Summary
Regulatory T cells (Tregs) prevail as a specialized cell lineage that has a central role in the dominant control of immunological tolerance and maintenance of immune homeostasis. Thymus-derived Tregs (tTregs) and their peripherally-induced counterparts (pTregs) are imprinted with a unique Foxp3-dependent and independent transcriptional and epigenetic characteristics that bestows on them the ability to suppress disparate immunological and non-immunological challenges. Thus, unidirectional commitment and the predominant stability of this regulatory lineage is essential for their unwavering and robust suppressor function and has clinical implications for the use of Tregs as cellular therapy for various immune pathologies. However, recent studies have revealed considerable heterogeneity or plasticity in the Treg lineage, acquisition of alternative effector or hybrid fates, and promotion rather than suppression of inflammation in extreme contexts. In addition, the absolute stability of Tregs under all circumstances has been questioned. Since these observations challenge the safety and efficacy of human Treg therapy, the issue of Treg stability versus plasticity continues to be enthusiastically debated. In this review, we assess our current understanding of the defining features of Foxp3+ Tregs, the intrinsic and extrinsic cues that guide development and commitment to the Treg lineage, and the phenotypic and functional heterogeneity that shapes the plasticity and stability of this critical regulatory population in inflammatory contexts.
Keywords: regulatory T Cells, Foxp3, plasticity, stability, heterogeneity, epigenetics
Foxp3+ Tregs in immune tolerance
The immune system has developed elaborate mechanisms to mount effective responses against a broad array of pathogens. Immune responses are protective due to the diversity of lymphocyte antigen receptors generated by somatic gene rearrangements. While this process allows the host to effectively respond to rapidly evolving pathogens and numerous challenges, it also creates the danger of mounting harmful immune responses to innocuous self (e.g. tissue antigens) and non-self components (e.g. non-pathogenic gut-resident bacteria and food antigens). Cell-intrinsic mechanisms of tolerance in the thymus and periphery ensure the deletion or functional inactivation of self-reactive T cells (1). However, these mechanisms are often insufficient and are reinforced by complementary peripheral, cell-extrinsic tolerance mechanisms mediated by regulatory subsets acting in trans (2). Suppressive potential has been ascribed to a number of lymphoid and non-lymphoid subsets (e.g. CD4+ and CD8+ T-cell subsets, regulatory B cells, myeloid-derived suppressor cells and tolerogenic dendritic cells) (3–6). This review focuses on the role of thymus-derived CD4+Foxp3+ regulatory T cells (tTregs) and their peripherally induced counterparts (pTregs) in the control of immune tolerance (7–10). Although initially identified as key players in dominant immune tolerance (7), Tregs have now been convincingly shown to suppress inflammatory responses in diverse anatomical locations, such as mucosal interfaces that are constantly exposed to air and food-borne allergens (11), commensal gut microbiota (12, 13), transplanted organs (14), pathogenic infections (12) and tumors (15). Recent studies have also suggested a role for Tregs in alternate contexts, such as adipose tissue-resident Tregs controlling metabolic disorders (16, 17), skeletal muscle Tregs promoting muscle repair (18), and Tregs limiting organ rejection and atherosclerosis (19, 20). In certain cases, however, the suppressive function of Tregs limits beneficial host effector responses against tumors and chronic infections (21–24). Thus, the activities of this critical suppressive population needs to be finely tuned to strike the right balance between restraining deleterious inflammatory and autoimmune insults, while facilitating protective responses against infections and tumors.
Early observations of fatal autoimmune symptoms resembling the human disease IPEX (immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome) in Foxp3 (forkhead box P3)-deficient Scurfy mice and the demonstration that ectopically expressed Foxp3 was able to induce suppressor potential in conventional T cells, led to the identification of Foxp3 as the ‘lineage-specifying’ transcription factor for Tregs (25, 26). Treg-restricted, high level expression of Foxp3 confers a major component of the Treg transcriptome, including constitutive expression of CD25 (IL-2Rα), which is essential for their survival and proliferation, high expression of suppressor genes and repression of effector cytokines of Th1, Th2, and Th17 lineages (8, 10, 27, 28). Foxp3 stabilizes Treg lineage features, directly and indirectly, by regulating distinct cell surface and signaling molecules, interacting with a myriad of transcription factors, inducing miRNAs and modulating epigenetic machinery to maintain Treg identity, function and stability in response to diverse environmental cues (28–32). However, the notion that Foxp3 is the ‘sole requisite’ transcription factor required to define the Treg lineage has been challenged by numerous studies. While Foxp3 is indispensable for the majority of the Treg transcriptional and functional landscape, Treg fate specification is also influenced by contributions from TCR, IL-2, and TGFβ signaling pathways (33, 34). Foxp3 transduction by itself does not completely recapitulate the Treg transcriptional profile (34). These observations are in line with studies employing Tregs with non-functional Foxp3, which demonstrated that not all Foxp3+ T cells are functional Tregs and that part of the Treg signature can be induced in the absence of Foxp3 (28, 31, 35–38). This issue is particularly relevant in the case of human Tregs, as activated human T cells transiently express Foxp3 without the acquisition of suppressor potential (39–41). Stable Foxp3 expression in Tregs is subject to higher order regulation by epigenetic modifications of the conserved non-coding sequences (CNS) in the Foxp3 locus (35, 42, 43), adding to the complexity in this canonical Foxp3-centric scheme of Treg differentiation.
Recent reports of heterogeneity within this regulatory lineage have added an additional layer of complexity. Different Treg sub-populations have been identified in diverse anatomical and pathological conditions (44–46). Heterogeneity within the Foxp3+ Treg lineage is characterized by (i) differential expression of lineage-defining transcription factors and miRNAs that mediate functional specialization to control different types of immune responses (47–53), (ii) differential expression of chemokine receptors that enable Treg trafficking into diverse lymphoid and non-lymphoid compartments (44, 45, 53), and (iii) differential expression of suppressor modules to control diverse target cell types in distinct environmental and disease settings (54–59). We define this form of heterogeneity as ‘Treg plasticity’, since the core Treg identity (Foxp3 expression and suppressive capacity) is maintained, but their malleable nature allows phenotypic/functional adaptation to suppress a plethora of immune responses (60).
In contrast, Tregs can also become unstable, lose Foxp3 expression and their suppressive capacity, and acquire features reminiscent of effector T-cell in response to environmental cues (61–64). While there is compelling evidence for the stability of Tregs in healthy immune settings (28, 43), several studies have suggested that inflammatory scenarios are associated with downregulation/loss of Foxp3, secretion of effector cytokines and the generation of so-called ‘ex-Tregs’ (65–67). Fate mapping of Foxp3+ Tregs using lineage-tracing mouse models has revealed an uncommitted Treg population that loses Foxp3 in lymphopenic/inflamed settings, with the potential to differentiate into effector T cells (61, 64, 65). Heterogeneity linked to Foxp3 expression is particularly evident with human T cells, where FOXP3hi T cells display potent suppressive activity while FOXP3lo T cells secrete pro-inflammatory cytokines (41). This type of Treg heterogeneity raises the issue of ‘Treg stability’. Loss of the key lineage-specifying transcription factor Foxp3 would imply that Treg identity is not permanently imprinted and that Tregs can be reprogrammed into inflammatory cells in response to microenvironment cues. On one hand, this may represent the extreme adaptability of Tregs to provide an immediate effector-type response to keep the enemy at bay. However, considering that potentially self-reactive TCRs could be expressed by Tregs (68), such events could also have harmful consequences to the host. Thus, while Treg plasticity and stability could be categorized as ‘two sides of the same coin’, these two fates markedly impact the suitability of Tregs for immunotherapy. Here we review the literature documenting this remarkable heterogeneity of the Treg lineage, discuss evolving views on Treg plasticity versus stability, take a deeper look into the regulatory modules and pathways that guide commitment to a stable suppressor lineage and assimilate the existing literature in a unifying model that will highlight the dynamic adaptability of this regulatory subset and facilitate its therapeutic utility.
Treg plasticity
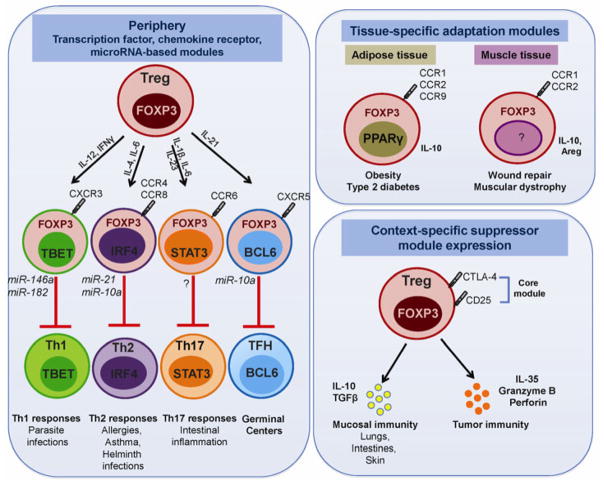
Considerable progress has been made over the past several years in delineating the molecular basis of the dynamic immune-regulation afforded by Tregs. Treg-mediated suppression can been attributed to the four broad categories of contact-dependent and contact-independent mechanisms: (i) suppression by inhibitory cytokines (IL-10, IL-35, TGFβ), (ii) suppression by cytolysis, (iii) suppression by metabolic disruption, and (iv) suppression via modulation of dendritic cell maturation or function (56, 69, 70). In most scenarios, the primary mechanism employed by Tregs likely depends on the disease setting, the target cell type, the local inflammatory milieu and the anatomical location. However, considering the growing list of distinct immunological and non-immunological responses that are influenced by this dominant regulatory subset, it is not surprising that distinct Treg sub-populations have been identified (45, 46, 71). These observations suggest that Tregs are not terminally differentiated, but retain the developmental plasticity to differentiate into specialized hybrid fates in sync with their local milieu for effective control of immune responses (72). This notion is synonymous with the proposed two-module model of Treg function wherein Treg-specific expression of Foxp3 encodes the expression of the core Treg suppressor module (increased CD25, CTLA-4), while their adaptability to the changing environment leads to induction of additional suppressive modules (transcription factors, miRNAs, chemokine receptors, suppressive pathways) for optimal immune regulation (73).
The emerging paradigm of paired differentiation between Tregs and effector T cells indicates that Tregs undergo functional specialization in the periphery by co-opting the transcriptional program of specific effector T cells they suppress (72). Thus, Tregs expressing the Th1 transcription factor Tbet and Th1-associated chemokine receptor CXCR3 are poised to adopt the Th1 program and accumulate at sites of Th1 inflammatory responses (e.g. mycobacterium tuberculosis infection) (47). Tbet+ Tregs can also be induced by IL-27 signaling during T. gondii infection and notable differences were observed in the transcriptional profiles of IFNγ-versus IL-27-induced Tbet+ Tregs (74).
Similar to Tbet, expression of the Th2 differentiation factor, IRF4, endows Tregs with the ability to control Th2 responses (48). Mice with Treg-specific deletion of IRF4 develop spontaneous Th2-mediated autoimmune lesions in the pancreas, stomach and kidneys, and exhibit increased Th2 and plasma cells, spontaneous germinal centers, and elevated serum IgG and IgE antibodies (Th2 isotypes). The master Th2 transcription factor, GATA3, is also highly expressed in Tregs (75, 76). However, unlike the selective defect in Th2 control exhibited by IRF4-deficient Tregs, GATA3-deficient Tregs have reduced expression of Foxp3 and other Treg suppressive genes and exhibit a broader defect in control of Th1, Th2, and Th17 responses.
