Abstract
Elevation of amyloid β-peptide (Aβ) is critically associated with Alzheimer’s disease (AD) pathogenesis. Aβ-induced synaptic abnormalities, including altered receptor trafficking and synapse loss, have been linked to cognitive deficits in AD. Recent work implicates a lipid critical for neuronal function, phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2], in Aβ-induced synaptic and behavioral impairments. Synaptojanin 1 (Synj1), a lipid phosphatase mediating the breakdown of PI(4,5)P2, has been shown to play a role in synaptic vesicle recycling and receptor trafficking in neurons. Heterozygous deletion of Synj1 protected neurons from Aβ-induced synaptic loss and restored learning and memory in a mouse model of AD. Thus, inhibition of Synj1 may ameliorate Aβ-associated impairments, suggesting Synj1 as a potential therapeutic target. To this end, we developed a screening assay for Synj1 based on detection of inorganic phosphate liberation from a water-soluble, short chain PI(4,5)P2. The assay displayed saturable kinetics and detected Synj1’s substrate preference for PI(4,5)P2 over PI(3,4,5)P3. The assay will enable identification of novel Synj1 inhibitors which have potential utility as chemical probes to dissect the cellular role of Synj1 as well as potential to prevent or reverse AD-associated synaptic abnormalities.
Keywords: Lipid phosphatase; phosphatidylinositol-4,5-bisphosphate; Synaptojanin 1; src homology 2 domain-containing inositol-5-phosphatase 2 (SHIP2); small molecule screening; Alzheimer’s disease; drug discovery
INTRODUCTION
There are still no effective prophylactic or ameliorative therapeutics for AD, therefore novel strategies and cellular and molecular targets are required. The major focus for developing AD therapeutics has been targeting amyloid β-peptide (Aβ).1 Emphasis in the field has been to target Aβ biogenesis through inhibition or modulation of the amyloidogenic pathway. However these studies yielded extremely limited successes, indicating that new strategies and therapeutic targets are required.1 Current evidence indicates that soluble oligomeric forms of Aβ are associated with synaptic deficits including alterations in synaptic plasticity and spine morphology.2 Aβ-induced alterations in synaptic plasticity are believed to be associated with changes in receptor trafficking, such as cell surface expression of AMPA receptors and NMDA receptors as well as altered intracellular signaling cascades.2,3 Further, Aβ also induces synapse and dendritic spine loss. 2–4 Thus, ameliorating Aβ-associated synaptic alterations is an attractive strategy toward AD intervention.
It is increasingly evident that phosphoinositides (i.e., phosphorylated derivatives of phosphatidylinositol) play a significant role in AD.5 Phosphatidylinositol-(4,5)-bisphosphate, PI(4,5)P2, is a signaling lipid involved in ion channel regulation, exocytosis, endocytosis, actin cytoskeleton rearrangement and cell signaling.6 PI(4,5)P2 dephosphorylation at the synapse is mediated primarily by Synaptojanin 1 (Synj1), a type II polyphosphate-5-phosphatase encoded by a gene present on human chromosome 21. Synj1 is expressed and enriched at the synapse and has been shown to play a role in endocytosis, presynaptic vesicle recycling as well as postsynaptic receptor trafficking.6–9 Synj1 belongs to a large enzyme family which includes src homology 2 domain-containing inositol-5-phosphatase 2 (SHIP2), the mammalian homolog of the yeast suppressor of actin (SAC) domain PI(3,5)P2 5-phosphatase Factor induced gene (Fig 4) and Oculocerebrorenal Syndrome of Lowe (ORCL).10 Synj1 contains a central 5-phosphatase domain, which can dephosphorylate PI(4,5)P2 and PI(3,4,5)P3 on the 5′ position of the inositol ring. Synj1 also has an NH2-terminal Sac1-like domain responsible for 3-, 4- and 5-phosphatase activity towards substrates as diverse as PI3P, PI4P, PI5P and PI(3,5)P2.11 Previous mouse genetic studies have indicated that the predominant physiological substrate of Synj1 is PI(4,5)P2, but enzymatic activity has also been reported towards PI(3,4,5)P311–13.
Fig. 4. Optimization of Synj1 activity assay.
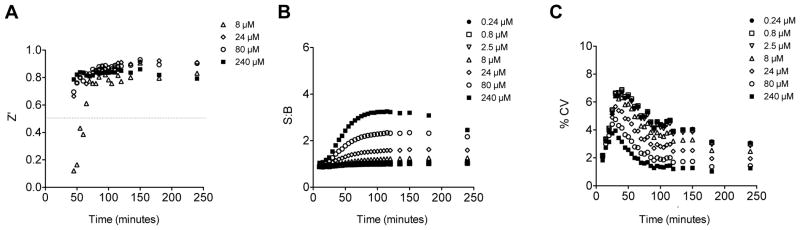
(A) Z′ factor calculated for each time point for specific activity of 100 ng Synj1 at differing lipid concentrations. (B) Signal to Background (S:B) is shown for Synj1 activity compared to background fluorescence in control without enzyme (C) % CV is shown for variance of Synj1 activity compared to specific Synj1 activity. Results are representative of at least two independent experiments.
