments were measured with a laboratory-constructed crossbeam apparatus, essentially as described (17), but for pigments with fast photocycles data were acquired with a GaGe Octopus digitizer board (model CS8327, DynamicSignals LLC, Lockport, IL) at the sampling rate up to 40 ns/point. Data analysis was performed with pClamp 10.2 (Molecular Devices) and OriginPro 7 (OriginLab, Northampton, MA) software.
Structural modeling
Homology models of WT CaChR1 and its mutants were built by the Swiss-Model server using the C1C2 structure (3ug9) (8) as a template. The PROPKA 3.1 method (33) was used for empirical prediction of pKa values for these models.
Results
Replacement of the photoactive site carboxylic acid residues with nonionizable residues causes opposite spectral shifts in two groups of ChRs
In all tested ChRs substitution of the photoactive site carboxylic residues with nonionizable residues reduces the amplitude of channel currents (4,8,31). Here, we examined the influence of such mutations on color tuning in ChRs by action spectroscopy of photoinduced electrical currents in HEK293 cells. This approach allowed us to assess properties of ChRs in their functional state in biological membranes and rapidly screen numerous mutants. Channel currents were recorded in response to short light pulses of variable wavelength in the linear range of the stimulus-response curve, and action spectra were determined as described in the Materials and Methods and (34).
In low-efficiency CaChR1 mutations of the residues corresponding to Asp-85 and Asp-212 of BR (E169Q and D299N) caused pronounced blue shifts as compared to the WT (Fig. 1 A). The same spectral dependence on the presence of the carboxylic residues in the photoactive site was found in another low-efficiency ChR, DsChR1. This protein has an Ala residue in the position of Asp-85, where glutamate is found in all other known ChRs (35). The action spectrum of channel currents generated by the WT DsChR1, which contains only the Asp-212 homolog (Glu-309), was blue-shifted with respect to that of the DsChR1_A178E mutant, in which both photoactive site carboxylic acid residues are in place (Fig. 1 B, circles and squares); see also (35). The E309Q mutation introduced in the DsChR1_A178E background (i.e., with glutamate in the Asp-85 position) also caused a blue shift of the action spectrum (Fig. 1 B, triangles).
Figure 1.
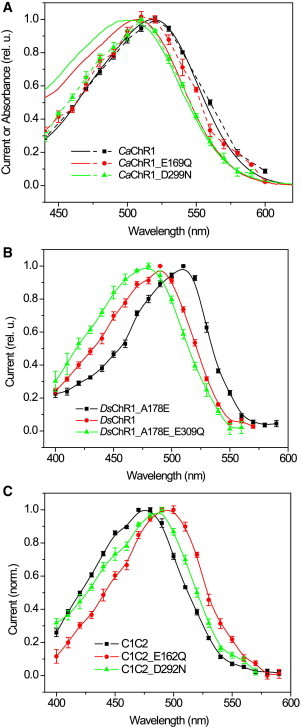
The action spectra of channel currents (symbols and dashed lines in A, symbols and solid lines in B and C) generated in HEK293 cells by: (A) CaChR1 and its E169Q and D299N mutants; (B) WT DsChR1 and its A178E and A178E_E309Q mutants; (C) C1C2 and its E162Q and D292N mutants. Currents generated by the mutants of CaChR1 were measured at pH 5.4 because at pH 7.4 their amplitude was at the noise level. In all panels squares show the data from the pigments in which both photoactive site carboxylic acid residues are present, circles from the pigments in which the Asp-85 homolog is absent, and triangles from the pigments in which the Asp-212 homolog is absent. The solid lines in (A) show absorption spectra of detergent-purified WT CaChR1 and its E169Q and D299N mutants at pH 5.4. To see this figure in color, go online.
In contrast, the corresponding mutations in relatively high-efficient CrChR2 and PsChR caused red spectral shifts (17,29), as in most other microbial rhodopsins. Similar results were observed in the C1C2 chimera, the only ChR for which high-resolution structural data are available (8). Mutation of either of the photoactive site carboxylic residues to nonionizable residues caused red spectral shifts in C1C2 (Fig. 1 C), as predicted by computational studies (36,37). Although this chimera mostly consists of CrChR1 (helices 1–5), in this respect it resembled more CrChR2 than CrChR1, in the latter of which practically no spectral shift was reported upon neutral residue substitution of the Asp-85 homolog (38). Mutation of the Asp-85 homolog caused a larger red shift than that of the other carboxylic residue in high-efficiency C1C2 and PsChR, whereas mutation of the Asp-212 homolog caused a larger blue shift in low-efficiency CaChR1. Numerical values of spectral maxima are summarized in Table 1.
Table 1.
Peak wavelength (nm) of action spectra for channel currents generated by WT and mutant ChRs in HEK293 cells (residue numbers are according to the C1C2 sequence)
| WT | E162Q | D292N | K132Fa | K132Fa_E162Q | K132Fa_D292N | R159A | |
|---|---|---|---|---|---|---|---|
| CaChR1 | 519 | 513 | 509 | 482 | 518 | 519 | 510 |
| C1C2 | 476 | 493 | 485 | 489 | 485 | 476 | 478 |
| PsChR | 442 | 473b | 459b | 455 | 452 | 442 | 442 |
In CaChR1 a Phe residue was mutated to Lys, and in C1C2 and PsChR a Lys residue was mutated to Phe.