Similar to control of Th1 and Th2 responses by functionally specialized Treg populations, expression of the Th17 transcription factor STAT3 is essential for control of Th17 responses (49). Deletion of STAT3 in Tregs provokes spontaneous intestinal inflammation with increased numbers of infiltrating Th17 cells, highlighting their selective failure in control of Th17 responses. In addition to STAT3, co-expression of Foxp3 and the other Th17 transcription factor, RORγt, in Tregs has been extensively reported, even in humans (77–79).
Expression of the Tfh transcription factor Bcl6 in Tregs has been demonstrated to be essential for Treg control of aberrant germinal center responses and autoantibody formation (80, 81). These T-follicular regulatory (Tfr) cells share phenotypic characteristics with conventional Foxp3+ Tregs and Tfh cells, derive from Foxp3+ tTreg precursors and are dependent on Bcl6 to adopt the Tfh cell development program. In addition to its role in Tfr cells, Bcl6, also functions to limit Th2 conversion of Tregs, analogous to its role in repressing Th2 differentiation of T cells (82). Bcl6-deficient Tregs exhibit elevated Th2 genes and a selective failure in control of Th2 inflammatory responses in vivo.
Tregs deficient in the Ikaros family transcription factor Eos are reprogrammed into helper T cells that are capable of priming CD8+ T-cell responses (83). This reprogramming is mediated by IL-6 such that the Eos-labile Treg subset secretes the pro-inflammatory cytokines (IL-2 and IL-17), upregulates CD40L, and can license DCs for antigen-presentation and CD8+ T-cell priming. Lastly, BLIMP1 expression in Tregs marks a distinct effector Treg population with increased expression of IL-10, ICOS, CCR6 and Bcl2 (84). Thus, variegated expression of transcription factors (Tbet, IRF4, GATA3, STAT3, RORγt, Bcl6, Eos, BLIMP1) defines functionally specialized sub-phenotypes of Tregs that each control distinct immune responses, suggesting that Tregs differentiate in the periphery alongside effector T cells in response to local environmental and inflammatory cues.
In addition to expressing distinct transcription factor modules, Tregs in diverse settings express distinct microRNA signatures. miR-146a is critical for Treg control of Th1 inflammatory responses by targeting Stat1 (50). Tregs deficient in this miRNA exhibit increased Stat1 phosphorylation, acquire a Th1-bias and fail to control Th1 responses. miR-21 expression results in a Th2-bias in Tregs by targeting the negative regulator of MAPK pathway, Sprouty1, and thereby indirectly augments GATA3 expression in Tregs (51). MiR-10a blocks Bcl6 expression in Tregs and thereby prevents Treg conversion to T-follicular helper cells (85). miRs-182 and -10a function as ‘regulatory hubs’ and are responsible for inducing distinct Treg populations during Th1 (L. major) and Th2 (S. mansoni) pathogen challenges (52). IL-12/IFNγ signaling activates the miR-10a-driven regulatory hub that controls IFNγ production in Th1-associated Tregs, while IL-4 signaling induces the miR-182-driven regulation thereby restricting IL-2 production by Th2-associated Tregs. Thus, different miRs are preferentially induced in Tregs to restrict effector conversion of Tregs while enabling efficient control of the corresponding immune response.
Synonymous to other T-cell populations, Tregs also express a diverse array of chemokine receptors and adhesion molecules that enable their trafficking to lymphoid and non-lymphoid compartments, in the presence or absence of overt inflammation (44). Expression of CD62L and CCR7 is required for Treg migration and retention in lymphoid tissues, as demonstrated by failure of CCR7-deficient Tregs to control colitis (86). Expression of P- and E-selectin ligands directs Treg homing to the skin (87), CD103 to gut-associated lymphoid tissues (88), CCR9 to the small intestine (89), CXCR6 to the liver (90) and CXCR4 to bone marrow, Payer’s patches and tumors (90–92). Differential chemokine receptor expression also guides the migration of Tregs to effectively control diverse immune responses – CXCR3 for control of Th1 responses (47), CCR4 and CCR8 for Th2 responses (48), CCR6 for Th17 responses (49), and CXCR5 for control of germinal center responses (80, 81). Visceral adipose tissue (VAT) Tregs display a unique chemokine receptor expression pattern (elevated CCR1, CCR2, CCR9 and CXCL10 and low CXCR3) that is guided by PPARγ, the transcription factor that regulates accumulation, phenotype, and function of this subset (53). More recently, Tregs have been demonstrated to accumulate in skeletal tissues following acute injury and contribute to muscle repair. Muscle-resident Tregs exhibit a distinct transcriptional profile characterized by expression of the anti-inflammatory cytokine IL-10, chemokine receptor CCR1, and growth factors, namely, plate-derived growth factor (PDGF) and amphiregulin (Areg), that likely aid in their healing and regenerative capabilities (18). Thus, Treg priming in response to tissue-specific antigens can guide their differential expression of chemokine receptors and thus migration to restrain immune responses in distinct anatomical compartments.
Tregs also tend to exhibit remarkable plasticity in their usage of key suppressor modules and exhibit control mechanisms to compensate for the loss of these key modules (56, 69, 93). Tregs at mucosal interfaces like the lungs, gut and skin exhibit increased expression of IL-10 and TGFβ (57, 94, 95), while the tumor environment is enriched in IL-35, although its source is being actively investigated (96, 97). Perforin and granzyme-expressing Tregs are rare in the periphery, but are abundant in the tumor microenvironment (59). Tregs deficient in both IL-10 and Ebi3 (and thus IL-35) are fully functional due to a compensatory increase in Cathepsin E expression, which in turn enhances TRAIL (Tnfsf10)-mediated expression and suppression (93). The genetic background of the mice may also differentially regulate utilization of suppressor modules, with B6 and Balb/c mice appearing to be more dependent on IL-10/IL-35- and TRAIL-dependent mechanisms, respectively (93).
While more examples of variegated Treg function and fate are likely to accumulate; what has been shown thus far illustrates their considerable adaptability. However, it also raises the question of their stability in the face of changing microenvironments. Thus, understanding how Tregs are able to exhibit phenotypic and functional heterogeneity while retaining their intrinsic suppressor potential is essential for their therapeutic application.
Treg stability
In addition to the Treg sub-populations detailed above that illustrate the remarkable cellular plasticity amidst retention of the core Treg program (intact Foxp3 expression and suppressive capacity), there is also evidence for Treg instability, loss of Foxp3 expression, and acquisition of an effector phenotype (the resultant cells being referred to as ‘exTregs’). The earliest evidence for loss of Treg stability came from studies that showed that incubation of CD4+CD25+ Tregs with IL-6 and some other dendritic cell-derived factors led to loss of Treg suppressor function (98). These initial observations were bolstered by in vitro experiments demonstrating loss of Foxp3 expression following exposure of sorted GFP+ Tregs isolated from Foxp3 reporter mice to T-helper polarizing cytokines (e.g. IL-4 and IL-6) (65, 67, 99). Since then, extensive literature has accumulated documenting that pro-inflammatory cytokines of Th1, Th2, and Th17 lineages present in diverse inflammatory settings mediate antagonistic down-regulation of Foxp3 and in some cases, conversion of Tregs to effector T cells. The transcription factors Tbet, Gata3, and RORγt provide a negative cross-regulatory mechanism for Treg differentiation (67, 100–102). The inflammatory cytokine IL-6 in conjunction with IL-1 and IL-23 induces RORγt and downregulates Foxp3 leading to concomitant production of IL-17 by the Foxp3− T cells (67, 103). Similar in vitro observations of human FOXP3+ T cells acquiring a Th17 phenotype have also been reported (104). Even in the absence of inflammation, continued TCR stimulation and constitutive activation of the PI3K/mTOR pathway has been shown to downregulate Foxp3 expression (105–107).
Transfer of highly purified sorted Foxp3+ Tregs into lymphopenic hosts resulted in a dramatic loss of Foxp3 expression, and these Foxp3− T cells reactivated their potential to express effector cytokines thereby failing to mediate immune suppression (65, 108). In some scenarios, these cells regained Foxp3 expression following TCR activation in vitro or following secondary transfers to lymphopenic mice (65). Loss of Foxp3 also occurs, albeit to a lesser extent, when effector T cells are co-transferred with Tregs into lymphopenic recipients (65). The propensity for Foxp3 down-regulation seems to be particularly enriched within the CD25−Foxp3+ Treg population, such that these Tregs differentiate into follicular T-helper cells in the gut-associated lymphoid tissues of lymphopenic mice (65, 66).
Induction of autoimmunity following reduced Foxp3 expression has been reported in a number of autoimmune conditions (109, 110). IL-2 deficiency within the pancreatic islets is associated with dramatic loss of Foxp3 expression and reduced Treg numbers (109, 111). Treg numbers also decline following T. gondii-driven Th1-inflammation as a consequence of reduced IL-2 production (112).
Most of the studies noted above reported reduced/loss of Foxp3 expression using Tregs sorted from Foxp3-GFP reporter mouse strains. These studies were plagued by the concern that the GFP− T cells that emerged in lymphopenic or inflammatory settings might have derived from small numbers of contaminating non-Tregs in the sorted Treg preparations that underwent homeostatic proliferation in vivo (113). Further, it has always been difficult to differentiate between Tregs that lost Foxp3 over those that did not express Foxp3 in the first place. This led to the emergence of Foxp3 lineage tracing mouse models to definitively probe the issue of stability and the subsequent outcome for Tregs that downregulate Foxp3 in inflammatory contexts.
Three different types of Foxp3 lineage tracing mouse strains have been reported (61, 63, 64). In one study (61), NOD BAC transgenic mice expressing GFP-Cre under control of the Foxp3 promoter were crossed with Rosa-LSL-YFP reporter mouse. Thus, in Foxp3+ Tregs, the activated Cre will excise the stop cassette flanked by the LoxP sites, thereby permanently labeling these cells with YFP. Thus, even if Foxp3-GFP expression is lost, these ‘GFP−YFP+ exTregs’ will still retain YFP expression, allowing for an effective strategy to distinguish Tregs that have lost Foxp3 from those that never expressed it. These dual reporter mice helped to identify a sizeable fraction of exTregs (10–15%) present in peripheral lymphoid organs even under homeostatic conditions, and this population increased significantly (~30%) in the pancreatic lymph nodes and islets of autoimmune diabetes-prone NOD mice. This suggested that exTreg generation involved autoantigen-driven reprogramming rather than a stochastic event. These exTregs were reported to exhibit an activated-memory phenotype, with production of effector cytokines, IFNγ and IL-17A, based on the microenvironment. Moreover, BDC2.5 TCR Tg-specific exTregs were capable of inducing diabetogenic pancreatic inflammation upon transfer into lymphopenic mice, demonstrating their pathogenic potential. In a recent study from the same group, testing Treg stability in an experimental autoimmune encephalomyelitis (EAE) model (114), exTregs were noted to develop during the peak of the antigen-driven CNS inflammatory response from stable bona fide Tregs (CD25hiFoxp3hi with a completely demethylated TSDR/CNS2, discussed later). Almost a third of the antigen-specific Tregs were shown to have to lost Foxp3, relative to the polyclonal Treg pool, reinforcing the notion that loss of Foxp3 is antigen-driven. Interestingly, stable Foxp3 expression was regained during the resolution phase of the inflammation or if mice received IL-2/anti-IL-2 complexes during the antigen priming phase.