Relative to kinases, phosphatases are considered more difficult targets for the discovery of selective and potent small molecule inhibitors.14,15 However, ample target validation studies have revealed the biological significance of phosphatases in various human disease.10,14–19 For example, the tyrosine phosphatase STEP has been suggested as a candidate therapeutic target in AD.20 Among lipid phosphatases, SHIP2 which like Synj1 is a type II polyphosphate-5-phosphatase, is considered a promising pharmaceutical target for type-2 diabetes.10,17 Further, selective small molecule inhibitors for SHIP2 have been characterized.21,22. Investigational small molecule inhibitors for other lipid phosphatases including SHIP1 and PTEN (the 3′-phosphatase, protein phosphatase and tensin homolog deleted on chromosome ten) have also been identified as potential anticancer therapies.10 Therefore, lipid phosphatases that metabolize phosphoinositides, such as Synj1, represent a promising and potentially druggable class of pharmaceutical targets in various human diseases.10,17
A potential link between AD and Synj1 was first suggested by studies demonstrating that familial Alzheimer’s disease (FAD) mutations in the PS1 and PS2 genes result in a perturbation of the metabolism of PI(4,5)P2. Consequently, overexpression of membrane-targeting Synj1 led to an increase in Aβ production.23 Thus, an imbalance in PI(4,5)P2 may underlie some aspects of FAD pathogenesis which can be modulated by Synj1. Treatment of primary neurons with oligomeric preparations of Aβ42 decreased the levels of PI(4,5)P2 and concordantly caused loss of a PI(4,5)P2 probe from the plasma membrane in a neuronal cell line.24 Accordingly, PI(4,5)P2 levels are reduced in a synapse enriched fraction of brain of a mouse model of AD, Tg2576. Genetically reducing Synj1 ameliorated the PI(4,5)P2 deficit as well as performance in learning and memory tasks.4 Recent systems level analysis of transcriptional changes revealed that Synj1 expression is selectively altered in a mouse model and in AD patients.25,26 Additionally, one of the yeast orthologs of Synj1, INP52, was identified in an unbiased genome wide screen for modifiers of Aβ toxicity in yeast.27 Based on these studies, reduction or pharmacological inhibition of Synj1 may ameliorate Aβ42 oligomer induced synaptic defects. Additionally, a recent study has pinpointed deficits in PI(4,5)P2 due to normal aging, which led to cognitive decline.28 Finally, overexpression of Synj1 in a mouse model of Down syndrome (DS) has been shown to cause a deficiency of PI(4,5)P2 in the brain as well as cognitive deficits.12 Thus, preventing PI(4,5)P2 catabolism through pharmacological modulation of Synj1 has potential benefits for AD, DS, and aging.
In order for identification of small molecule inhibitors of Synj1 activity, we developed a 5-phosphatase activity assay using recombinant Synj1 and water-soluble lipid substrate for detection of free inorganic phosphate. This assay was able to detect Synj1 activity towards PI(4,5)P2 but has the potential to be optimized for additional phosphoinositide phosphatases. We furnish a reliable, sensitive and robust screening assay for identification of small molecule inhibitors of Synj1. Small molecules identified with this assay have potential for use as chemical probes for experimentally dissecting the role of PI(4,5)P2 in AD. Further, promising small molecules may eventually lead to pharmacological tools for AD intervention.
MATERIALS AND METHODS
Materials
Lipids were obtained from Echelon (Salt Lake City, UT). The diC8 lipids were D-myo-Phosphatidylinositol 4,5-bisphosphate [(PI(4,5)P2] and D-myo-Phosphatidylinositol 3,4,5-trisphosphate [(PI(3,4,5)P3] and were solubilized in water at 3 mM. M2 anti-FLAG antibody, anit-FLAG affinity resin and FLAG peptide were from Sigma, (St. Louis, MO). GFP antibody was from Roche (Indianapolis, IN). G418 was from Calbiochem (Billerica, MA). SHIP2 lipid phosphatase was purchased from Echelon and resuspended at 500ng/ul and stored at −80°C. SHIP2 inhibitor, AS 1949490, was from Tocris Biosciences (Minneapolis, MN). The PiPer Phosphate Assay Kit was purchased from Molecular Probes (Life Technologies, Grand Island, NY) and used as per manufacturer protocol except for the modifications described below.
Expression and Purification of Synj1
FLAG-human synaptojanin1-145 (Synj1-FLAG) construct was obtained from Pietro De Camilli, Yale University. Synj1 constructs were stably expressed in human embryonic kidney (HEK) cells under selection of G418 (CalBiochem) for production of Synj1-FLAG as previously described.29 Synj1-FLAG and Synj1-GFP-FLAG were detected using antibodies recognizing the epitope tags, M2 anti-FLAG (Sigma) and anti-GFP (Roche). Synj1-FLAG was purified using anti-FLAG M2 affinity resin (Sigma), eluted with FLAG peptide, resuspended at 86ng/ul and stored at −80°C in storage buffer (50mM Tris-HCl, pH 8.0, 200mM NaCl). Synj1-GFP-FLAG was expressed in HEK cells, purified and resuspended at a concentration of 75 ng/uL in storage buffer and stored at −80°C. Protein concentration was determined by comparing the eluted protein with bovine serum albumin protein standards on a commassie stained gel using quantitative densitometry with LI-COR Odessey quantitative fluorescence imaging system.
Synj1 Activity Assay Using BODIPY-Labeled Lipids
Synj1 activity was determined with BODIPY-labeled lipids as previously described.29 Briefly, water soluble, short chain (C-6) BODIPY-labeled PI(4,5)P2 was incubated with purified recombinant enzyme for 15 min at 37°C in assay buffer (25mM HEPES, pH 7.4, 100mM KCl, 1mM EGTA, 1mM MgCl2). Lipids were separated by thin layer chromatography using silica plates (EMD Chemicals) using the solvent mixture chloroform:acetone:methanol:acetic acid:water (64:30:24:30:13). Fluorescent lipids were visualized by UV range excitation and recorded using the Bio Doc-It system (UVP). Lipids were identified by co-migration with individual BODIPY-labeled lipid standards and quantified using Image J software (NIH). Rat brain cytosol (RBC) was used as a positive control for conversion of BODIPI-PI(4,5)P2 to BODIPI-PI(4)P.