Data are from (17).
One possible explanation for the observed blue-shifted spectra of the CaChR1_E169Q and D299N mutants would be a larger fraction of 13-cis retinal than in the WT, which is responsible for a blue spectral shift after dark adaptation of BR (39) and light adaptation of Anabaena sensory rhodopsin (40). When probed by preresonance Raman scattering (785-nm excitation), the fraction of 13-cis retinal was found to be slightly increased in the E169Q mutant compared to the WT, but the magnitude of this change was too small to explain the blue shift we observe (personal communication from Prof. Kenneth J. Rothschild, Boston University).
Another possibility we considered was that the measured blue spectral shifts of the CaChR1_169Q and D299N mutants reflect binding of a Cl− ion, as occurs, for example, in the BR_D85N_D212N double mutant, the absorption maximum of which is close to that of WT BR (41). However, no spectral changes were observed in the CaChR1 mutants after their transfer from a 100 mM Cl− solution to that of SO42-(data not shown).
A Lys residue in the second helix determines the direction of the spectral shift in ChRs
Sequence alignment revealed that ChRs in which mutations of the photoactive site carboxylic residues cause red spectral shifts contain a conserved Lys residue in the position 132 (CrChR1/C1C2 numbering), whereas in those that show blue spectral shifts upon corresponding mutations this position is occupied by a different residue (Fig. 2, left). According to the C1C2 structure (8), the side chain of Lys-132 is located within hydrogen bonding distance from oxygen atoms of both photoactive site carboxylic acid residues (Fig. 2, right).
Figure 2.
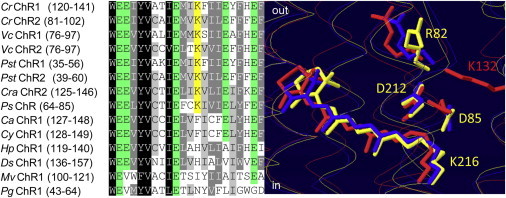
(Left) A Clustal W alignment of helix B regions in reported ChRs from the following algal species (abbreviated by the first two or three letters in the sequence title): Chlamydomonas reinhardtii, Volvox carteri, Pleodorina starrii, C. raudensis, Platymonas subcordiformis, C. augustae, C. yellowstonensis, Haematococcus pluvialis, Dunaliella salina, Mesostigma viride, and Pyramimonas gelidicola. Lysine residues corresponding to Lys-132 (CrChR1/C1C2 numbering, because from the N-terminus to beyond the shown portion of the C1C2 derives from CrChR1) are highlighted yellow, and glutamate residues green. (Right) An overlay of photoactive site structures of BR (1c3w, yellow), NpSRII (1jgj, blue), and C1C2 ChR (3ug9, red) generated by DeepView software.
To test the role of the Lys-132 homolog in the direction of spectral shifts induced by mutation of the carboxylic acid residues at the photoactive site, we generated three series of double mutants. In low-efficiency CaChR1 this position is occupied by Phe. In the CaChR1_F139K mutant background replacement of either of the carboxylic acid residues with nonionizable residues led to red shifts (Fig. 3 A). On the other hand, when the Lys-132 homolog was mutated to Phe in C1C2 and PsChR, the corresponding mutations of the carboxylic acid residues led to blue shifts (Fig. 3, B and C).
Figure 3.
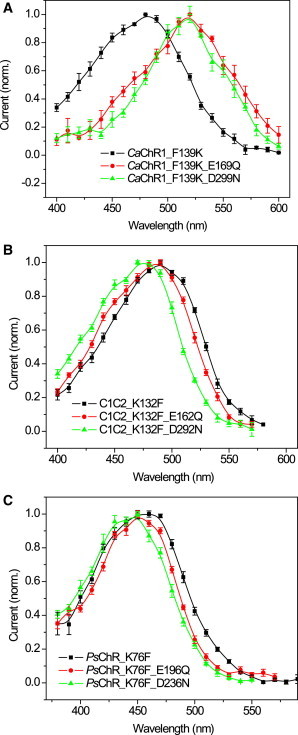
Action spectra of channel currents generated in HEK293 cells by ChRs with the modified Lys-132 position and their carboxylic acid residue mutants: (A) CaChR1_F139K; (B) C1C2_K132F; (C) PsChR_K76F. In all panels squares show the data from the pigments in which both photoactive site carboxylic acid residues are present, circles in which the Asp-85 homolog is absent, and triangles in which the Asp-212 homolog is absent. To see this figure in color, go online.
Interestingly, in all three examined ChRs (low-efficiency CaChR1 and high-efficiency C1C2 and PsChR), the pigments with Phe at the Lys-132 position showed red-shifted spectra relative to the corresponding pigments with Lys, regardless of whether this form was the WT or obtained by mutagenesis, although in CaChR1 the shift was larger than in C1C2 and PsChR (Fig. 4). The absorption spectrum of Pichia-expressed detergent-purified CaChR1_F139K was also blue-shifted with respect to the WT and showed a pronounced fine structure similar to that of WT CrChR2 (Fig. 4 A).
Figure 4.