This notion of Treg instability was challenged in a subsequent study (63) that took advantage of an inducible Foxp3 lineage tracing mouse model, wherein a GFP-Cre fusion with an estrogen receptor mutant (GFP-CreERt2) was inserted into the Foxp3 locus and these mice were subsequently crossed with a Rosa-LSL-YFP reporter mouse. In this model, Cre is only active following tamoxifen treatment. Thus, this system allowed for the temporal, inducible tagging of Tregs (63). This study demonstrated that ~96% of Tregs remained stable when tracked over 5 months and there was minimal Foxp3 downregulation in immune-deficient settings. The frequency of GFP−YFP+ exTregs was not increased following exposure to sub-lethal dose of X-ray irradiation, infection with L. monocytogenes or Th1-driven inflammation mediated by CD40 ligation. In addition, they demonstrated that the double-purified diabetogenic BDC2.5 or arthritogenic K/BxN TCR-expressing Foxp3+ Tregs did not develop into exTregs following transfer into prediabetic or prearthritic recipients, respectively. While they did note a ~30% decrease in total Foxp3+ Treg numbers and a modest decrease in Foxp3 protein following anti-IL-2 treatment, this did not result in expression of pro-inflammatory cytokines (IFNγ, IL-17, IL-2, TNFα) in these GFP−YFP+ exTregs. Therefore this study concluded that Tregs predominantly constitute a stable suppressor population.
In an attempt to reconcile the discrepancies over Treg stability, a third group employed a similar Foxp3 knock-in mouse model wherein GFP-Cre fusion protein was inserted into the 3′-UTR of the endogenous Foxp3 locus and these mice were subsequently crossed with Rosa-LSL-RFP reporter mice (64). Interestingly, this study also reported the development of GFP−RFP+ exTregs under steady-state conditions that showed low or heterogeneous expression of classical Treg markers (CD25, CD103, OX40, GITR, Helios) and acquired an effector-memory phenotype associated with inflammatory cytokine production (IL-2, IFNγ, IL-4, IL-17 and IL-21). This unstable Treg population developed during ontogeny and accumulated through adult life. However, this study noted that this unstable exTreg population was actually derived from non-regulatory Foxp3− T cells that transiently acquire Foxp3 expression under lymphopenic and inflammatory conditions. Thus, while this study supported the presence of a small population (less than 5%) of unstable/exTregs in vivo similar to the BAC transgene study (61), their observations also demonstrated that majority of the Tregs (~95%) exhibit stable Foxp3 expression consistent with the Foxp3-CreERT2 study (63). The tendency to lose Foxp3 was confined to a small population of RFPneg/lowCD25neg/low GFP+ T cells, constituting less than 5% of the total Foxp3+ Treg pool, with these cells retaining the capacity to regain Foxp3 expression and function upon optimal stimulation (64).
Lineage-tracing mice with Treg-specific deletion of the mTOR-signaling regulator, TSC1 (Foxp3YFPCreTSC1fl/fl x Rosa-LSL-RFP) identified the generation of exTregs with reduced Foxp3 expression and acquisition of Th17 effector features in inflammatory settings (115). Thus, the advent of Foxp3 lineage tracing systems will likely prime future investigations into the role of distinct transcription factors/signaling modules in the intrinsic stability of Foxp3 expression in Tregs under homeostatic and distinct inflammatory settings.
Reconciling emerging viewpoints on the issue of Treg stability These dual Foxp3 reporter lineage-tracing studies have initiated a contentious debate regarding Treg stability (60, 62, 116–119). A number of explanations have been proposed to reconcile these disparate observations. The initial notion that the apparent Foxp3 instability was due to the outgrowth of contaminating Foxp3− T cells in the adoptively transferred Foxp3+ Tregs has been ruled out by studies evaluating Treg stability using Foxp3 lineage tracing reporters (113). Furthermore, it has been shown that a small number of congenically-marked Foxp3− T cells spiked into adoptively transferred Tregs failed to proliferate in lymphopenic hosts, suggesting that the observed exTregs were likely derived from the transferred Tregs (108).
A second possibility was that the BAC Foxp3 transgene used in lineage-tracing studies might have been inherently less stable due to its chromosomal integration site resulting in Foxp3 instability due to aberrant Cre expression (61). However, exTregs were also reported in another study (64) that used a direct Foxp3 gene targeting approach (insertion into the 3′-UTR of the endogenous Foxp3 locus). The presence of normal Treg numbers and lack of any aberrant immune regulation in the Foxp3 lineage tracing mouse strains also limited the possibility of toxic effects caused by the expression of non-self-proteins (GFP, RFP, Cre recombinase) linked to exTreg generation.
One of the more likely and intriguing explanations for the differences observed stems from the different time windows during which Foxp3+ Tregs were labeled in the fate mapping studies. In Foxp3-Cre BAC transgenic mice, Tregs are labeled from birth, while in Foxp3-CreERT2 mice, Tregs are only labeled during tamoxifen treatment (61, 63, 64). Thus, the continuous labeling of Tregs may include those cells that transiently turned on Foxp3, but didn’t develop into Tregs, while the limited expression of Cre in Foxp3-CreERT2 mice is likely to only label bona fide Tregs (63).
Another possible explanation for the discrepancies noted may be due to differences in the severity and/or type of inflammatory insults used to evaluate Treg stability. The Foxp3-CreERT2 study used sub-lethal irradiation as one measure to assess Treg stability in lymphopenic conditions (63); unlike TCR-deficient hosts used in the Foxp3-BAC-transgene study (61). Since Foxp3− T cells limit the loss of Foxp3 expression in Tregs (108) and sub-lethal irradiation does not completely wipe out all the T cells, this difference can also contribute to the maintenance of Foxp3 expression in the Foxp3-CreERT2 study (63).
It is important to note that despite disparate interpretations, all the Foxp3 lineage-mapping studies reported a comparable, small population (1–5%) of Tregs that downregulated Foxp3 (61, 63, 64). Thus, the reported controversies may stem from different interpretations of the same observation by the different groups. While the focus of the Foxp3-CreERT2 study was that the majority of Tregs (~95%) are stable (63), results in full agreement with the data from the other two lineage tracing studies (61, 64); the Foxp3-BAC-transgene study was specifically highlighting the small population (~5%) of exTregs that were observed even under homeostatic conditions (61). This suggests that exTregs are indeed a true Treg population that downregulate Foxp3 in response to lymphopenic and inflammatory cues and can potentially be pathogenic, considering the self-reactive TCRs on Tregs (114).
Developmental commitment to a stable Treg lineage
Prior to determining the cellular precursors for these unstable exTregs, the intrinsic and extrinsic cues that actively maintain Treg stability or the environmental mediators that drive Treg instability, it is essential to understand the developmental events that guide commitment to a stable suppressor lineage and whether these early events prime the generation of stable versus transient, uncommitted Tregs.
Developmental commitment to a stable tTreg lineage begins during thymopoiesis, primarily in the SP CD4+ T cells (27, 120). Thymic Treg development relies on recognition of self-antigen presented by thymic medullary APCs in the context of MHC class II molecules (121–123). Treg fate specification involves a multistep orderly interplay of key signals (TCR, γ chain cytokines, Foxp3, and epigenetic factors) sculpting the Treg-destined thymocytes into functional and stably committed Tregs. Here we primarily focus on the contributions of Foxp3 and epigenetic regulation, and discuss how these two events individually and complementarily prime intrinsic commitment to a stable Treg fate.
Foxp3 in Treg fate specification
Foxp3 plays a pivotal role in the development and function of Tregs and confers the canonical features of Tregs (increased CD25 and Treg suppressor genes, and repression of effector genes) (10, 27, 28). The Treg transcriptional and functional landscape relies on high and sustained expression of Foxp3 in Tregs, as mice with attenuated Foxp3 protein expression (referred as ‘FILIG’ mice) develop fatal autoimmune disease, resembling Foxp3-deficient Scurfy mice (29). FILIG Tregs exhibit impaired suppressor potential, due to decreased Treg signature genes, and secrete Th2 cytokines. Also Cre-mediated Foxp3 deletion in mature Tregs results in loss of function and conversion to pro-inflammatory cytokine-producing effector T cells, suggesting that stable Treg commitment requires sustained Foxp3 expression through life (30). Genome wide analysis combining ChIP and tiling arrays have shown that Foxp3 directly binds to 20–30% of the Foxp3-dependent genes, and functions both as a transcriptional activator and repressor, mediating distinct regulation in the thymus and periphery (32, 124, 125). This dual regulation is possible due to Foxp3-dependent specific histone modifications at binding sites in its target genes. Thus, Foxp3-bound sites are enriched for permissive (H3K4me3) and inhibitory (H3K27me3) histone marks in activated and repressed genes, respectively. Foxp3 target genes in the thymus primarily encode factors implicated in gene regulation and chromatin remodeling, while those shared in the periphery include Treg surface markers and intracellular signaling regulators. In addition, Foxp3 also establishes its developmental and functional program indirectly in conjunction with other transcription factors (PRDM1, CREM, IRF6, ZFPN1A2, STAT5, STAT4, STAT3, TBET, IRF4, HIF1α). Foxp3 has been demonstrated to mediate its functions by interacting with a number of transcription factors, like NFAT, AML1/Runx1, HAT/HDAC complex, and NFκB (126, 127). The Foxp3-NFAT interaction occurs via the forkhead (FKH) domain of Foxp3 and blocks NFAT-mediated transcription of IL-2, IL-4 and induces CTLA-4 and CD25 expression, thus conferring suppressor potential in activated T cells (128). Interaction of AML1/Runx1 with the N-terminal region of Foxp3 between the FKH and leucine zipper is important for repression of IL-2 and suppressive activity of Tregs (129). Acetylation of Foxp3 by HATs such as TIP60 increases Foxp3 binding to the IL-2 promoter, thereby facilitating optimal repression of IL-2 (130). Mice selectively lacking optimal Foxp3-HAT/HDAC interactions exhibit an altered transcriptional landscape and reduced Foxp3-driven gene repression, particularly at the IL-2 promoter, leading to reduced Treg numbers and increased susceptibility to autoimmunity in disease-prone environments (131). Foxp3 can also regulate the Treg transcriptional and functional program indirectly by targeting microRNAs. Indeed, Treg-specific deletion of miRNA-processing enzymes results in fatal autoimmunity resembling Foxp3-deficient Scurfy mice and the resultant Tregs lose suppressor activity and gain effector characteristics (132, 133). Foxp3 was shown to directly bind miR-155, which is critical for Treg homeostasis (134). Thus, Foxp3 expression sculpts the transcriptional and epigenetic landscape required to establish a heritable Treg differentiation program.