Detection of Inorganic Phosphate Release from diC8 Lipids
The PiPer Phosphate Assay Kit (Life Technologies, Grand Island, NY) was used for detection of the inorganic phosphate liberated from dephosphorylation of diC8-PI(4,5)P2 or diC8-PI(3,4,5)P3 by Synj1-FLAG or SHIP2. The dephosphorylation of PI4P to PIP did not contribute to the phosphate detected due to the loss of the Sac domain activity during purification.11,29 PiPer assay was performed according to manufacturer’s instructions except an assay buffer of 0.1 M NaCl, 1 mM MgCl, 0.25 mM EDTA, and 20 mM HEPES, pH 7.4, was used as in previous reports indicating these conditions are optimal for Synj1 activity.30 The Synj1 assay buffer did not interfere with the detection of inorganic phosphate by the PiPer assay (data not shown). All assays were performed in Black/Clear Bottom 96-well assay plates (BD Falcon). All plates were incubated at 37°C in the dark and fluorescence was read in a TECAN infinite M200 microplate reader fluorometer (Tecan, Switzerland) at excitation/emission wavelengths of 544/590 nm at a constant temperature of 37°C with intermittent mixing.
Data Analysis and Statistical Methods
Data were analyzed using Prism 5.0 software (GraphPad Software, San Diego, CA) for kinetic analysis. Standard curve for determination of phosphate concentration relative to fluorescence was analyized by Graphpad Prism software with linear fit. Z′ factor was determined using the standard equation:31
The equation for Z′ compares the standard deviation of positive control (σ+) and negative control (σ−) to the difference in the averages of positive and negative controls (μ+ and μ−, respectively). The positive control reactions included both substrate and enzyme and the negative control reactions lacked either substrate (no Subst) or enzyme (no Enz) as indicated. Signal to background (S:B) was calculated by dividing signal from experimental reactions (containing Synj1, di-C8 lipid and reaction reagents) by background fluorescence obtained from the reactions lacking substrate. Coefficient of variation (%CV) was calculated by dividing fluorescent signal from experimental reactions by background fluorescence obtained from the reaction reagents alone and multiplying by 100. Standard deviation is shown of representative experiments except where indicated.
RESULTS
Active Synj1 Expression
To develop an assay for identifying small molecule inhibitors of Synj1, we expressed and purified recombinant human Synj1 and examined whether the recombinant human Synj1 harbored PI(4,5)P2 5′-phosphatase activity.29 In order to purify a sufficient amount of catalytically active Synj1 for biochemical analysis, we established a stable HEK293 cell line expressing full-length human Synj1 containing GFP and FLAG (Synj1-GFP-FLAG) epitope tags. To determine if Synj1 was expressed in the stable cell line, antibodies recognizing the epitope tags were used in immunocytochemistry to label fixed cells. Both anti-FLAG and anti-GFP antibodies detected Synj1 protein which higher magnification indicates was localized to the cytosol (Fig. 1A). Cell lysates were harvested from the stable cell line and were then subjected to FLAG-affinity chromatography. Human Synj1-GFP-FLAG appeared as a ~170 kDa single band in SDS-PAGE followed by coommassie staining (Fig. 1B). The identity of this band was further confirmed by Western blot analysis using anti-Synj1 and anti-FLAG antibodies (Fig. 1C). These analyses indicated that Synj1 was highly enriched in the elution fraction. In order to determine whether the purified Synj1 harbored catalytic activity (conversion of PI(4,5)P2 into PI4P), Synj1-FLAG was incubated with PI(4,5)P2 labeled with a BODIPY fluorescent tag [BODIPY-PI(4,5)P2]. Incubation of recombinant Synj1 with BODIPY-PI(4,5)P2 resulted in increased accumulation of PI4P, indicating that the 5-phosphatase domain of Synj1 is catalytically active (Fig. 1D)29. Since significant lipid phosphatase activity is associated with rat brain cytosol (RBC), it was used as a positive control for conversion of PI(4,5)P2 to PI(4)P (Fig. 1D). In addition to 5-phosphatase activity, Synj1 also has 4-phosphatase activity mediated by the Sac domain, however, 4-phosphatase activity associated with the Sac domain was not detectable after purification, consistent with the propensity of the catalytic cysteine to rapidly oxidize (Fig. 1D).11,29 Synj1 was stored at −80°C without significant loss of 5-phosphatase enzyme activity (data not shown). This indicates that recombinant Synj1 is catalytically active and can be purified without loss of 5-phosphatase activity.
Fig. 1. Expression and purification of active Synj1 from HEK-239T cells.
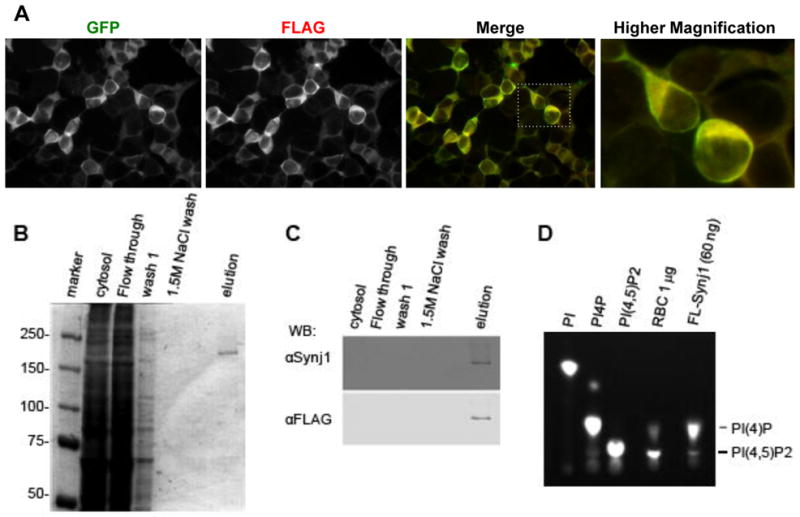
(A) HEK-293T cells stably expressing Synj1-GFP-FLAG labeled with indicated antibodies. Image was acquired with a 40x objective on an Olympus IX81 microscope using image analysis software Slidebook version 5.0. (B) Commassie stained gel showing aliquots from purification scheme of Synj1-GFP-FLAG. (C) Western blot detection of Synj1-GFP-FLAG using Synaptojanin 1 and FLAG specific antibodies. (D) Purified Synj1-GFP-FLAG phosphatase activity was detected using BODIPY labeled lipids and a positive control for lipid phosphatase activity, rat brain cytosol (RBC).