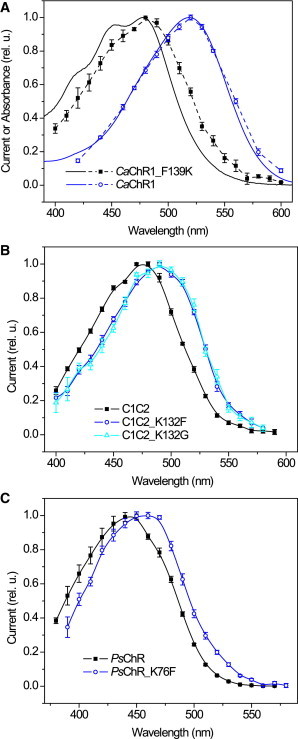
(A) The action spectra of channel currents (symbols and dashed lines in A, symbols and solid lines in B and C) generated in HEK293 cells by: (A) CaChR1 and its F139K mutant; (B) C1C2 chimera and its K132F and K132G mutants; (C) WT PsChR and its K76F mutant. The solid lines in (A) show the absorption spectra of detergent-purified pigments. In all panels solid squares show the data from the pigments in which the Lys-132 homolog is present, and open circles and triangles in which it is absent. To see this figure in color, go online.
Vertical excitation energy calculation by a combined quantum mechanics/molecular mechanics (QM/MM) approach predicted a blue shift upon neutralization of Lys-132 in C1C2 (36,37). In the latter study the spectral shift was specifically computed for the K132G mutant. As different substitutions of the same residue can sometimes cause different effects, we also tested this particular mutant in addition to C1C2_K132F. The spectra of currents generated by the two mutants were indistinguishable and showed a 13-nm bathochromic shift relative to the WT (Fig. 4 B), i.e., in the opposite direction than predicted by the calculations.
The pH titration of the absorption spectrum of CaChR1 provides additional evidence for the existence of two alternative acceptors of the Schiff base proton
WT CaChR1 and its mutants could be expressed and purified from Pichia, which allowed us to measure the dependence of their spectra on pH over a wider range than HEK cells can tolerate. The absorption maximum of CaChR1 progressively shifted to longer wavelengths within the entire tested range from pH 10 to 2 (Fig. 5, squares), which is typical for neutralization of the Schiff base counterion in all other microbial rhodopsins. Three spectral transitions of unequal amplitudes could be identified in the titration curve of WT CaChR1. The largest transition occurred at pH∼9, which is consistent with the results obtained by action spectroscopy of channel currents measured at pH 7.4 and 9 (32), although the magnitude of the spectral shift was smaller in cells than in the purified pigments likely due to the influence of the membrane lipids and/or a different ionic composition of the cell interior. The spectral transitions observed at acidic pH showed much smaller amplitudes, but nevertheless could be fit to yield pKas 5.5 and 2.7.
Figure 5.
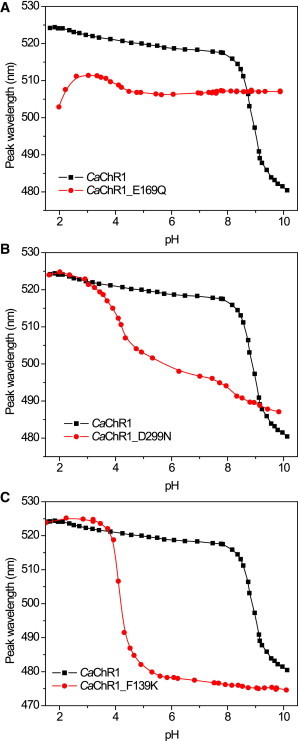
pH titration of the absorption maximum of detergent-purified WT CaChR1 (squares, the same data are shown in all three panels for reference) and its mutants (circles): E169Q (A), D299N (B), and F139K (C). To see this figure in color, go online.
When the Asp-85 homolog was replaced with a nonionizable residue in the CaChR1_E169Q mutant, the major transition with pKa∼9 disappeared (Fig. 5 A, circles), indicating that the transition reflects mainly protonation/deprotonation of this residue. The results of empirical pKa calculations supported this assignment. The PROPKA3.1 algorithm (42) identified the Asp-85 and Asp-212 homologs in CaChR1 as noncovalently coupled residues, and the pKa of the former predicted taking this into account was very close to our experimental value (Table 2).
Table 2.
PROPKA calculated pKa values of the photoactive site residues in WT CaChR1 and its mutants (residue numbers are according to the CaChR1 sequence)
| WT | E169Q | D299N | F139K | R166A | |
|---|---|---|---|---|---|
| Glu-169 | 10.89 (8.85)a | N/A | 5.77 | 5.57 (3.52) | 10.97 (8.94) |
| Asp-299 | 4.9 (6.93) | 4.54 | N/A | 2.91 (4.94) | 5.13 (7.17) |
| Lys-139 | N/A | N/A | N/A | 13.99 | N/A |
The numbers in parentheses were calculated using a program option for coupled residues (42).