There have been several studies that challenged the Foxp3-centric scheme of Treg differentiation and suggested that Foxp3 may not be sufficient to stably maintain Treg identity and function. A cross-sectional analysis attempting to tease out the contributions of Foxp3 to the Treg gene signature by comparing transcriptional profiles of Foxp3+ tTregs versus iTregs induced by Foxp3 transduction or cytokine-treatment revealed that the Treg signature is derived from influences of TCR, IL-2 and TGFβ signaling that are distinct from Foxp3-mediated effects (33, 34). Foxp3 transduction alone could not completely recapitulate the transcriptional profile of tTregs. These observations supported earlier studies, which demonstrated that part of the Treg signature could be induced even in the absence of functional Foxp3 (28, 31). Foxp3 has been demonstrated to actively engage linage-specific transcription factors for effective suppression of the corresponding effector T-cell responses [Tbet for suppressing Th1 effectors (47), IRF4 for Th2 effectors (48), STAT3 for Th17 effectors (49) and Bcl6 for Tfh effectors (80, 81)]. Also Foxp3 functions as a late-acting differentiation factor, while the early Treg fate commitment is regulated by the Forkhead box O (Foxo) family of transcription factors (135–137). Proteome profiling in Tregs demonstrated that Foxp3 is part of a large transcriptional complexes, comprising several hundred interacting partners (36). The majority of these interacting partners are proteins implicated in transcriptional regulation (NFATc2, Runx1, Foxp1, GATA3, Stat3, Ikaros, Aiolos, Ets, Cnot3), are direct Foxp3 targets (Gata3, Stat3, Runx1), and also play a role in regulating Foxp3 expression by directly binding to its promoter and intronic enhancers in tTregs and pTregs (Runx1, NFATc2, Bcl11b, Gata3). More recently, computational simulation of the complex regulatory network in Tregs has led to the identification of a ‘quintet’ of transcription factors (IRF4, Eos, Lef1, Gata1 and Satb1), that in conjunction with Foxp3, can reproduce the Treg gene signature in conventional T cells (38). In addition, high throughput DNAseI hypersensitivity analysis revealed that chromatin accessibility of Foxp3-bound enhancers is similar in Tregs and conventional T cells (37). The other structurally related forkhead family transcription factor, Foxo1, actually functions as a place-holder for Foxp3 in precursor cells. Thus, the fact that only a small proportion of Foxp3-bound enhancers (2%) are Treg-specific suggests that Foxp3 does not shape the chromatin landscape, but rather acts via pre-exisiting enhancers already bound by cofactors (37). Taken together, these studies have strengthened the notion that Foxp3 acts in an opportunistic fashion, exploiting the pre-existing transcriptional and chromatin landscape shaped by other factors, acting either upstream or in parallel to Foxp3- mediated effects.
Epigenetic programming in Treg fate specification
In addition to high Foxp3 expression, Treg fate specification involves an orderly epigenetic process of chromatin remodeling, nucleosome positioning, and DNA hypomethylation, for initiating progressive acquisition of a stable suppressor profile (42, 43, 138, 139). Most of the thymic Treg emigrants possess a completely demethylated TSDR (Treg-associated demethylated region), suggesting that the thymic microenvironment is sufficient to mediate demethylation and generate stable Tregs that subsequently migrate to the periphery (35, 140). Demethylation of the Foxp3 locus is initiated at the CD4+ SP stage with the immature CD24hi thymocytes displaying a completely methylated TSDR and gets progressively established as the cells migrate to the periphery. Signaling via IL-2 and other gamma-chain cytokines is required during the early stages to initiate TSDR demethylation prior to Foxp3 induction, as CD25+Foxp3− thymocytes exhibit Treg-specific epigenetic marks (140–142). Thus, thymic Treg precursors that have upregulated CD25 and have acquired the epigenetic marks become poised for subsequent Foxp3 induction.
Epigenetic profiling of Tregs versus Tconv cells has revealed differential patterns of DNA methylation and histone modifications at several loci in both humans and mice (35, 42, 43, 139). In particular, tTregs display stable Foxp3 expression that is associated with specific demethylation of an evolutionarily conserved element in the CNS2 (conserved non-coding region 2) of the Foxp3 locus (also referred to as the TSDR) (43, 143–145). Complete demethylation of the TSDR in tTregs is a prerequisite for permanent commitment to a stable suppressor lineage, as TGFβ-induced iTregs and Tconv cells display a partial and completely methylated TSDR, respectively (43). Indeed, human CD4+CD25lo T cells that transiently upregulate Foxp3 upon activation display only a partial methylation pattern (146). Genetic deletion in CNS2-deficient mice resulted in the gradual loss of Foxp3 expression in Tregs following cell division (145). Thus, demethylation within Foxp3 promoter marks a committed Treg population in both mice and humans. Importantly, the methylation status of the CpG motifs within the Foxp3 TSDR also controls its transcriptional activity. Key transcription factors involved in molecular regulation of Foxp3 expression (CREB/ATF, NFκ B, Ets-1) can only bind to the demethylated TSDR (144, 147, 148). Disruption of Ets-1 binding sites within the TSDR dramatically reduced its transcriptional enhancer activity (147). Furthermore, Foxp3-Runx complexes could only bind to the demethylated TSDR, which is key to stabilizing Foxp3 expression (145).
In addition to demethylation of the Foxp3 promoter, CpG hypomethylation of a limited number of Treg loci, referred as ‘Treg cell representative regions’ (Foxp3 intron 1 corresponding to CNS2, Ctla4 exon 2, Gitr exon 5 and Eos intron 1b), is exclusively imprinted in Tregs and remains stable under various activation conditions (35). These key Treg suppressor genes (Foxp3, Ctla4, Eos) are present in loci enriched with CpG hypomethylation, abundance of active histone H3K4me3 marks and chromatin accessibility. Interestingly, developing Tregs from Foxp3-null mice (that lack functional Foxp3) also exhibit a similar pattern and kinetics of acquisition of DNA hypomethylation and display the classical Treg gene signature similar to Foxp3-sufficient Tregs (35, 149). These findings revealed that Treg-specific hypomethylation is imprinted in Tregs in a Foxp3-independent manner and these epigenetic marks impart the genomewide gene expression profile of Tregs. In addition, these epigenetic marks are key to acquisition of suppressor potential and lineage stability of Tregs. Thus, Foxp3 expression and CpG hypomethylation complement each other for complete specification of the Treg lineage, but they also exhibit a distinct division of labor (e.g. repression of Il2, Ifng, Zap70 gene expression in Tregs is attributable to Foxp3 while expression of some Treg-specific genes such as Helios and Eos is CpG hypomethylation-dependent) (35).
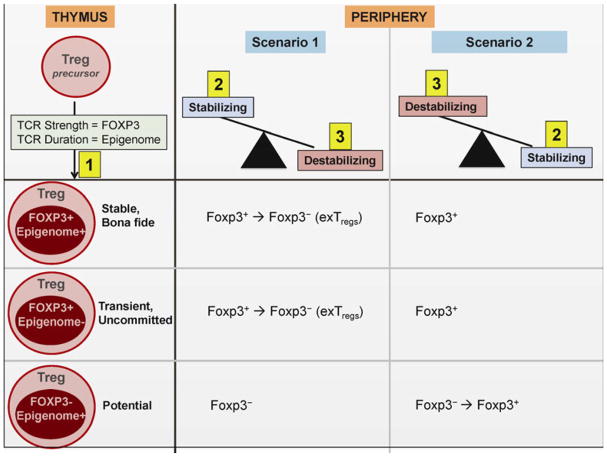
The Treg cell transcriptome and epigenome orchestrate a highly synchronized developmental act in specifying the Treg-cell gene expression portfolio, suppressor function, and stability. Commitment is initiated when developing CD4+ SP immature thymocytes receive TCR signals of the appropriate strength and duration. TCR signals that recognize self-ligands with relatively high intensity, but below the threshold for negative selection favor Foxp3 induction (122, 150, 151). Thymocytes that receive too strong signals are deleted during negative selection by apoptosis, while those that receive too weak signals and are unable to recognize self-ligands fail during positive selection. Thus, while Foxp3 expression is directly tied to the strength of TCR signaling (122), CpG hypomethylation relies on the duration of TCR signaling (35). Thus, the status of the transcriptome and epigenome encodes the generation of stable versus uncommitted Tregs (35, 152). Only those thymocytes that receive TCR signals of a certain strength (Foxp3+) and duration (Epigenome+) will eventually develop into stable committed Tregs. Thymocytes that receive the appropriate duration of TCR signaling (epigenome+), but fail on the TCR strength scale (Foxp3−) possess the epigenetic machinery to encode the Treg cell specific gene expression and develop into Tregs at later time points in the context of signals that induce Foxp3 (potential Tregs). Peripherally induced pTregs could belong to this category of developing thymocytes. In contrast, thymocytes that develop the opposite phenotype (Foxp3+epigenome−), transiently turn on Foxp3 but lack the epigenome that encodes long-term Treg stability (unstable Tregs). These thymocytes are predisposed to losing Foxp3 expression and could generate exTregs in conditions that antagonize Treg fate (inflammatory cytokines, absence of survival factors, etc.) in the periphery (64).
Cellular precursors to exTreg generation
This brings us to the critical question of what actually are the cellular precursors of these unstable exTregs – stable fully committed bona fide Foxp3+ Tregs, transient uncommitted Tregs, or the peripherally-induced pTregs? Literature supporting all three of these possibilities exists. While some studies suggest that all Foxp3+ Tregs can undergo lineage reprogramming (61, 108), there is also evidence that Tregs represent a heterogeneous population of stably committed and uncommitted cells (65). Analysis of the TCR sequences of exTregs generated in the BDC2.5 TCR transgenic mice indicated a significant overlap of these cells with both Foxp3+ Tregs and Foxp3− T-cell lineages during ontogeny (61); however, there is also evidence suggesting little to no inter-conversion between these populations for MOG-specific Foxp3+ and Foxp3− T cells infiltrating into the CNS in an experimental EAE model (153). This could mean that the pressures of environment-induced Treg cell reprogramming can also play into creating more diversity in the TCR repertoires of the resulting exTregs, making it difficult to trace their true precursors. In any situation, the generation of exTregs may depend upon both the cellular precursors of these exTregs and the type and severity of environmental pressures that counteract the inherent stability of Foxp3+ Tregs.
Bona fide stable Treg conversion to exTregs
The issue of bona fide Foxp3hi Tregs converting to exTregs in lymphopenic/inflammatory settings is very controversial, since it fails to explain the robustness of Treg function and their uncanny ability to maintain immune tolerance under such circumstances. A sizable conversion rate could have catastrophic consequences to the host considering the self-reactive specificity of Treg TCRs. These stably committed Tregs have received the right strength and duration of TCR signals (Foxp3+Epigenome+) and hence have the perfect transcriptomic and epigenomic signature endowing them with the ability to sustain their identity in demanding scenarios [the term ‘Epigenome’ was used here by the authors to indicate whether the Foxp3 CNS2 and other Treg related promoter CpG regions are hypomethylated; i.e. Epigenome+ = hypomethylated] (152). However, the conversion of stable Foxp3hi Tregs into exTregs was very elegantly demonstrated in a recent report in the experimental EAE disease model (114). During the inflammatory autoimmune response in the CNS, a significant percentage of antigen-specific Tregs exhibiting the canonical Treg signature (CD25hiFoxp3hi, demethylated TSDR) downregulated Foxp3 at both the transcript and protein level and acquired effector characteristics (production of IFNγ and pathogenic potential to induce EAE). Importantly, exTregs were only generated when bona fide and truly committed MOG-specific CD25hiFoxp3hi Tregs with completely demethylated TSDR were adoptively transferred into mice with ongoing EAE (~39% of transferred MOG-specific Tregs lost Foxp3 versus ~10% of polyclonal Tregs). Transfer of MOG-specific Foxp3−GFP− Tconv cells did not result in transient Foxp3 upregulation, thus demonstrating that exTregs derive from bona fide, antigen-primed Tregs and not from transient Foxp3-expressing uncommitted Tregs or Tconv cells. Interestingly, the percentage of MOG-specific exTregs was lower in the CNS during the resolution phase of EAE. Thus, Foxp3 expression returned in these exTregs when the local inflammatory milieu reduced. Similar reduction in the proportion and numbers of MOG-specific exTregs was noted with IL-2/anti-IL-2 complex treatment early during the disease onset. IL-2 deficit linked to Foxp3 downregulation and Treg instability has also been reported in the case of strong Th1 responses during parasite infections (112) and for the islet-infiltrating Tregs in the diabetic setting (109, 111). Thus, IL-2 deficiency may be a key-destabilizing signal for affecting the stability of bona fide stably committed antigen-specific Tregs and may have detrimental consequences for tissue-specific tolerance in autoimmune scenarios.