Fluorescent Assay for Synj1 Activity
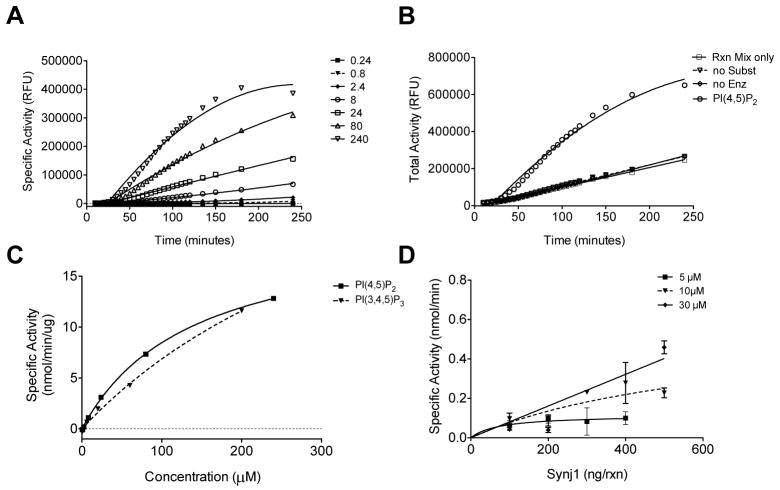
To detect Synj1 enzymatic activity a fluorescent phosphatase detection reagent kit (PiPer, Molecular Probes, Life Technologies) was used to detect the loss of phosphate from PI(4,5)P2. This assay detects free phosphate liberated by phosphatase activity through a series of oxidative reactions leading to the formation of the fluorescent product, resorufin. Maltose phosphorylase converts maltose into glucose 1-phosphate and glucose in the presence of free phosphate. Glucose is then oxidized by glucose oxidase to form gluconolactone and H2O2 which drives the formation of resorufin from the non-fluorescent Amplex Red reagent using horseradish peroxidase (HRP) as a catalyst (Supplemental Fig. 1). The Synj1 assay reaction included purified recombinant Synj1-FLAG and water-soluble short chain lipid species, either diC8-PI(4,5)P2 or diC8- PI(3,4,5)P3. Phosphate liberation was proportional to the fluorescence of resorufin. Assay signal was normalized to the total fluorescence elicited by inclusion of 10μM H2O2 in a control reaction which drives generation of resorufin (Supplemental Fig. 1) Phosphate content of reactions was quantified by comparison to a standard curve of potassium phosphate (Supplemental Fig. 2). Incubation of Synj1 with soluble lipid substrate and phosphatase assay reagents resulted in an increase in fluorescence over time which was dependent on the lipid concentration (Fig. 2A). Specific activity was expressed in Relative Fluorescent Units (RFU) after subtracting the background fluorescence of reactions lacking either substrate (no Subst), enzyme (no Enz) or both substrate and enzyme (Rxn Mix only) shown in Fig. 2B. At 100 minutes after assay start, specific activity of 100ng Synj1 was determined for known substrates PI(4,5)P2 and PI(3,4,5)P3. The relative apparent Km for Synj1 acting on PI(4,5)P2 was determined to be 138.5 (± 8.6) μM (Fig. 2C).
Fig. 2. Detection of inorganic phosphate release from PI(4,5)P2 by Synj1.
(A) Detection of inorganic phosphate release from PI(4,5)P2 by purified Synj1 shown in specific activity relative fluorescent units (RFU) after background fluorescence is subtracted. (B) Background fluorescence in reactions without lipid substrate (no Subst), without enzyme (no Enz) or without both substrate and enzyme (Rxn Mix only) compared to total activity detected from Synj1 and 240 μM PI(4,5)P2. (C) Selectivity of phosphatase activity of Synj1 for PI(4,5)P2 over PI(3,4,5)P3 shows Michaelis-Menten kinetic analysis for increasing concentration of substrate. (D) Dependence of specific activity on increasing concentrations of Synj1 at three different concentrations of PI(4,5)P2. Results are representative of at least two independent experiments.
We next wanted to determine if the assay could detect substrate selectivity profiles for Synj1 which has reduced activity toward PI(3,4,5)P3 relative to PI(4,5)P2.11,12,30 Synj1 was incubated in the presence of increasing amount of diC8-PI(3,4,5)P3 and background fluorescence (no substrate) was subtracted to obtain specific activity. The relative apparent Km for Synj1 acting on PI(3,4,5)P3 in this assay system was determined to be 438.5 (±130.4) μM (Fig. 2C). The substrate specificity constant kcat/Km for PI(4,5)P2 was 3.6×105 M−1s−1 while for PI(3,4,5)P3 it was slightly lower at 2.1×105 M−1s−1, indicating PI(4,5)P2 is a better substrate for Synj1.
To analyze the assay parameters for dependence on lipid and enzyme concentration, we incubated increasing amounts of enzyme with varying concentration of diC8-PI(4,5)P2. These experiments indicated that assay was dependent on concentration of the Synj1 enzyme (Fig. 2D). At substrate concentrations less than 30 μM, Synj1 activity was limited by substrate concentration. At a substrate concentration of 30 μM, Synj1 activity was no longer dependent on the concentration of substrate and was linear with concentration of Synj1 indicating substrate was in excess (Fig. 2D). Our data indicate that the assay is able to detect Synj1 activity recapitulating a relative substrate selectivity profile for PI(4,5)P2 and PI(3,4,5)P3.30 The assay is also dependent on Synj1 enzyme concentration and is linear at concentrations of substrate in excess of 30 μM. This indicates that the assay shows dependence on substrate and enzyme concentration which are critical parameters for measuring enzymatic properties.