Our earlier study (31) showed that in CaChR1 the Asp-212 homolog serves as an alternative proton acceptor. Therefore, we expect one of the minor acidic transitions to be attributable to its protonation/deprotonation. We hypothesized that it is the transition with pKa 5.5 because in the CaChR1_E169Q mutant the amplitude of the first acidic transition increased and its pKa shifted from 5.5 to 3.9 (Fig. 5 A, circles), as expected after elimination of one of the two alternative acceptors. Our interpretation was confirmed by measurements of the pH dependence of the M intermediate amplitude. The pKa of a strong decrease in the M amplitude upon acidification was very close to that of the first acidic spectral transition (Fig. S1 in the Supporting Material). The experimentally determined value 5.5 was between those predicted for the Asp-212 homolog (Asp-299) by using PROPKA software with and without the coupling option (Table 2).
Previously, we had shown that the rates of M intermediate formation and outward transfer of the Schiff base proton are slowed in the E169Q mutant and accelerated in the D299N mutant of CaChR1, suggesting that the Asp-212 homolog acts as a less efficient (secondary) proton acceptor in this pigment (31). The smaller amplitude of spectral transition that we assign to protonation of Asp-299 than that of Glu-169 corroborates this conclusion. A weaker influence of the Asp-212 homolog protonation state on the absorption spectrum in CaChR1 may indicate its position more distant from the Schiff base and/or a weaker interaction of this residue with the Schiff base than that of the Asp-85 homolog. If this interpretation is correct, the active site of CaChR1 is more similar in this respect to that of BR than to that of the C1C2 chimera. Note that given their pKa values, our data indicate that in CaChR1 the Asp-212 homolog is the predominant Schiff base proton acceptor in the typically near-neutral pH conditions used in optogenetic applications. An FTIR study reports a similar conclusion for CrChR2 (30).
The pKa of the Asp-85 homolog in CaChR1 is controlled by the Asp-212 homolog and the Lys-132 homolog
When the Asp-212 homolog was neutralized in the CaChR1_D299N mutant, the major spectral transition shifted to pH∼4 (Fig. 5 B, circles). Our interpretation of this result would be a decrease in pKa of Glu-169 due to a reduction of the local negative charge or hydrogen bonding (43) resulting from mutation of Asp-299. This shift in pKa of Glu-169 was much larger than that of Asp-299 measured after mutation of Glu-169. Such asymmetry was also predicted by empirical calculation of pKa values using homology models (Table 2).
Introduction of a lysine residue in the position of Lys-132 in CaChR1 caused an even more pronounced shift of the titration curve to lower pH values evident in the F139K mutant (Fig. 5 C, circles), indicating an even greater decrease in the pKa of Glu-169 than upon neutral residue replacement of Asp-299, also predicted by empirical calculations (Table 2). Furthermore, introduction of the Lys-132 homolog in CaChR1 caused even stronger acceleration of M formation (Fig. 6 A) and of outward proton transfer current (Fig. 6 B) relative to the WT, than was previously observed in the CaChR1_D299N mutant (31).
Figure 6.
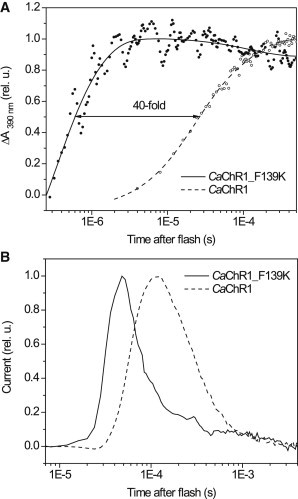
(A) Absorbance changes monitored at 390 nm in WT CaChR1 (open circles, dashed line) and its F139K mutant (solid circles, solid line) in response to a 6-ns laser flash (532 nm). The arrow shows the difference in the time of the half-maximal amplitude of the signal. (B) Outward proton transfer currents recorded upon laser excitation from WT CaChR1 (dashed line) and its F139K mutant (solid line) expressed in HEK293 cells at the reversal potential for channel currents to minimize their contribution to the signal kinetics. The current kinetics is limited by the time resolution of the measuring system. In both panels the signals were normalized to their maximal values.
The Lys-132 homolog regulates channel current kinetics
It has previously been reported that alanine substitution for Lys-132 in the C1C2 chimera did not change the amplitude of channel currents, but led to acceleration of their kinetics and an increase in inactivation under continuous illumination (8). We observed similar changes, as compared to the WT, in PsChR when the corresponding Lys-76 was replaced with Phe found at this site in CaChR1 (Fig. 7 A). Introducing a lysine at the respective position in CaChR1 (the F139K mutant) caused a very dramatic decrease in the current decay rate and reduction of inactivation (Fig. 7 B). Slower current decay in the CaChR1_F139K mutant correlates with a significant slowing of the photocycle (see Fig. S3).
Figure 7.
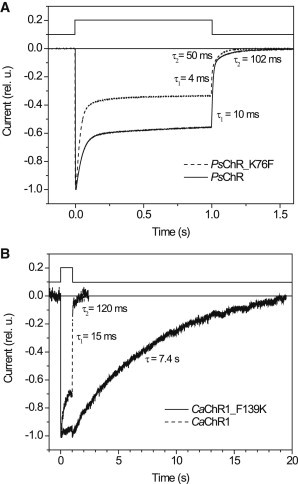
Photocurrents generated in HEK293 cells by: (A) WT PsChR (solid line) and its K76F mutant (dashed line); (B) WT CaChR1 (dashed line) and its F139K mutant (solid line). Excitation wavelengths were (in nm): 440 for PsChR, 450 for PsChR_K76F, 520 for CaChR1, and 470 for CaChR1_F139K. In both panels the traces were normalized to their maximal value. Light excitation protocol is shown at the top. The numbers are time constants (τ) of current decay generated by its fitting with exponential functions.