Transient or uncommitted Treg conversion to exTregs
Two different scenarios can be envisioned leading to the generation of transient uncommitted Tregs: (i) cells that were destined to a Foxp3+ Treg fate during ontogeny but did not develop into fully committed Tregs, and (ii) Foxp3− T cells that acquired transient Foxp3 expression in lymphopenic/inflammatory settings. In both cases, the transient Foxp3+ cells never gain the transcriptional and epigenetic signature characteristic of bona fide committed Foxp3+ Tregs and hence are highly prone to Foxp3 downregulation in demanding environments (reduced Foxp3 stabilizing cytokines and increased pro-inflammatory cytokines). Generation of exTregs from cells that transiently expressed Foxp3 but aborted commitment to Treg fate (scenario 1) has been noted as one of the major difference underlying the discrepancies in the reported Foxp3 lineage tracing studies (61, 63). These transient Foxp3-expressing cells were labeled in the Foxp3-BAC transgenic mice where Cre activity was present from birth (61) but failed to be labeled in the inducible Cre system used in the Foxp3-CreERT2 study (63). Further, developmental plasticity has been extensively reported between Tregs and Th17 cells in both the mouse and human settings often leading to a TGFβ-induced Foxp3+RORgt+ intermediary state (77–79). In Th17-priming conditions (e.g. inflammatory cytokines IL-1β, IL-6, IL-23), this transient Foxp3+RORγt+ population can abort the Treg fate and develop into Foxp3−RORγt+ Th17 cells.
Generation of exTregs from non-regulatory Foxp3− T cells (scenario 2) was demonstrated in one of the three Foxp3 lineage tracing studies (64, 65), showing that the CD25−/low T cells acquired Foxp3 expression in response to lymphopenia/inflammatory cytokines; however, these uncommitted Tregs readily lost Foxp3 expression, while the CD25hiFoxp3+ Tregs were stable and persisted. These CD25−Foxp3+ Tregs were also shown to lose Foxp3 and convert into follicular T-helper cells in the Payer’s patches of the intestine (66). While the above-mentioned two scenarios may not be mutually exclusive, they point to a heterogeneity model (65) wherein the Foxp3+ Tregs are heterogeneous and comprise primarily of stable committed Tregs with a minor population of transient uncommitted Tregs that bear the potential to be reprogrammed into exTregs. Such transient or uncommitted Tregs likely develop from Foxp3+Epigenome− thymocytes (152). Thus, despite turning on Foxp3, they lack the epigenetic signature that encodes long-term Treg stability. This fits with the notion that commitment to a stable Treg lineage involves multi-step, orderly events orchestrated by Foxp3 and higher-order regulation afforded by epigenetic stabilization of Foxp3 expression.
pTreg conversion to exTregs
Although the general consensus is that tTregs are critical for curtailing systemic autoimmunity and pTregs are induced to restrain more localized inflammatory responses, both complement each other in maintaining immune homeostasis (154). In a recent study, it was suggested that tTregs alone were not sufficient to limit autoimmune inflammation in Scurfy mice, in the absence of pTregs (155). Transfer of Foxp3− T cells with tTregs was required to induce the pool of pTregs to maintain tolerance. However, a number of studies directly comparing the suppressive abilities of tTregs and TGFβ-induced pTregs have reported reduced efficiency and poor stability of pTregs in vivo (34, 145, 155, 156). In lymphoreplete settings, pTregs effectively curtailed the islet-specific diabetogenic response in NOD.CD28-deficient mice that lack tTregs (157). However, when these pTregs were transferred along with naive T cells in lymphopenic RAG-deficient mice, they not only failed to control EAE (157) but also lost Foxp3 expression. Neuropilin-1 was used as the marker to distinguish the two Treg subsets in these studies, with tTregs being Nrp-1hi and pTregs being Nrp-1lo (157, 158). Nrp expression on Tregs has been shown to confer functional superiority by enhancing Treg-DC interactions and by converting latent TGFβ into its active form (159, 160). Our group has also demonstrated that Sema4a:Nrp-1 ligation through the PTEN-Akt-Foxo axis augments Treg suppressor function by increasing expression of suppressor modules and by providing a survival advantage, particularly at inflammatory sites such as tumors and colitic mucosa (161). Thus reduced Nrp-1 expression on pTregs relative to tTregs could contribute to their reduced functional capabilities and stability in homeostatic versus lymphopenic/inflammatory settings. Preferential loss of Foxp3 in the pTreg subset relative to the tTregs has also been reported in the MBP-TCR transgenic 1B3 mouse model that can only develop pTregs and lack tTregs when crossed onto the Rag-deficient background (162). When 1B3.Rag−/− mice were crossed with Foxp3GFP-Cre x Rosa26LSL-YFP mice, the generation of GFP−YFP+ exTregs was increased relative to WT or 1B3.Rag+/− mice. This increased tendency to lose Foxp3 and generate exTregs in the pTreg subset makes teleological sense since these Tregs are preferentially induced to restrict local inflammatory responses and these regulatory cells can wane and/or revert back to effector T cells following resolution (62, 64, 163).
The increased exTreg generation potential in the pTreg fraction may be inherent in their TCR activation and epigenetic status. Although not experimentally verified, we propose that pTregs could belong to the category of developing thymocytes that are Foxp3−epigenome+ (152). Thus, while these thymocytes failed to obtain a sufficiently strong TCR signal to induce Foxp3, they possess the epigenetic machinery to encode the Treg-specific gene expression pattern and develop into Tregs at later time points in the context of antigenic stimulation or cytokines (e.g. TGFβ) promoting Treg differentiation in the periphery. Despite bearing the epigenetic marks that confer stability, the pattern of epigenetic marks in pTregs is very different from that exhibited by stable tTregs, which may underlie the basis for their inherent bias towards exTreg generation (145, 164, 165). The CNS2/TSDR is completely demethylated in tTregs, but bears partial/incomplete methylation marks in pTregs (43, 145). This lack of complete demethylation in pTregs disrupts the positive autoregulatory loop to stabilize Foxp3 expression driven by Foxp3 itself. In addition, the pTreg-inducing cytokine, TGFβ, uses a distinct conserved region in the Foxp3 locus (CNS1) to induce Foxp3 (145). Thus differences in expression of stability modules (e.g. Nrp-1 pathway) and epigenetic signature between the tTregs and pTregs can contribute to their differential functionality and stability in diverse settings.
Intrinsic and extrinsic modulators of Treg stability
Recent studies have identified many transcription factors, signaling modules and regulatory elements that positively and negatively regulate Treg stability. The mediators on the two sides of the Treg stability equation can act by either modulating the expression of (i) Foxp3, (ii) the effector T-cell programs that normally need to be repressed in Tregs, and/or (iii) the survival/quiescence factors that need to be enhanced in Tregs (60). It is highly likely that many of these mediators modulate more than one aspect, thus making it difficult to segregate them into different categories. Understanding how these cooperative and counteractive factors impinge on Treg stability, both individually and complementarily, under homeostatic and inflammatory contexts can guide the development of methodologies to inhibit/boost Treg stability for therapeutic benefits.
Considering the pivotal role of Foxp3 in the Treg lineage, its expression needs to be under tight control to allow sustained Treg stability in the face of changing circumstances. Foxp3 expression in Tregs is subject to multi-tier regulation. Firstly, there are positive and negative extracellular stimuli, such as TCR signaling, CD28 costimulation, cytokines and downstream signaling pathways (e.g. NFAT, NFκ B, AP1, CREB, ATF) that impinge on Foxp3 expression (32, 166–168). Continued TCR stimulation and constitutive activation of PI3K/mTOR pathway downregulates Foxp3 expression (105, 169). The cytokines IL-2 and TGFβ, acting via STAT5 and SMAD phosphorylation respectively (166, 170), are important for stable Foxp3 expression in Tregs, while pro-inflammatory cytokines of the effector lineages (IFNγ, IL-4, IL-6) downregulate Foxp3 expression via STAT1, STAT4, and STAT6 signaling modules (100, 101, 171). Transcription factors T-bet, Gata3, and RORγt afford negative cross-regulation for Treg differentiation (102). Gata3 and STAT6 inhibit iTreg differentiation by direct binding to regulatory elements in the Foxp3 locus (101, 171). On the other hand, transcription factor Runx1 and its cofactor Cbfβ are indispensible for optimal Foxp3 expression in Tregs (172). Mice harboring Runx1 or Cbfβ-deficient Tregs exhibit attenuated Foxp3 expression, reduced Treg suppressor genes, secretion of the effector cytokine IL-4, and development of autoimmunity associated with splenomegaly and lymphadenopathy, serum autoantibodies and hyper-IgE production. The Th2 master regulator Gata3, expressed in Tregs in an IL-4/STAT6-independent manner, is also vital for optimal induction and maintenance of Foxp3 (75, 76). Gata3 directly binds to a regulatory region in the Foxp3 locus and promotes the activity of cis-acting elements in the Foxp3 gene, thereby exerting its positive influence on Foxp3 and Treg suppression capacity.
Higher order regulation of Foxp3 expression is mediated by epigenetic modifications of the Foxp3 locus. Besides the promoter, three evolutionarily conserved non-coding sequences (CNS) have been identified in the Foxp3 locus that are critical for optimal Foxp3 expression in tTregs and pTregs (145). CNS3 acts as a pioneering element and facilitates Foxp3 induction during thymic and peripheral Treg differentiation by recruiting c-Rel (145). On the other hand, CNS1 functions as a TGFβ sensor and is primarily essential for peripheral induction of Foxp3, acting via SMAD3 (145, 173). In complete contrast, CNS2 (also known as TSDR) is dispensable for Foxp3 induction, but functions as a cellular memory module and is responsible for heritable Foxp3 expression in dividing mature Tregs (143, 145). Importantly, Foxp3 drives an auto-regulatory feedback loop amplifying its own expression (28). In addition to Foxp3, demethylated CNS2 also contains CpG motifs that serve as binding sites for Runx and CREB/ATF and Ets-1 complexes (144, 145, 147, 148). Thus TCR-driven recruitment of these transcription factor complexes to the demethylated Foxp3 CNS2 guides maintenance of Foxp3 expression in Tregs.