Lipid Phosphatase SHIP2 Substrate Selectivity
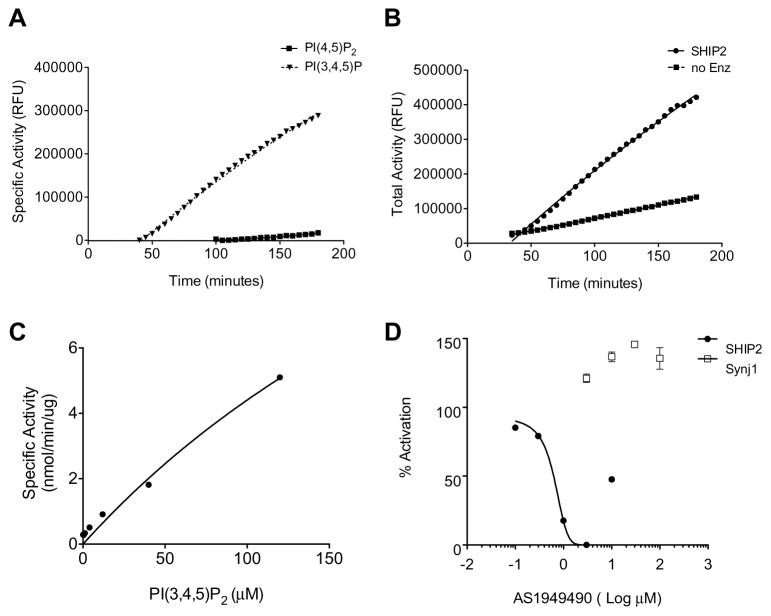
Lipid phosphatases can catalyze the 5′-phosphatase reaction with different lipid species as substrates. Previous studies indicate that SHIP2 activity toward PI(4,5)P2 is minor compared to activity toward its preferred substrate PI(3,4,5)P3.32 To test whether this assay system could detect SHIP2 activity and its substrate selectivity, recombinant SHIP2 was incubated with PI(3,4,5)3 or PI(4,5)P2. The assay was able to detect activity of SHIP2 for its preferred substrate PI(3,4,5)P3. Detection of phosphate liberation from PI(3,4,5)P3 by SHIP2 was apparent 40 minutes after assay start and increased with time (Fig. 3A). Additionally, a low level of fluorescence was detected from reactions containing PI(4,5)P2 and SHIP2 in this assay but did not accumulate to detectable levels (over background fluorescence from reactions containing PI(4,5)P2, but no enzyme) until a much later time point, 100 minutes (Fig. 3B). Since we observed only minor signal strength it is plausible that this is indicative of phosphate liberation from PI(4,5)P2 by SHIP2 activity which has been previously described as lower than its activity with PI(3,4,5)P3.32 For specific activity determination, background fluorescence from reactions lacking Synj1 enzyme (no Enz) was subtracted from the total fluorescent signal from experimental reactions (Fig 3B). The specific activity of SHIP2 toward PI(3,4,5)P3 yielded a relative apparent Km of 383.3 (±190.6) μM and kcat/Km for PI(3,4,5)P3 was 9.5×104 M−1s−1 (Fig. 3C). This indicates that the assay is amenable to study of SHIP2 as well as Synj1 and has potential to be adapted for use with additional lipid phosphatases.
Fig. 3. Lipid phosphatase assay using SHIP2.
(A) SHIP2 phosphatase activity with 120 μM preferred substrate PI(3,4,5)P3 compared to minimal activity toward 120 μM PI(4,5)P2 with respect to time. Specific activity in relative fluorescent units is shown for which background fluorescence has been subtracted at each time point. (B) Total activity of SHIP2 toward 120 μM PI(3,4,5)P3 and background fluorescence of reaction without enzyme (no Enz). (C) Specific activity with respect to increasing concentration of lipid substrate at 100 minutes after assay start. (D) SHIP2 and Synj1 activity in presence of AS1949490 shown as % SHIP2 activation after normalization to phosphate standard curve and calculation of phosphate liberated per minute (μM/min). Error bars represent standard error of the mean.
To determine if the assay was able to detect small molecule-mediated inhibition of lipid phosphatase activity, we wanted to test a known lipid phosphatase inhibitor. Since there are currently no inhibitors of Synj1 activity, we used a well characterized selective inhibitor of SHIP2 enzymatic activity, AS1949490.22 SHIP2 was incubated with AS1949490 in the presence of excess lipid substrate, 100μM PI(3,4,5)P3, resulting in reduction of SHIP2 activity. SHIP2 activity was inhibited with a Km of 0.625 (±0.12) μM, comparable to the published Km for AS1949490 of 0.62 μM (Fig. 3D).22 It has been previously reported that this SHIP2 selective inhibitor was unable to inhibit Synj1 activity at concentrations up to 50μM.22 In order to determine if the selectivity of this compound is reflected in this assay, we incubated Synj1 with excess lipid substrate, 100μM PI(4,5)P2, in the presence of up to 100μM AS1949490. There was no inhibition of Synj1 activity at any concentration (Fig. 3D). We also determined if AS1949490 was able to inhibit any components of the enzymatic detection of phosphate liberation. AS1949490 up to 100μM did not inhibit detection of 100μM free phosphate by assay enzymes (data not shown). This indicates that the assay is able to accurately detect inhibition of lipid phosphatases.