To summarize, regardless of whether this lysine residue was initially present in the WT or introduced by mutation, slower currents were observed with the Lys than without it in each of the three tested ChRs, although the degree of this effect was much greater in CaChR1 than in C1C2 and PsChR. Introduction of the Lys-132 homolog in CaChR1 did not significantly change the amplitude of channel currents, i.e., it did not convert CaChR1 into a high-efficiency ChR. Therefore, although this Lys is found mostly in ChRs capable of generating greater channel currents, the residue itself is not sufficient for high efficiency.
The role of the Arg-82 homolog in spectral tuning in ChRs is negligible
Finally, we tested a possible role of the Arg-82 homolog in color tuning in CaChR1, C1C2, and PsChR by generation of their alanine replacement mutants (CaChR1_R166A, C1C2_R159A, and PsChR_R103A). In CaChR1 and PsChR the substitution led to a strong reduction of channel currents as reported for C1C2 (8) and CrChR2 (44). However, the influence of alanine replacement of the Arg-82 homolog on action spectra was very minor in all three tested ChRs (Fig. S2) as predicted by PROPKA in CaChR1 (Table 2).
Discussion
In the unphotolyzed state of BR and NpSRII the photoactive site carboxylates are unprotonated, and their interaction with the positively charged Schiff base makes the largest contribution to spectral tuning (20–22). The residue corresponding to Asp-212 in BR is conserved as a carboxylic acid residue in all known ChRs, and the position of Asp-85 is occupied by Glu in all ChRs except DsChR1 (in which a glutamate residue is found in the Asp-212 position). From comparative analysis of M formation and intramolecular proton transfer currents, we concluded previously that the Asp-85 and Asp-212 homologs act as alternative acceptors of the Schiff base proton in low-efficiency ChRs such as CaChR1 (31). The pH titration data presented here show that in WT CaChR1 the Asp-85 homolog has pKa ∼9. The pH of algal cultures reach values as high as 10–11 without growth inhibition ((45) and our unpublished observations), under which condition the more efficient Asp-85 homolog would be expected to act as the main Schiff base proton acceptor. However, under neutral or acidic conditions the Asp-85 homolog is neutral and the Schiff base proton is transferred to an alternative acceptor—the Asp-212 homolog.
The protonation states of the photoactive site carboxylic acid residues have been probed by vibrational spectroscopy in CrChR2, the most popular ChR used as an optogenetic tool. A time-resolved FTIR study identified the Asp-212 homolog (Asp-253) rather than the Asp-85 homolog (Glu-123) as an acceptor of the Schiff base proton at neutral pH (30). In contrast to BR, in which neutralization of either Asp-85 or Asp-212 resulted in a dramatic inhibition of M formation (46), the D253N mutation hardly affected it in CrChR2 (30), which argues for the existence of an alternative proton acceptor.
In BR neutralization of positively charged Arg-82 leads to a considerable red shift, as do neutralization of the photoactive site carboxylic acid residues, indicating that their effective charge is modulated by Arg-82 (47–49). The shift is smaller in NpSRII (50,51), where the side chain of the corresponding Arg-72 is directed away from the Schiff base. The position of this side chain is stabilized in NpSRII by a salt bridge between the positively charged guanidine moiety and the negatively charged Asp-193, a residue absent in BR (20). We found only a small influence of the Arg-82 homolog on wavelength regulation in the C1C2 chimera, in which a similar outward direction of the corresponding side chain was shown by x-ray crystallography. Our results confirm computational predictions for this residue in C1C2 (36,37) and suggest its similar geometry in CaChR1 and PsChR, for which crystal structures are not available.
In the crystallized C1C2 chimera the side chain of Lys-132 is located within hydrogen bonding distance of the homologs of Asp-85 and Asp-212 that form a complex counterion in BR (8). Our mutant analysis provides direct evidence for the role of the Lys-132 homolog in wavelength regulation in ChRs: its substitution with a neutral residue caused a red spectral shift in both C1C2 chimera and PsChR, whereas its substitution for native Phe in CaChR1 caused a large blue shift. Furthermore, this lysine also determined the direction of the spectral shifts observed upon mutation of the photoactive site carboxylic acid residues. When the Lys-132 homolog was present, whether naturally or introduced by mutagenesis, red shifts were detected in neutral residue substitutions mutants of the Asp-85 and Asp-212 homologs, as expected from a weakened electrostatic interaction with the Schiff base; when the Lys-132 homolog was absent, atypical blue shifts were measured.
The following scheme can be suggested to explain the blue shifts measured in the CaChR1_E169Q and D299N mutants and corresponding mutants of other ChRs that lack the Lys-132 homolog. Neutralization of Asp-299 decreases the pKa of Glu-169 that strongly interacts with retinal and is neutral in the WT near neutral pH. The resulting generation of a negative charge near the Schiff base causes a blue spectral shift. Similarly, neutralization of Glu-169 decreases the pKa of Asp-299. This decrease is smaller and the interaction of this residue with retinal is weaker, and consequently the resultant blue shift is smaller than upon mutation of Asp-299. Our results indicate that the Lys-132 homolog controls the effective charge of the photoactive site carboxylic acid residues in tested ChRs in a manner similar to that of Arg-82 in BR (47–49).