A third regulatory mechanism is afforded by microRNA, highlighted by the fatal spontaneous multi-organ autoimmunity, which resembles the Foxp3-deficient Scurfy phenotype, in mice with Treg-specific loss of the key microRNA-processing enzymes, Dicer and Drosha (132, 133). Tregs that are unable to process microRNA exhibit reduced Foxp3 expression, reduced suppressor function and stability, and skew towards effector lineages in inflammatory contexts. Indeed, miRNA-10a was recently shown to positively regulate Foxp3 expression and mark stable Tregs, although genetic ablation of this miRNA did not affect Treg numbers or Foxp3 expression (174). Foxp3 was also shown to maintain Treg identity and functionality by mediating miRNA-driven repression of the genome organizer, SATB1 (175). A cluster of five miRNAs (miR-7, miR-18a, miR-21, miR-34a, and miR-155) was noted to target the 3′-UTR of SATB1, thus preventing acquisition of effector T-cell program in Tregs.
Another critical checkpoint on Foxp3 expression is mediated by survival or quiescence factors, whose levels are often imbalanced in inflammatory scenarios, thereby impinging on the Treg/Teff balance. Deficiency of IL-2 linked to defective Treg survival and function has been reported in a number of settings (inflamed islets in the diabetic settings, strongly Th1-polarizing environments during T. gondii infection) (109, 112). In fact, administration of low doses of IL-2 or IL-2/anti-IL-2 complexes has been demonstrated to increase Treg numbers and also restore the loss of Foxp3 expression in exTregs (114).
Our group has identified another novel regulatory pathway impinging on Treg survival and stability involving cognate interactions between the immune-cell expressed ligand, Semaphorin-4a (Sema4a) and the Treg-expressed receptor, Neuropilin-1 (Nrp-1) (161). Sema4a:Nrp-1 interaction restrained Akt-mTOR signaling via PTEN recruitment to the immunological synapse, allowing increased nuclear localization of the transcription factor Foxo3a, thus potentiating Treg function. Nrp-1 ligation induced a transcriptional profile characterized by increased expression of modules regulating Treg survival (increased IL-2 and IL-7-driven transcripts, increased anti-apoptotic protein Bcl2), stability (Helios and reduced effector transcription factors, RORγt, IRF4, Eomes and cytokines), function (increased suppressor genes–CD73), and quiescence (stabilization of KLF2 and its targets). A striking feature of the Nrp-1:Sema4a axis is that it is entirely dispensable for immune homeostatic control and prevention of autoimmunity but is essential for Treg-mediated prevention of antitumor immunity and control of chronic colitogenic inflammation. Thus, this pathway offers a rare and unique opportunity to selectively target Treg stability and function in the tumor microenvironment without inducing autoimmunity.
It is highly likely that these different cell-intrinsic and extrinsic regulators/signaling pathways are differentially expressed or utilized by Tregs in different contexts, disease states or anatomical locations (e.g. dicer-deficient Tregs exhibit normal suppressive activity during homeostasis, but lose their function in inflammatory contexts) (132). Similarly, the Sema:Nrp1 pathway may only be active in tumors, and not autoimmune inflammation, based on differential availability of the ligand or receptor expression (161). Thus, understanding the differential utilization of the Foxp3 stabilizing and destabilizing modules/pathways in different inflammatory diseases can offer rational approaches to maintain stable Treg identity for therapeutic benefits.
Proposed model for Treg plasticity/stability and future considerations
While there is ample evidence supporting functional and phenotypic heterogeneity/plasticity exhibited by Foxp3+ Tregs, there is also evidence for Treg reprogramming associated with loss of Foxp3. Some have suggested that potential reprogramming is a feature of all Foxp3+ Tregs, while others believe in the existence of distinct stable and unstable Treg sub-populations. There are also suggestions that Teff cells can transiently acquire Treg potential.
Accounting for all these reported discrepancies in Treg behavior, coupled with recent insights, we present a unified model that integrates these different facets of Treg cell fate: (i) Tregs have evolved as a suppressor lineage to maintain immune homeostasis and tolerance, (ii) activation of effector capacity in these suppressive cells bearing self-reactive TCRs can trigger autoimmune responses, and (iii) acquisition of suppressor potential in effector T cells can limit the beneficial effector responses required to limit infections and tumors.
At any point in time, Tregs have been shown to possess a distinct intrinsic signature, dependent on their Foxp3 expression and epigenomic status (demethylation extent of the CNS2/TSDR and Treg cell representative regions). This signature is reflective of their antigenic exposure; specifically, the strength and duration of TCR signaling and costimulation). Thus, the Treg pool is a composite of stably committed bona fide Tregs (Foxp3+Epigenome+), potential Tregs (Foxp3−Epigenome+), and uncommitted/transient Tregs (Foxp3+Epigenome−). The stability or plasticity of Tregs in any given environmental milieu is influenced by the balance between three components: (i) their intrinsic Foxp3 expression/epigenomic signature, (ii) a set of signals constantly striving to maintain Treg stability (Stabilizing signals), and (iii) a set of signals that by-pass the stability mechanisms to drive Treg instability (Destabilizing signals). The convergence of these three key parameters is likely to be distinct, based on (1) developmental, environmental, and homeostatic history, (ii) the type of inflammatory challenge (infection, autoimmunity, cancer), and (iii) temporal aspects of inflammation (initiation versus resolution), thereby creating different thresholds to drive Treg stability or instability.
Inflammatory scenarios associated with a higher concentration of destabilizing signals relative to stabilizing ones could drive Treg instability and exTreg generation owing to the local presence of pro-inflammatory cytokines and deficiency of survival and quiescence factors. The strong environmental pressures generated by these destabilizing signals can drive instability, even in the bona fide Treg population (Foxp3+Epigenome+). In such settings, the uncommitted Treg population (Foxp3+Epigenome−) is highly prone to losing Foxp3 expression, while the potential Treg population (Foxp3−Epigenome+) is less likely to acquire Foxp3 and Treg fate. Such environment-induced reprogramming of Tregs to Teff cells may be advantageous to the host in certain scenarios (e.g. peak of an infection by making increased effectors available to counteract the challenge). What remains unknown is the fate of these converted Tregs – whether they undergo apoptosis, continue as effector T cells, or retain their ‘Treg memory’ and revert back to stable Tregs. Indeed, reconverting back to Tregs may be beneficial to restore immune homeostasis once the impending challenge is curtailed.
Contrary to the above, environments associated with a higher concentration of stabilizing relative to destabilizing signals are favorable for inducing and maintaining Treg identity since the local milieu is enriched with anti-inflammatory cytokines (TGFβ, IL-10), survival, quiescence, and remodeling signals. Thus, the stably committed bona fide Tregs (Foxp3+Epigenome+) will maintain their Foxp3 expression while potential Tregs (Foxp3−Epigenome+) and transient Tregs (Foxp3+Epigenome−) are likely to acquire Foxp3 expression owing to presence of Foxp3 inducing signals (TGFβ). Some autoimmune diseases exhibit an oscillating nature (e.g. relapsing-remitting multiple sclerosis) and transient Foxp3 upregulation in these settings may be beneficial for the host during the remission phase, allowing these cells to revert back to effector T cells in case of a future relapse.
This model is consistent with the multifaceted behavior of Tregs as discussed above, but also highlights the importance of continued discussion concerning the safety of current and future Treg-based therapies. However, on a more positive note, the model also raises the possibility that one could influence the fate of Treg-targeted therapies in order to enhance stability or instability depending on the desired outcome.
While we have gained considerable insights into Treg heterogeneity and fate, it is clear that additional studies will be required. First, further fate-mapping studies to assess Treg lineage fates and their durability, in diverse inflammatory settings, and during different phases of immune response are warranted to gain a comprehensive understanding of Treg plasticity and stability. Second, we need to gain a greater understanding of all the factors and signaling pathways that maintain stability or drive instability in Tregs. Third, there is a clear need for the development of innovative approaches to assess the epigenetic status of Treg-relevant loci at the single cell level and also to develop better models to unequivocally distinguish between tTregs and pTregs. Fourth, we need to coalesce and utilize all this information to develop optimal therapies that either undermine Tregs in cancer or chronic viral infections, enhances Tregs in autoimmune or inflammatory diseases, or facilitates the development of robust, stable and effective Tregs for adoptive immunotherapy. Knowledge gained from these studies will help to optimally channel the dynamic adaptability of this regulatory cell lineage for successful therapeutic interventions.
Fig. 1. Plasticity of Tregs.
(Left) Tregs differentiate in parallel to effector T cells in response to micro-environmental cues in the periphery. Thus, Tregs adopt distinct transcription factor, chemokine receptor and microRNA-signatures to suppress effector T cells. (Right top) Tregs primed in response to tissue-specific antigens express chemokine receptors guiding their migration to restrain immune responses in distinct tissues. (Right bottom) Tregs exhibit plasticity in their differential expression and usage of suppressor modules.
Fig. 2. The three signal-hypothesis of Treg stability.
Tregs in the periphery bear distinct intrinsic signatures (signal 1) based on their Foxp3 expression and epigenetic status, resulting in generation of stable bona fide, transient uncommitted and potential Tregs. The stability of these differentially committed Tregs in any given microenvironment is set by their intrinsic signature and the balance of two other signals – Stabilizing signals (signal 2) and Destabilizing signals (signal 3). Stabilizing signals include survival and growth factors, quiescence factors and remodeling factors, while destabilizing signals include pro-inflammatory cytokines and mediators that antagonize Treg fate. Thus, inflammatory scenario 1 is likely to tip the balance towards the destabilizing factors, resulting in loss of Treg stability. On the contrary, scenario 2 is likely to tip the balance towards the stabilizing factors, thus maintaining stable Treg lineage.
Acknowledgments
We thank members of the Vignali laboratory for critical reading of the manuscript. This work was supported by the National Institutes of Health (R01 AI091977 to D.A.A.V.), NCI Comprehensive Cancer Center Support CORE grant (CA21765, to D.A.A.V.), and ALSAC (to D.A.A.V.).