Assay Optimization and Miniaturization
Assay parameters of Synj1 activity toward PI(4,5)P2 were first examined in the 96-well plate platform to gage potential adaptability for high throughput screening (HTS). Synj1-FLAG was incubated with increasing concentrations of diC8-PI(4,5)P2. Using lipid substrate concentrations greater than 8 μM yielded Z′-factor (an assay sensitivity factor which accounts for standard deviation of negative and positive controls) of greater than 0.5, indicating that the assay is suitable for HTS (Fig. 4A).31 We addressed additional metrics of assay performance and sensitivity which include signal to background (S:B) and coefficient of variation (%CV) (see methods for calculations). Typically, an assay with S:B greater than 2-fold and %CV value <15% is considered a robust assay suitable for performance in HTS.31 Both of these metrics validated assay parameters (Fig. 4B–C).
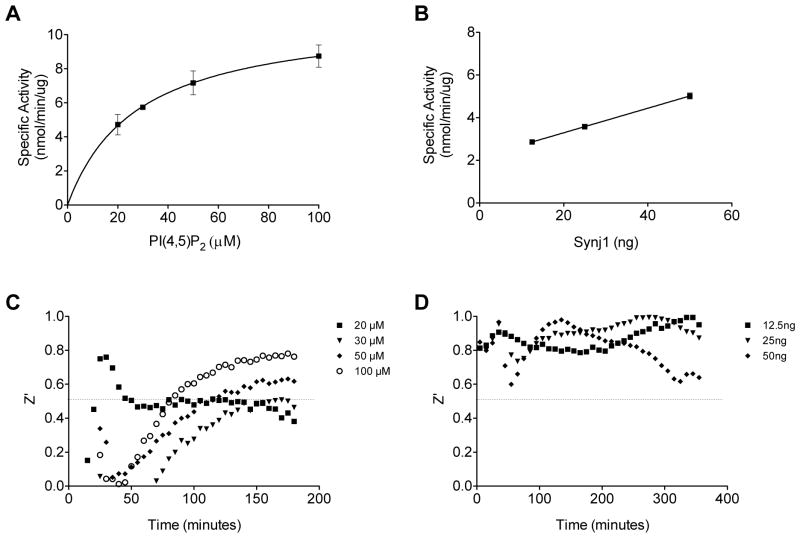
For potential use in 384-well platform, the assay volume was miniaturized to 50 μl, a volume which is amenable to 384-well platform (Fig. 5A–D). Specific activity displayed Michaelis-Menten enzyme kinetics with an apparent Km of 27.8 (±5.2) μM (Fig. 5A). The reduced Km compared to that obtained in the larger volume (Fig. 2) may be due to improved mixing in the smaller volume or reduced absorption of lipid substrate or enzyme to well surface area since both volumes were assayed in 96-well plates. Enzyme activity was determined in the miniaturized volume with lipid substrate in excess (100 μM) and found to display linear activity dependence on enzyme concentration (Fig. 5B).
Fig. 5. Miniaturization of Synj1 activity assay.
(A) Synj1 specific activity with increasing concentrations of PI(4,5)P2 in miniaturized reaction with 50 ng of Synj1 in 50μL reaction volume. (B) Specific activity of increasing concentrations of Synj1 enzyme in a 50 μl final reaction volume with 100 μM PI(4,5)P2. (C–D) Corresponding Z′ factors for varied lipid concentration in reactions with excess enzyme and (C) and varied enzyme concentrations in reactions with excess substrate (100 μM). Results are representative of at least two independent experiments.
The Z′ factors were calculated for each time point for the miniaturized experiments with increasing substrate concentration as well as increasing Synj1 enzyme amount (Fig. 5A–B). Calculated Z′ factor of miniaturized volumes showed dependence on substrate concentration (Fig. 5C). However, when substrate was in excess at 100 μM and only the amount of Synj1 was varied (Fig. 5B), the Z′ factor did not show dependence on enzyme concentration and was greater than 0.5 for all concentrations at each time point assayed (Fig. 5D). These results indicate that Synj1 activity is highly dependent on substrate concentration which should be used in excess for a screening assay. Therefore, this miniaturized screening assay is suitable for the identification of small molecule inhibitors of Synj1.
Since many compounds which are often used in HTS are dissolved in DMSO as a solvent, we next determined the assay sensitivity to DMSO. DMSO up to 5% did not inhibit assay component enzymes from detecting 100μM free phosphate (Supplemental Fig. 3). However, DMSO inhibited Synj1 activity approximately 10–15% at concentrations typically used for HTS 0.1–1% (Supplemental Fig. 3). Future studies will require careful consideration for inclusion of the same concentration of DMSO in control and experimental conditions.
DISCUSSION
To date there are no known small molecule inhibitors of Synj1 phosphatase activity. Successful identification of small molecule inhibitors for SHIP2, a lipid phosphatase target for type-2 diabetes,21–22 suggests that Synj1, also a type II polyphosphate-5-phosphatase, might also be chemically tractable. Our current studies establish a convenient and reliable screening assay which will enable discovery of small molecule inhibitors of Synj1 activity.
In order to effectively develop and miniaturize an assay for lipid modifying enzymes suitable for HTS, the lipid substrates are a critical determinant.33 The water-soluble short chain di-C8 lipid substrates used in this assay have advantages over the prevalent use of radiolabeled substrate precluding HTS. Further, the commonly used BODIPY labeled substrate has the potential to lead to steric interactions with small molecule inhibitors or to interfere with enzyme kinetics as well as being cost prohibitive. Additionally, as opposed to malachite green, which requires absorbance reading from a quenched, end-point assay, the use of the real time fluorescent probe in this assay, resorufin, allows multiple fluorometric reads at multiple times points within one assay. It also uniquely enables normalization to total resorufin fluorescence driven by H2O2 for each assay plate. Thus, this strategy for lipid phosphatase activity is an improvement over prior assays such as use of radiolabeled substrate or malachite green and is uniquely suited for HTS.