In a previous study (31) we detected outward proton transfers only in ChRs that generate relatively small net channel currents (low-efficiency ChRs). Moreover, within this group an anticorrelation between the amplitude of channel current and fast outward proton transfer was observed, which led us to suggest that channel function evolved in ChRs at the expense of their putative ancestors’ proton transfer function. Although the presence of the Lys-132 homolog is more typical of native ChRs capable of generating greater net channel currents, the results of our mutant analysis showed that this residue itself is not solely responsible for their high efficiency. Nor does the presence of the Lys-132 homolog guarantee the lack of a detectable fast outward proton transfer in a particular ChR variant, as exemplified by VcChR1 that does contain this residue and yet exhibits fast outward proton transfer current (31).
It should be noted that color tuning in ChRs does not appear to be confined to residues in the retinal binding pocket. For example, Glu-87 was identified as a key residue that controls pH-dependence of wavelength regulation in CrChR1 (52). Using homology modeling, the authors suggested that Glu-87 corresponds to Met-20 in BR and forms part of the complex counterion in CrChR1. However, the C1C2 crystal structure (8) revealed that in fact this residue is located close to the outside membrane surface. Computational analysis of C1C2 structure also predicted significant contributions of two other outside surface residues, Lys-209 and Arg-213, to color tuning (36), but experimental tests of these predictions have not been reported.
The absorption spectrum of detergent-purified WT CaChR1 matched the action spectrum of channel currents generated by this protein at the same bath pH ((32) and this work). However, for several CaChR1 mutants we observed a small blue shift of the absorption spectra as compared to the respective action spectra. Similar observations have been reported previously for WT CrChR1 (53) and PsChR (17). One explanation could be the influence of membrane lipids and/or the difference in the ionic composition on spectral properties of the pigments. Alternatively, this difference may reflect the existence of a substantial nonfunctional pigment fraction (e.g., containing 13-cis retinal) that contributes to the absorption spectrum of the purified preparation, but not to the action spectrum of photocurrents.
Interestingly, the action spectrum of CrChR1-mediated photoreceptor currents measured in intact C. reinhardtii cells peaks at 510 nm (1), but that of channel currents in heterologous systems only at 497 nm even at nonphysiological pH 5.5 (53). The absence of the C-terminal domain in expression constructs does not influence CrChR1 spectral properties (32), and factors responsible for red shifting the spectrum in native algal cells remain to be identified.
Despite ever growing interest in ChRs for optogenetic applications, our understanding of the relationship between their structure and function is still very limited. Determination of factors that contribute to wavelength regulation in these pigments is not only of fundamental importance for basic photoscience, but is also needed for rational design of optogenetic tools. Extension of ChR absorption to longer wavelengths is desirable to minimize scattering by biological tissue, and availability of molecules with nonoverlapping spectra is required for multimodal optical control.
Acknowledgments
The authors thank Dr. Brandon Goblirsch for help with PROPKA software and useful discussions and Cassie Lane for her expert technical assistance.
This work was supported by grant R01GM027750 from the National Institute of General Medical Sciences, grant R21MH098288 from the National Institute of Mental Health, the Hermann Eye Fund, and Endowed Chair AU-0009 to J.L.S. from the Robert A. Welch Foundation.
Footnotes
Hai Li and Elena G. Govorunova contributed equally to this work.
Supporting Material
References
- 1.Sineshchekov O.A., Jung K.-H., Spudich J.L. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 2002;99:8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Govorunova E.G., Jung K.H., Spudich J.L. Chlamydomonas sensory rhodopsins A and B: cellular content and role in photophobic responses. Biophys. J. 2004;86:2342–2349. doi: 10.1016/S0006-3495(04)74291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagel G., Ollig D., Hegemann P. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 4.Nagel G., Szellas T., Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deisseroth K. Optogenetics. Nat. Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow B.Y., Boyden E.S. Optogenetics and translational medicine. Sci. Transl. Med. 2013;5:177ps5. doi: 10.1126/scitranslmed.3003101. [DOI] [PubMed] [Google Scholar]
- 7.Yawo H., Asano T., Ishizuka T. Optogenetic manipulation of neural and non-neural functions. Dev. Growth Differ. 2013;55:474–490. doi: 10.1111/dgd.12053. [DOI] [PubMed] [Google Scholar]