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 2.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang H, Chess L. An integrated view of suppressor T cell subsets in immunoregulation. J Clin Invest. 2004;114:1198–1208. doi: 10.1172/JCI23411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mann MK, Ray A, Basu S, Karp CL, Dittel BN. Pathogenic and regulatory roles for B cells in experimental autoimmune encephalomyelitis. Autoimmunity. 2012;45:388–399. doi: 10.3109/08916934.2012.665523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinman RM, Hawiger D, Nussenzweig MC. Tolerogenic dendritic cells. Annu Rev Immunol. 2003;21:685–711. doi: 10.1146/annurev.immunol.21.120601.141040. [DOI] [PubMed] [Google Scholar]
- 7.Sakaguchi S. Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- 8.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 9.Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- 10.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 11.Curotto de Lafaille MA, Lafaille JJ, Graca L. Mechanisms of tolerance and allergic sensitization in the airways and the lungs. Curr Opin Immunol. 2010;22:616–622. doi: 10.1016/j.coi.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belkaid Y, Tarbell K. Regulatory T cells in the control of host-microorganism interactions. Annu Rev Immunol. 2009;27:551–589. doi: 10.1146/annurev.immunol.021908.132723. [DOI] [PubMed] [Google Scholar]
- 13.Demengeot J, Zelenay S, Moraes-Fontes MF, Caramalho I, Coutinho A. Regulatory T cells in microbial infection. Springer Semin Immunopathol. 2006;28:41–50. doi: 10.1007/s00281-006-0024-5. [DOI] [PubMed] [Google Scholar]
- 14.Kendal AR, Waldmann H. Infectious tolerance: therapeutic potential. Curr Opin Immunol. 2010;22:560–565. doi: 10.1016/j.coi.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi T, Sakaguchi S. Regulatory T cells in immune surveillance and treatment of cancer. Semin Cancer Biol. 2006;16:115–123. doi: 10.1016/j.semcancer.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Cipolletta D, Kolodin D, Benoist C, Mathis D. Tissular T(regs): a unique population of adipose-tissue-resident Foxp3+CD4+ T cells that impacts organismal metabolism. Semin Immunol. 2011;23:431–437. doi: 10.1016/j.smim.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Feuerer M, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burzyn D, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vudattu NK, Herold KC. Delayed anti-CD3 therapy in a mouse heart transplant model induced tolerance and long-term survival of allograft: achieving tolerance. Immunotherapy. 2013;5:1173–1176. doi: 10.2217/imt.13.113. [DOI] [PubMed] [Google Scholar]
- 20.Ait-Oufella H, et al. Natural regulatory T cells control the development of atherosclerosis in mice. Nat Med. 2006;12:178–180. doi: 10.1038/nm1343. [DOI] [PubMed] [Google Scholar]
- 21.Rouse BT, Sarangi PP, Suvas S. Regulatory T cells in virus infections. Immunol Rev. 2006;212:272–286. doi: 10.1111/j.0105-2896.2006.00412.x. [DOI] [PubMed] [Google Scholar]
- 22.Belkaid Y. Regulatory T cells and infection: a dangerous necessity. Nat Rev Immunol. 2007;7:875–888. doi: 10.1038/nri2189. [DOI] [PubMed] [Google Scholar]
- 23.Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Annu Rev Immunol. 2009;27:313–338. doi: 10.1146/annurev.immunol.021908.132657. [DOI] [PubMed] [Google Scholar]
- 24.Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- 25.Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 26.Bennett CL, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 27.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 28.Gavin MA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 29.Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–770. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 30.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 31.Lin W, et al. Regulatory T cell development in the absence of functional Foxp3. Nat Immunol. 2007;8:359–368. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- 32.Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 33.Sugimoto N, et al. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int Immunol. 2006;18:1197–1209. doi: 10.1093/intimm/dxl060. [DOI] [PubMed] [Google Scholar]
- 34.Hill JA, et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Ohkura N, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37:785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 36.Rudra D, et al. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat Immunol. 2012;13:1010–1019. doi: 10.1038/ni.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samstein RM, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu W, et al. A multiply redundant genetic switch ‘locks in’ the transcriptional signature of regulatory T cells. Nat Immunol. 2012;13:972–980. doi: 10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allan SE, et al. Activation-induced FOXP3 in human T effector cells does not suppress proliferation or cytokine production. Int Immunol. 2007;19:345–354. doi: 10.1093/intimm/dxm014. [DOI] [PubMed] [Google Scholar]
- 40.d’Hennezel E, Yurchenko E, Sgouroudis E, Hay V, Piccirillo CA. Single-cell analysis of the human T regulatory population uncovers functional heterogeneity and instability within FOXP3+ cells. J Immunol. 2011;186:6788–6797. doi: 10.4049/jimmunol.1100269. [DOI] [PubMed] [Google Scholar]
- 41.Miyara M, et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity. 2009;30:899–911. doi: 10.1016/j.immuni.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 42.Schmidl C, et al. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res. 2009;19:1165–1174. doi: 10.1101/gr.091470.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Floess S, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huehn J, et al. Developmental stage, phenotype, and migration distinguish naive- and effector/memory-like CD4+ regulatory T cells. J Exp Med. 2004;199:303–313. doi: 10.1084/jem.20031562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campbell DJ, Koch MA. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat Rev Immunol. 2011;11:119–130. doi: 10.1038/nri2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feuerer M, Hill JA, Mathis D, Benoist C. Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nat Immunol. 2009;10:689–695. doi: 10.1038/ni.1760. [DOI] [PubMed] [Google Scholar]
- 47.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaudhry A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu LF, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sawant DV, Wu H, Kaplan MH, Dent AL. The Bcl6 target gene microRNA-21 promotes Th2 differentiation by a T cell intrinsic pathway. Mol Immunol. 2013;54:435–442. doi: 10.1016/j.molimm.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelada S, et al. miR-182 and miR-10a are key regulators of Treg specialisation and stability during Schistosome and Leishmania-associated inflammation. PLoS Pathog. 2013;9:e1003451. doi: 10.1371/journal.ppat.1003451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cipolletta D, et al. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bettini M, Vignali DA. Regulatory T cells and inhibitory cytokines in autoimmunity. Curr Opin Immunol. 2009;21:612–618. doi: 10.1016/j.coi.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collison LW, et al. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450:566–569. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- 56.Vignali D. How many mechanisms do regulatory T cells need? Eur J Immunol. 2008;38:908–911. doi: 10.1002/eji.200738114. [DOI] [PubMed] [Google Scholar]
- 57.Rubtsov YP, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 58.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26:579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 59.Cao X, et al. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity. 2007;27:635–646. doi: 10.1016/j.immuni.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 60.Sakaguchi S, Vignali DA, Rudensky AY, Niec RE, Waldmann H. The plasticity and stability of regulatory T cells. Nat Rev Immunol. 2013;13:461–467. doi: 10.1038/nri3464. [DOI] [PubMed] [Google Scholar]
- 61.Zhou X, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol. 2009;10:1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bailey-Bucktrout SL, Bluestone JA. Regulatory T cells: stability revisited. Trends Immunol. 2011;32:301–306. doi: 10.1016/j.it.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rubtsov YP, et al. Stability of the regulatory T cell lineage in vivo. Science. 2010;329:1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miyao T, et al. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 2012;36:262–275. doi: 10.1016/j.immuni.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 65.Komatsu N, Mariotti-Ferrandiz ME, Wang Y, Malissen B, Waldmann H, Hori S. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc Natl Acad Sci USA. 2009;106:1903–1908. doi: 10.1073/pnas.0811556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsuji M, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science. 2009;323:1488–1492. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 67.Yang XO, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsieh CS, Liang Y, Tyznik AJ, Self SG, Liggitt D, Rudensky AY. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 69.Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30:636–645. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 70.Vignali DA. Mechanisms of T(reg) Suppression: Still a Long Way to Go. Front Immunol. 2012;3:191. doi: 10.3389/fimmu.2012.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li L, Boussiotis VA. Molecular and functional heterogeneity of T regulatory cells. Clin Immunol. 2011;141:244–252. doi: 10.1016/j.clim.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barnes MJ, Powrie F. Hybrid Treg cells: steel frames and plastic exteriors. Nat Immunol. 2009;10:563–564. doi: 10.1038/ni0609-563. [DOI] [PubMed] [Google Scholar]
- 73.Wing JB, Sakaguchi S. Multiple Treg suppressive modules and their adaptability. Front Immunol. 2012;3:178. doi: 10.3389/fimmu.2012.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hall AO, et al. The cytokines interleukin 27 and interferon-gamma promote distinct Treg cell populations required to limit infection-induced pathology. Immunity. 2012;37:511–523. doi: 10.1016/j.immuni.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35:337–348. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wohlfert EA, et al. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J Clin Invest. 2011;121:4503–4515. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Osorio F, et al. DC activated via dectin-1 convert Treg into IL-17 producers. Eur J Immunol. 2008;38:3274–3281. doi: 10.1002/eji.200838950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beriou G, et al. IL-17-producing human peripheral regulatory T cells retain suppressive function. Blood. 2009;113:4240–4249. doi: 10.1182/blood-2008-10-183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Voo KS, et al. Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc Natl Acad Sci USA. 2009;106:4793–4798. doi: 10.1073/pnas.0900408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chung Y, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Linterman MA, et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sawant DV, et al. Bcl6 controls the Th2 inflammatory activity of regulatory T cells by repressing Gata3 function. J Immunol. 2012;189:4759–4769. doi: 10.4049/jimmunol.1201794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sharma MD, et al. An inherently bifunctional subset of Foxp3+ T helper cells is controlled by the transcription factor eos. Immunity. 2013;38:998–1012. doi: 10.1016/j.immuni.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cretney E, et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol. 2011;12:304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 85.Takahashi H, et al. TGF-beta and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat Immunol. 2012;13:587–595. doi: 10.1038/ni.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schneider MA, Meingassner JG, Lipp M, Moore HD, Rot A. CCR7 is required for the in vivo function of CD4+ CD25+ regulatory T cells. J Exp Med. 2007;204:735–745. doi: 10.1084/jem.20061405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dudda JC, Perdue N, Bachtanian E, Campbell DJ. Foxp3+ regulatory T cells maintain immune homeostasis in the skin. J Exp Med. 2008;205:1559–1565. doi: 10.1084/jem.20072594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Suffia I, Reckling SK, Salay G, Belkaid Y. A role for CD103 in the retention of CD4+CD25+ Treg and control of Leishmania major infection. J Immunol. 2005;174:5444–5455. doi: 10.4049/jimmunol.174.9.5444. [DOI] [PubMed] [Google Scholar]
- 89.Guo Z, et al. CD4+CD25+ regulatory T cells in the small intestinal lamina propria show an effector/memory phenotype. Int Immunol. 2008;20:307–315. doi: 10.1093/intimm/dxm143. [DOI] [PubMed] [Google Scholar]
- 90.Lim HW, Broxmeyer HE, Kim CH. Regulation of trafficking receptor expression in human forkhead box P3+ regulatory T cells. J Immunol. 2006;177:840–851. doi: 10.4049/jimmunol.177.2.840. [DOI] [PubMed] [Google Scholar]
- 91.Grauer OM, et al. CD4+FoxP3+ regulatory T cells gradually accumulate in gliomas during tumor growth and efficiently suppress antiglioma immune responses in vivo. Int J Cancer. 2007;121:95–105. doi: 10.1002/ijc.22607. [DOI] [PubMed] [Google Scholar]
- 92.Wald O, et al. CD4+CXCR4highCD69+ T cells accumulate in lung adenocarcinoma. J Immunol. 2006;177:6983–6990. doi: 10.4049/jimmunol.177.10.6983. [DOI] [PubMed] [Google Scholar]
- 93.Pillai MR, et al. The plasticity of regulatory T cell function. J Immunol. 2011;187:4987–4997. doi: 10.4049/jimmunol.1102173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen W, Perruche S, Li J. CD4+CD25+ T regulatory cells and TGF-beta in mucosal immune system: the good and the bad. Curr Med Chem. 2007;14:2245–2249. doi: 10.2174/092986707781696591. [DOI] [PubMed] [Google Scholar]