One potential limitation of the assay is enzyme-dependent detection of free inorganic phosphate opposed to chemical detection of free phosphate in the malachite green based assay. Small molecules have the potential to inhibit one or both of the reactions subsequent to Synj1 liberation of free inorganic phosphate leading to false positive hits. In order to rule out potential false positives, a counter screen without Synj1 is required to determine if small molecules which are potential hits will also inhibit either maltose phosphorylase or glucose oxidase which drive the formation of the fluorescent resorufin (Supplemental Fig. 1).
In addition to AD, Synj1 inhibitors might also have utility in Down Syndrome (DS) and other neurological diseases. Synj1 maps to human chromosome 21 and is overexpressed in trisomic models of DS, a condition characterized by severe mental retardation that is invariably associated with AD pathology in middle-aged individuals.12 Overexpression of Synj1 in transgenic mice caused a decrease in brain PI(4,5)P2 levels, which correlated with cognitive deficits.12 Therefore, small molecules able to selectively and potently inhibit Synj1 activity have implications in both AD as well as Down Syndrome. Beneficially, the phosphatase activity assay could be adapted to target other lipid phosphatases, including ORCL and Fig 4. Loss of ORCL function is implicated in oculocerebrorenal syndrome of Lowe and Dent 2 disease10 while Fig 4 is mutated in a novel form of Charcot-Marie-Tooth disorder, CMT4J.34 Fig 4 is also mutated in the sporadic and familial Amyotrophic Lateral Sclerosis.35 Unlike AD and DS, these disease targets may benefit from agonists of enzyme activity which could also be identified by this assay. Despite this apparent need, pharmacological interventions targeting phosphoinositide phosphatases are currently limited.
Selectively targeting protein tyrosine phosphatases (PTP) with high affinity has been challenging due to the high degree of structural similarity of the active sites.14 In contrast to PTP, the inositol 5-phosphatase catalytic (IPP5C) domain, common to Synj1 and 5-phosphatases, is composed of an active site His and Asp pair coordinating a cation (typically Mg2+) resembling the active site of serine/threonine-protein phosphatases.15,16 However, the catalytic domain lacks the classical CX5R(T/S) motif present in other protein and lipid phosphatases.36,37 Despite the common IPP5C shared by 5′-phosphatases, SHIP2 inhibitors display 30-fold higher affinity for SHIP2 over SHIP1 and Synj1.22 As such, the challenges of targeting protein phosphatases may not hinder discovery of selective and potent small molecules for Synj1 inhibition.
Selective and potent small molecules can be used as chemical probes to better temporally modulate cellular processes in a dose dependent manner. They are vital and invaluable tools for discerning cellular and molecular pathways underlying complex and multi-factorial phenotypes such as Synj1-mediated modulation of synaptic pathobiology relevant to AD. Development and validation of this assay is the first critical step toward identifying candidate small molecules with promise for targeting neurodegenerative diseases in a novel way by harnessing lipid phosphatases.
Supplementary Material
Acknowledgments
This research was supported by grants from NIH P50 AG080702 and the Alzheimer’s Association to L.B.J.M. and NIH R01 grants NS074536 and NS056049 to T.W.K. and G.D.P., respectively. B.C.I was supported by NIH/NRSA fellowship F31 NS054607. This publication was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant Number UL1 TR000040, formerly the National Center for Research Resources, Grant Number UL1 RR024156. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank Dr. Pietro De Camilli, Yale University, for the Synj1-FLAG construct, and Austin Cavelli for the help with the generation of the stable Synj1-overexpressing cell lines.
References
- 1.Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheng M, Sabatini BL, Südhof TC. Synapses and Alzheimer’s disease. Cold Spring Harb Perspect Biol. 2012:4. doi: 10.1101/cshperspect.a005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pozueta J, Lefort R, Shelanski ML. Synaptic changes in Alzheimer’s disease and its models. Neuroscience. 2012 doi: 10.1016/j.neuroscience.2012.05.050. in press. [DOI] [PubMed] [Google Scholar]
- 4.McIntire LB, Berman D, Myaeng J, et al. Synaptojanin1 haploinsufficiency ameliorates learning and memory impairments in a mouse model of Alzheimer’s disease. J Neurosci. 2012;32:15271–15276. doi: 10.1523/JNEUROSCI.2034-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Paolo G, Kim T-W. Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat Rev Neurosci. 2011;12:284–296. doi: 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 7.McPherson PS, Garcia EP, Slepnev VI, et al. A presynaptic inositol-5-phosphatase. Nature. 1996;379:353–357. doi: 10.1038/379353a0. [DOI] [PubMed] [Google Scholar]