- 8.Kato H.E., Zhang F., Nureki O. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luecke H., Schobert B., Lanyi J.K. Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 10.Belrhali H., Nollert P., Pebay-Peyroula E. Protein, lipid and water organization in bacteriorhodopsin crystals: a molecular view of the purple membrane at 1.9 A resolution. Structure. 1999;7:909–917. doi: 10.1016/s0969-2126(99)80118-x. [DOI] [PubMed] [Google Scholar]
- 11.Luecke H., Schobert B., Spudich J.L. Crystal structure of sensory rhodopsin II at 2.4 angstroms: insights into color tuning and transducer interaction. Science. 2001;293:1499–1503. doi: 10.1126/science.1062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Royant A., Nollert P., Navarro J. X-ray structure of sensory rhodopsin II at 2.1-A resolution. Proc. Natl. Acad. Sci. USA. 2001;98:10131–10136. doi: 10.1073/pnas.181203898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawson M.A., Zacks D.N., Spudich J.L. Retinal analog restoration of photophobic responses in a blind Chlamydomonas reinhardtii mutant. Evidence for an archaebacterial like chromophore in a eukaryotic rhodopsin. Biophys. J. 1991;60:1490–1498. doi: 10.1016/S0006-3495(91)82184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegemann P., Gärtner W., Uhl R. All-trans retinal constitutes the functional chromophore in Chlamydomonas rhodopsin. Biophys. J. 1991;60:1477–1489. doi: 10.1016/S0006-3495(91)82183-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi T., Yoshihara K., Nakanishi K. Photoisomerization of retinal at 13-ene is important for phototaxis of Chlamydomonas reinhardtii: simultaneous measurements of phototactic and photophobic responses. Biochem. Biophys. Res. Commun. 1991;178:1273–1279. doi: 10.1016/0006-291x(91)91031-7. [DOI] [PubMed] [Google Scholar]
- 16.Nack M., Radu I., Heberle J. The retinal structure of channelrhodopsin-2 assessed by resonance Raman spectroscopy. FEBS Lett. 2009;583:3676–3680. doi: 10.1016/j.febslet.2009.10.052. [DOI] [PubMed] [Google Scholar]
- 17.Govorunova E.G., Sineshchekov O.A., Spudich J.L. Characterization of a highly efficient blue-shifted channelrhodopsin from the marine alga Platymonas subcordiformis. J. Biol. Chem. 2013;288:29911–29922. doi: 10.1074/jbc.M113.505495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritter E., Piwowarski P., Bartl F.J. Light-dark adaptation of channelrhodopsin C128T mutant. J. Biol. Chem. 2013;288:10451–10458. doi: 10.1074/jbc.M112.446427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welke K., Frahmcke J.S., Elstner M. Color tuning in binding pocket models of the channelrhodopsins. J. Phys. Chem. B. 2011;115:15119–15128. doi: 10.1021/jp2085457. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi S., Tajkhorshid E., Schulten K. Structural determinants of spectral tuning in retinal proteins - bacteriorhodopsin vs. sensory rhodopsin II. J. Phys. Chem. B. 2001;105:10124–10131. [Google Scholar]
- 21.Hoffmann M., Wanko M., Elstner M. Color tuning in rhodopsins: the mechanism for the spectral shift between bacteriorhodopsin and sensory rhodopsin II. J. Am. Chem. Soc. 2006;128:10808–10818. doi: 10.1021/ja062082i. [DOI] [PubMed] [Google Scholar]
- 22.Sekharan S., Sugihara M., Buss V. Origin of spectral tuning in rhodopsin–it is not the binding pocket. Angew. Chem. Int. Ed. Engl. 2007;46:269–271. doi: 10.1002/anie.200603306. [DOI] [PubMed] [Google Scholar]
- 23.Mogi T., Stern L.J., Khorana H.G. Aspartic acid substitutions affect proton translocation by bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 1988;85:4148–4152. doi: 10.1073/pnas.85.12.4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson K.D., Zhang X.N., Spudich J.L. Residue replacements of buried aspartyl and related residues in sensory rhodopsin I: D201N produces inverted phototaxis signals. Proc. Natl. Acad. Sci. USA. 1995;92:3185–3189. doi: 10.1073/pnas.92.8.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu J., Spudich E.N., Spudich J.L. Effects of substitutions D73E, D73N, D103N and V106M on signaling and pH titration of sensory rhodopsin II. Photochem. Photobiol. 1997;66:788–791. doi: 10.1111/j.1751-1097.1997.tb03225.x. [DOI] [PubMed] [Google Scholar]
- 26.Chizhov I., Schmies G., Engelhard M. The photophobic receptor from Natronobacterium pharaonis: temperature and pH dependencies of the photocycle of sensory rhodopsin II. Biophys. J. 1998;75:999–1009. doi: 10.1016/S0006-3495(98)77588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dioumaev A.K., Brown L.S., Lanyi J.K. Proton transfers in the photochemical reaction cycle of proteorhodopsin. Biochemistry. 2002;41:5348–5358. doi: 10.1021/bi025563x. [DOI] [PubMed] [Google Scholar]
- 28.Wang W.-W., Sineshchekov O.A., Spudich J.L. Spectroscopic and photochemical characterization of a deep ocean proteorhodopsin. J. Biol. Chem. 2003;278:33985–33991. doi: 10.1074/jbc.M305716200. [DOI] [PubMed] [Google Scholar]