- 95.Jutel M, et al. IL-10 and TGF-beta cooperate in the regulatory T cell response to mucosal allergens in normal immunity and specific immunotherapy. Eur J Immunol. 2003;33:1205–1214. doi: 10.1002/eji.200322919. [DOI] [PubMed] [Google Scholar]
- 96.Wang Z, et al. Tumor-derived IL-35 promotes tumor growth by enhancing myeloid cell accumulation and angiogenesis. J Immunol. 2013;190:2415–2423. doi: 10.4049/jimmunol.1202535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Olson BM, Jankowska-Gan E, Becker JT, Vignali DA, Burlingham WJ, McNeel DG. Human prostate tumor antigen-specific CD8+ regulatory T cells are inhibited by CTLA-4 or IL-35 blockade. J Immunol. 2012;189:5590–5601. doi: 10.4049/jimmunol.1201744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pasare C, Medzhitov R. Toll pathway-dependent blockade of CD4+CD25+ T cell-mediated suppression by dendritic cells. Science. 2003;299:1033–1036. doi: 10.1126/science.1078231. [DOI] [PubMed] [Google Scholar]
- 99.Xu L, Kitani A, Fuss I, Strober W. Cutting edge: regulatory T cells induce CD4+CD25-Foxp3- T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J Immunol. 2007;178:6725–6729. doi: 10.4049/jimmunol.178.11.6725. [DOI] [PubMed] [Google Scholar]
- 100.Dardalhon V, et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(−) effector T cells. Nat Immunol. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mantel PY, et al. GATA3-driven Th2 responses inhibit TGF-beta1-induced FOXP3 expression and the formation of regulatory T cells. PLoS Biol. 2007;5:e329. doi: 10.1371/journal.pbio.0050329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wei J, Duramad O, Perng OA, Reiner SL, Liu YJ, Qin FX. Antagonistic nature of T helper 1/2 developmental programs in opposing peripheral induction of Foxp3+ regulatory T cells. Proc Natl Acad Sci USA. 2007;104:18169–18174. doi: 10.1073/pnas.0703642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lal G, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182:259–273. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koenen HJ, Smeets RL, Vink PM, van Rijssen E, Boots AM, Joosten I. Human CD25highFoxp3pos regulatory T cells differentiate into IL-17-producing cells. Blood. 2008;112:2340–2352. doi: 10.1182/blood-2008-01-133967. [DOI] [PubMed] [Google Scholar]
- 105.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205:565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gabrysova L, Christensen JR, Wu X, Kissenpfennig A, Malissen B, O’Garra A. Integrated T-cell receptor and costimulatory signals determine TGF-beta-dependent differentiation and maintenance of Foxp3+ regulatory T cells. Eur J Immunol. 2011;41:1242–1248. doi: 10.1002/eji.201041073. [DOI] [PubMed] [Google Scholar]
- 107.Hoffmann P, et al. Loss of FOXP3 expression in natural human CD4+CD25+ regulatory T cells upon repetitive in vitro stimulation. Eur J Immunol. 2009;39:1088–1097. doi: 10.1002/eji.200838904. [DOI] [PubMed] [Google Scholar]
- 108.Duarte JH, Zelenay S, Bergman ML, Martins AC, Demengeot J. Natural Treg cells spontaneously differentiate into pathogenic helper cells in lymphopenic conditions. Eur J Immunol. 2009;39:948–955. doi: 10.1002/eji.200839196. [DOI] [PubMed] [Google Scholar]
- 109.Tang Q, et al. Central role of defective interleukin-2 production in the triggering of islet autoimmune destruction. Immunity. 2008;28:687–697. doi: 10.1016/j.immuni.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Korn T, et al. Myelin-specific regulatory T cells accumulate in the CNS but fail to control autoimmune inflammation. Nat Med. 2007;13:423–431. doi: 10.1038/nm1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yamanouchi J, et al. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet. 2007;39:329–337. doi: 10.1038/ng1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Oldenhove G, et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 2009;31:772–786. doi: 10.1016/j.immuni.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Murai M, et al. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat Immunol. 2009;10:1178–1184. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bailey-Bucktrout SL, et al. Self-antigen-Driven Activation Induces Instability of Regulatory T Cells during an Inflammatory Autoimmune Response. Immunity. 2013;39:949–962. doi: 10.1016/j.immuni.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hori S. Regulatory T cell plasticity: beyond the controversies. Trends Immuno. 2011;32:295–300. doi: 10.1016/j.it.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 117.Hori S. Developmental plasticity of Foxp3+ regulatory T cells. Curr Opin Immunol. 2010;22:575–582. doi: 10.1016/j.coi.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 118.Liston A, Piccirillo CA. Developmental plasticity of murine and human Foxp3(+) regulatory T cells. Adv Immunol. 2013;119:85–106. doi: 10.1016/B978-0-12-407707-2.00003-5. [DOI] [PubMed] [Google Scholar]
- 119.Hori S. Stability of regulatory T-cell lineage. Adv Immunol. 2011;112:1–24. doi: 10.1016/B978-0-12-387827-4.00001-2. [DOI] [PubMed] [Google Scholar]
- 120.Fontenot JD, Dooley JL, Farr AG, Rudensky AY. Developmental regulation of Foxp3 expression during ontogeny. J Exp Med. 2005;202:901–906. doi: 10.1084/jem.20050784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Apostolou I, Sarukhan A, Klein L, von Boehmer H. Origin of regulatory T cells with known specificity for antigen. Nat Immunol. 2002;3:756–763. doi: 10.1038/ni816. [DOI] [PubMed] [Google Scholar]
- 122.Jordan MS, et al. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2:301–306. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 123.Aschenbrenner K, et al. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat Immunol. 2007;8:351–358. doi: 10.1038/ni1444. [DOI] [PubMed] [Google Scholar]
- 124.Birzele F, et al. Next-generation insights into regulatory T cells: expression profiling and FoxP3 occupancy in Human. Nucleic Acids Res. 2011;39:7946–7960. doi: 10.1093/nar/gkr444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marson A, et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bettelli E, Dastrange M, Oukka M. Foxp3 interacts with nuclear factor of activated T cells and NF-kappa B to repress cytokine gene expression and effector functions of T helper cells. Proc Natl Acad Sci USA. 2005;102:5138–5143. doi: 10.1073/pnas.0501675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lopes JE, et al. Analysis of FOXP3 reveals multiple domains required for its function as a transcriptional repressor. J Immunol. 2006;177:3133–3142. doi: 10.4049/jimmunol.177.5.3133. [DOI] [PubMed] [Google Scholar]
- 128.Wu Y, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 129.Ono M, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 130.Li B, et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci USA. 2007;104:4571–4576. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bettini ML, et al. Loss of epigenetic modification driven by the Foxp3 transcription factor leads to regulatory T cell insufficiency. Immunity. 2012;36:717–730. doi: 10.1016/j.immuni.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liston A, Lu LF, O’Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhou X, et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lu LF, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ouyang W, et al. Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature. 2012;491:554–559. doi: 10.1038/nature11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ouyang W, Beckett O, Ma Q, Paik JH, DePinho RA, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. 2010;11:618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- 137.Kerdiles YM, et al. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33:890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 139.Wei G, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Toker A, et al. Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J Immunol. 2013;190:3180–3188. doi: 10.4049/jimmunol.1203473. [DOI] [PubMed] [Google Scholar]
- 141.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Burchill MA, et al. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–121. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Huehn J, Polansky JK, Hamann A. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat Rev Immunol. 2009;9:83–89. doi: 10.1038/nri2474. [DOI] [PubMed] [Google Scholar]
- 144.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204:1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Baron U, et al. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol. 2007;37:2378–2389. doi: 10.1002/eji.200737594. [DOI] [PubMed] [Google Scholar]
- 147.Polansky JK, et al. Methylation matters: binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J Mol Med. 2010;88:1029–1040. doi: 10.1007/s00109-010-0642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mouly E, et al. The Ets-1 transcription factor controls the development and function of natural regulatory T cells. J Exp Med. 2010;207:2113–2125. doi: 10.1084/jem.20092153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Delgoffe GM, Bettini ML, Vignali DA. Identity crisis: it’s not just Foxp3 anymore. Immunity. 2012;37:759–761. doi: 10.1016/j.immuni.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol. 2006;7:401–410. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- 151.Bautista JL, et al. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat Immunol. 2009;10:610–617. doi: 10.1038/ni.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity. 2013;38:414–423. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 153.Liu X, et al. T cell receptor CDR3 sequence but not recognition characteristics distinguish autoreactive effector and Foxp3(+) regulatory T cells. Immunity. 2009;31:909–920. doi: 10.1016/j.immuni.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Curotto de Lafaille MA, Lafaille JJ. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity. 2009;30:626–635. doi: 10.1016/j.immuni.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 155.Haribhai D, et al. A requisite role for induced regulatory T cells in tolerance based on expanding antigen receptor diversity. Immunity. 2011;35:109–122. doi: 10.1016/j.immuni.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Samstein RM, Josefowicz SZ, Arvey A, Treuting PM, Rudensky AY. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell. 2012;150:29–38. doi: 10.1016/j.cell.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yadav M, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med. 2012;209:1713–1722. S1711–1719. doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Weiss JM, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med. 2012;209:1723–1742. S1721. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Glinka Y, Prud’homme GJ. Neuropilin-1 is a receptor for transforming growth factor beta-1, activates its latent form, and promotes regulatory T cell activity. J Leukoc Biol. 2008;84:302–310. doi: 10.1189/jlb.0208090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sarris M, Andersen KG, Randow F, Mayr L, Betz AG. Neuropilin-1 expression on regulatory T cells enhances their interactions with dendritic cells during antigen recognition. Immunity. 2008;28:402–413. doi: 10.1016/j.immuni.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Delgoffe GM, et al. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature. 2013;501:252–256. doi: 10.1038/nature12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yadav M, Stephan S, Bluestone JA. Peripherally induced Tregs - role in immune homeostasis and autoimmunity. Front Immunol. 2013;4:232. doi: 10.3389/fimmu.2013.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhou X, Bailey-Bucktrout S, Jeker LT, Bluestone JA. Plasticity of CD4(+) FoxP3(+) T cells. Curr Opin Immunol. 2009;21:281–285. doi: 10.1016/j.coi.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Polansky JK, et al. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38:1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 165.Schlenner SM, Weigmann B, Ruan Q, Chen Y, von Boehmer H. Smad3 binding to the foxp3 enhancer is dispensable for the development of regulatory T cells with the exception of the gut. J Exp Med. 2012;209:1529–1535. doi: 10.1084/jem.20112646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Antov A, Yang L, Vig M, Baltimore D, Van Parijs L. Essential role for STAT5 signaling in CD25+CD4+ regulatory T cell homeostasis and the maintenance of self-tolerance. J Immunol. 2003;171:3435–3441. doi: 10.4049/jimmunol.171.7.3435. [DOI] [PubMed] [Google Scholar]
- 167.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 168.Mantel PY, et al. Molecular mechanisms underlying FOXP3 induction in human T cells. J Immunol. 2006;176:3593–3602. doi: 10.4049/jimmunol.176.6.3593. [DOI] [PubMed] [Google Scholar]
- 169.Sauer S, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci USA. 2008;105:7797–7802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li MO, Sanjabi S, Flavell RA. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25:455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 171.Takaki H, et al. STAT6 Inhibits TGF-beta1-mediated Foxp3 induction through direct binding to the Foxp3 promoter, which is reverted by retinoic acid receptor. J Biol Chem. 2008;283:14955–14962. doi: 10.1074/jbc.M801123200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kitoh A, et al. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 2009;31:609–620. doi: 10.1016/j.immuni.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 173.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 174.Jeker LT, et al. MicroRNA 10a marks regulatory T cells. PLoS One. 2012;7:e36684. doi: 10.1371/journal.pone.0036684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Beyer M, et al. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat Immunol. 2011;12:898–907. doi: 10.1038/ni.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]