- 8.Irie F, Okuno M, Pasquale EB, et al. EphrinB-EphB signalling regulates clathrin-mediated endocytosis through tyrosine phosphorylation of synaptojanin 1. Nat Cell Biol. 2005;7:501–509. doi: 10.1038/ncb1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong LW, De Camilli P. Regulation of postsynaptic AMPA responses by synaptojanin 1. Proc Natl Acad Sci U S A. 2008;105:17561–17566. doi: 10.1073/pnas.0809221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCrea HJ, De Camilli P. Mutations in phosphoinositide metabolizing enzymes and human disease. Physiology (Bethesda) 2009;24:8–16. doi: 10.1152/physiol.00035.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo S, Stolz LE, Lemrow SM, et al. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- 12.Voronov SV, Frere SG, Giovedi S, et al. Synaptojanin 1-linked phosphoinositide dyshomeostasis and cognitive deficits in mouse models of Down’s syndrome. Proc Natl Acad Sci U S A. 2008;105:9415–9420. doi: 10.1073/pnas.0803756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woscholski R, Finan PM, Radley E, et al. Synaptojanin is the major constitutively active phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase in rodent brain. J Biol Chem. 1997;272:9625–9628. doi: 10.1074/jbc.272.15.9625. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S, Zhang ZY. PTP1B as a drug target: recent developments in PTP1B inhibitor discovery. Drug Discov Today. 2007;12:373–81. doi: 10.1016/j.drudis.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Vintonyak VV, Waldmann H, Rauh D. Using small molecules to target protein phosphatases. Bioorg Med Chem. 2011;19:2145–2155. doi: 10.1016/j.bmc.2011.02.047. [DOI] [PubMed] [Google Scholar]
- 16.Vintonyak VV, Antonchick AP, Rauh D, et al. The therapeutic potential of phosphatase inhibitors. Curr Opin Chem Biol. 2009;13:272–283. doi: 10.1016/j.cbpa.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 17.Lazar DF, Saltiel AR. Lipid phosphatases as drug discovery targets for type 2 diabetes. Nat Rev Drug Discov. 2006;5:333–342. doi: 10.1038/nrd2007. [DOI] [PubMed] [Google Scholar]
- 18.Hughes TE. Emerging therapies for metabolic diseases--the focus is on diabetes and obesity. Curr Opin Chem Biol. 2009;13:332–7. doi: 10.1016/j.cbpa.2009.04.622. [DOI] [PubMed] [Google Scholar]
- 19.Johnson TO, Ermolieff J, Jirousek MR. Protein tyrosine phosphatase 1B inhibitors for diabetes. Nat Rev Drug Discov. 2002;1:696–709. doi: 10.1038/nrd895. [DOI] [PubMed] [Google Scholar]
- 20.Kurup P, Zhang Y, Venkitaramani DV, et al. The role of STEP in Alzheimer’s disease. Channels (Austin) 2010;4:347–50. doi: 10.4161/chan.4.5.12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Annis DA, Cheng CC, Chuang CC, et al. Inhibitors of the lipid phosphatase SHIP2 discovered by high-throughput affinity selection-mass spectrometry screening of combinatorial libraries. Comb Chem High Throughput Screen. 2009;12:760–771. doi: 10.2174/138620709789104870. [DOI] [PubMed] [Google Scholar]
- 22.Suwa A, Yamamoto T, Sawada A, et al. Discovery and functional characterization of a novel small molecule inhibitor of the intracellular phosphatase, SHIP2. Br J Pharmacol. 2009;158:879–887. doi: 10.1111/j.1476-5381.2009.00358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landman N, Jeong SY, Shin SY, et al. Presenilin mutations linked to familial Alzheimer’s disease cause an imbalance in phosphatidylinositol 4,5-bisphosphate metabolism. Proc Natl Acad Sci U S A. 2006;103:19524–19529. doi: 10.1073/pnas.0604954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berman DE, Dall’Armi C, Voronov SV, et al. Oligomeric amyloid-beta peptide disrupts phosphatidylinositol-4,5-bisphosphate metabolism. Nat Neurosci. 2008;11:547–554. doi: 10.1038/nn.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alldred MJ, Duff KE, Ginsberg SD. Microarray analysis of CA1 pyramidal neurons in a mouse model of tauopathy reveals progressive synaptic dysfunction. Neurobio Dis. 2012;45:751–762. doi: 10.1016/j.nbd.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller JA, Oldham MC, Geschwind DH. A systems level analysis of transcriptional changes in Alzheimer’s disease and normal aging. J Neurosci. 2008;28:1410–1420. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treusch S, Hamamichi S, Goodman JL, et al. Functional links between Aβ toxicity, endocytic trafficking, and Alzheimer’s disease risk factors in yeast. Science. 2011;334:1241–1245. doi: 10.1126/science.1213210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trovò L, Ahmed T, Callaerts-Vegh Z, et al. Low hippocampal PI(4,5)P2 contributes to reduced cognition in old mice as a result of loss of MARCKS. Nat Neurosci. 2013;16:449–455. doi: 10.1038/nn.3342. [DOI] [PubMed] [Google Scholar]
- 29.Chang-Ileto B, Frere SG, Chan RB, et al. Synaptojanin 1-mediated PI(4,5)P2 hydrolysis is modulated by membrane curvature and facilitates membrane fission. Dev Cell. 2011;20:206–218. doi: 10.1016/j.devcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chi Y, Zhou B, Wang WQ, et al. Comparative mechanistic and substrate specificity study of inositol polyphosphate 5-phosphatase Schizosaccharomyces pombe Synaptojanin and SHIP2. J Biol Chem. 2004;279:44987–44995. doi: 10.1074/jbc.M406416200. [DOI] [PubMed] [Google Scholar]
- 31.Inglese J, Johnson RL, Simeonov A, et al. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 32.Vandeput F, Combettes L, Mills SJ, et al. Biphenyl 2,3′,4,5′,6-pentakisphosphate, a novel inositol polyphosphate surrogate, modulates Ca2+ responses in rat hepatocytes. FASEB J. 2007;21:1481–1491. doi: 10.1096/fj.06-7691com. [DOI] [PubMed] [Google Scholar]
- 33.Davis MI, Sasaki AT, Shen M, et al. A homogeneous, high-throughput assay for phosphatidylinositol 5-phosphate 4-kinase with a novel, rapid substrate preparation. PLoS One. 2013;8:e54127. doi: 10.1371/journal.pone.0054127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chow CY, Landers JE, Bergren SK, et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet. 2009;84:85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chow CY, Zhang Y, Dowling JJ, et al. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsujishita Y, Guo S, Stolz LE, et al. Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase. Cell. 2001;105:379–389. doi: 10.1016/s0092-8674(01)00326-9. [DOI] [PubMed] [Google Scholar]
- 37.Whisstock JC, Wiradjaja F, Waters JE, et al. The structure and function of catalytic domains within inositol polyphosphate 5-phosphatases. IUBMB Life. 2002;53:15–23. doi: 10.1080/15216540210814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.