- 29.Gunaydin L.A., Yizhar O., Hegemann P. Ultrafast optogenetic control. Nat. Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- 30.Lórenz-Fonfría V.A., Resler T., Heberle J. Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc. Natl. Acad. Sci. USA. 2013;110:E1273–E1281. doi: 10.1073/pnas.1219502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sineshchekov O.A., Govorunova E.G., Spudich J.L. Intramolecular proton transfer in channelrhodopsins. Biophys. J. 2013;104:807–817. doi: 10.1016/j.bpj.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou S.Y., Govorunova E.G., Spudich J.L. Diversity of Chlamydomonas channelrhodopsins. Photochem. Photobiol. 2012;88:119–128. doi: 10.1111/j.1751-1097.2011.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsson M.H.M., Sondergaard C.R., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pK(a) predictions. J. Chem. Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 34.Govorunova E.G., Spudich E.N., Spudich J.L. New channelrhodopsin with a red-shifted spectrum and rapid kinetics from Mesostigma viride. MBio. 2011;2:e00115-11. doi: 10.1128/mBio.00115-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang F., Vierock J., Deisseroth K. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamiya M., Kato H.E., Hayashi S. Structural and spectral characterizations of C1C2 channelrhodopsin and its mutants by molecular simulations. Chem. Phys. Lett. 2013;556:266–271. [Google Scholar]
- 37.Sneskov K., Olsen J.M., Kongsted J. Computational screening of one- and two-photon spectrally tuned channelrhodopsin mutants. Phys. Chem. Chem. Phys. 2013;15:7567–7576. doi: 10.1039/c3cp44350g. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe H.C., Welke K., Elstner M. Structural model of channelrhodopsin. J. Biol. Chem. 2012;287:7456–7466. doi: 10.1074/jbc.M111.320309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettei M.J., Yudd A.P., Stoeckenius W. Identification of retinal isomers isolated from bacteriorhodopsin. Biochemistry. 1977;16:1955–1959. doi: 10.1021/bi00628a031. [DOI] [PubMed] [Google Scholar]
- 40.Sineshchekov O.A., Trivedi V.D., Spudich J.L. Photochromicity of Anabaena sensory rhodopsin, an atypical microbial receptor with a cis-retinal light-adapted form. J. Biol. Chem. 2005;280:14663–14668. doi: 10.1074/jbc.M501416200. [DOI] [PubMed] [Google Scholar]
- 41.Marti T., Rösselet S.J., Khorana H.G. The retinylidene Schiff base counterion in bacteriorhodopsin. J. Biol. Chem. 1991;266:18674–18683. [PubMed] [Google Scholar]
- 42.Sondergaard C.R., Olsson M.H.M., Jensen J.H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 43.Pace C.N., Grimsley G.R., Scholtz J.M. Protein ionizable groups: pK values and their contribution to protein stability and solubility. J. Biol. Chem. 2009;284:13285–13289. doi: 10.1074/jbc.R800080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plazzo A.P., De Franceschi N., Mongillo M. Bioinformatic and mutational analysis of channelrhodopsin-2 protein cation-conducting pathway. J. Biol. Chem. 2012;287:4818–4825. doi: 10.1074/jbc.M111.326207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Proctor V.W. Studies of algal antibiosis using Hematococcus and Chlamydomonas. Limnol. Oceanogr. 1957;2:125–139. [Google Scholar]
- 46.Otto H., Marti T., Heyn M.P. Substitution of amino acids Asp-85, Asp-212, and Arg-82 in bacteriorhodopsin affects the proton release phase of the pump and the pK of the Schiff base. Proc. Natl. Acad. Sci. USA. 1990;87:1018–1022. doi: 10.1073/pnas.87.3.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stern L.J., Khorana H.G. Structure-function studies on bacteriorhodopsin. X. Individual substitutions of arginine residues by glutamine affect chromophore formation, photocycle, and proton translocation. J. Biol. Chem. 1989;264:14202–14208. [PubMed] [Google Scholar]
- 48.Balashov S.P., Govindjee R., Feng Y. Effect of the arginine-82 to alanine mutation in bacteriorhodopsin on dark adaptation, proton release, and the photochemical cycle. Biochemistry. 1993;32:10331–10343. doi: 10.1021/bi00090a008. [DOI] [PubMed] [Google Scholar]
- 49.Brown L.S., Bonet L., Lanyi J.K. Estimated acid dissociation constants of the Schiff base, Asp-85, and Arg-82 during the bacteriorhodopsin photocycle. Biophys. J. 1993;65:124–130. doi: 10.1016/S0006-3495(93)81064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ikeura Y., Shimono K., Kamo N. Arg-72 of pharaonis phoborhodopsin (sensory rhodopsin II) is important for the maintenance of the protein structure in the solubilized states. Photochem. Photobiol. 2003;77:96–100. doi: 10.1562/0031-8655(2003)077<0096:aoppsr>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 51.Ikeura Y., Shimono K., Kamo N. Role of Arg-72 of pharaonis Phoborhodopsin (sensory rhodopsin II) on its photochemistry. Biophys. J. 2004;86:3112–3120. doi: 10.1016/S0006-3495(04)74359-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsunoda S.P., Hegemann P. Glu 87 of channelrhodopsin-1 causes pH-dependent color tuning and fast photocurrent inactivation. Photochem. Photobiol. 2009;85:564–569. doi: 10.1111/j.1751-1097.2008.00519.x. [DOI] [PubMed] [Google Scholar]
- 53.Berthold P., Tsunoda S.P., Hegemann P. Channelrhodopsin-1 initiates phototaxis and photophobic responses in chlamydomonas by immediate light-induced depolarization. Plant Cell. 2008;20:1665–1677. doi: 10.1105/tpc.108.057919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


