Abstract
Nanoparticle-based drug delivery systems have considerable potential for improvement of drug stability, bioavailability, and reduced dosing frequency. Important technological advantages of nanoparticles include high carrier capacity across biological membranes and controlled drug release. Ultimately, success of nano-delivery systems depends on toxicologic issues associated with the understanding of the fate of nanocarriers and their polymeric constituents within the targeted cells. Here we describe a method for determining subcellular distribution of nanoparticles by isolation and identification of organelles that come in direct contact with these structures.
Keywords: Macrophage, Nanoparticle, Drug delivery, Endosome, Subcellular trafficking
1 Introduction
A wide variety of nanoparticles have been developed for the cellular delivery of various therapeutic compounds and the potential clinical benefits of these particles are great (1, 2). However, very little is known about the subcellular distribution of nanoparticles in the targeted cells. This information is necessary if we are to explain how nanoparticles function on a subcellular level and to identify any potential sources of cellular toxicity. In order to accomplish this, a method must be used that can simultaneously allow for the isolation and subsequent identification of proteins that interact with a nanoparticle while it is in a cell. Here, we demonstrate that the proteins that come into contact with a nanoparticle can be individually labeled, isolated, and then identified by liquid chromatography–mass spectrometry (LC-MS/MS). This relatively simple method involves four basic steps: (1) labeling of the nanoparticles with a visible dye, (2) treatment of cells with the nanoparticles, (3) isolation of nanoparticle-laden subcellular compartments on a sucrose gradient, and (4) identification of the proteomes of subcellular compartments by LC/MS-MS. This method provides the user with a broad view of the subcellular distribution of nanoparticles within the same experiment. It is appropriate for use by researchers who do not know the fate of their nanoformulations within the targeted cells or their mechanisms of release. It can also be used successfully to identify the subcellular trafficking pathways of crystalline antiretroviral nanoparticles in human monocyte-derived macrophages (3). Alternative approaches such as immunostaining and confocal imaging of every cellular organelle and internalized nanoparticles as well as measurement of their fluorescence overlap are time consuming and costly.
2 Materials
Prepare all solutions using ultrapure water (prepared by purifying deionized water to attain a sensitivity of 18 M Ω cm at 25°C) and analytical grade reagents. Prepare and store all reagents at room temperature (unless indicated otherwise). Diligently follow all waste disposal regulations when disposing waste materials.
2.1 Components to Label Nanoparticles
Crystalline nanoparticles (see Note 1).
Coomassie Brilliant Blue R250 (CBB) (see Note 2).
Sterile 1× phosphate buffered saline (PBS).
0.5 or 1.7 mL microcentrifuge tubes.
Microcentrifuge tube tumbler rotator.
Table-top refrigerated centrifuge that can reach 20,000 × g.
Sonicator with probe.
Method to measure nanoparticle size and charge (see Note 3).
2.2 Cellular Treatment Components
Cells in vitro (see Note 4).
Cell incubator.
Serum-free DMEM (or other appropriate serum-free cell culture medium).
Labeled nanoparticles.
Sterile PBS.
2.3 Homogenization of Nanoparticle-Loaded Cells
Homogenization buffer: 100 mM sucrose, 10 mM imidazole, pH 7.4.
Dounce homogenizer (7 mL).
15 mL centrifuge tubes.
Refrigerated centrifuge.
2.4 Enrichment of Nanoparticle-Laden Compartments
Homogenization buffer: 100 mM sucrose, 10 mM imidazole, pH 7.4.
Sucrose solutions: 10, 20, 35, and 60% weight/volume, 10 mM imidazole, pH 7.4 in sterile water.
Transparent ultracentrifuge tubes (12 mL; Beckman-Coulter).
Refrigerated ultracentrifuge that can reach 100,000 × g and swinging bucket.
3 mL syringe with an 18-gauge needle.
Sterile PBS.
2.5 Sample Processing for 1D Electrophoresis and Mass Spectrometry Analysis
Lysis buffer: 30 mM Tris-Cl, 7 M urea, 2 M thiourea, 4% (w/v) 3-((3-cholamidopropyl)dimethylammonio)-1-propane-sulfonate, 20 mM dithiothreitol, 1× protease inhibitor cocktail, pH 8.5 (Sigma-Aldrich) (see Note 11).
ReadyPrep™ 2D Cleanup Kit (Bio-Rad Laboratories, Inc.).
2D Quant kit (GE Healthcare).
Bis–Tris 4–12% and 7% Tris-Glycine gels (Invitrogen).
Fixation buffer: 10% methanol and 7% acetic acid in distilled–deionized water.
Colloidal coomassie (GE healthcare).
Destaining buffer: 20% methanol and 10% acetic acid in distilled–deionized water.
Single edge razor blades.
Sterile glass autosampler vials (Thermo-Fisher Scientific).
Vacuum concentrator centrifuge (SpeedVac) with cooling trap.
In-Gel Tryptic Digestion Kit (Thermo-Fisher Scientific).
μC18 ZipTip ® pipette tips (Millipore; see Note 12).
Resuspension buffer: 0.5% trifluoroacetic acid (TFA; Sigma-Aldrich).
Wetting solution: 100% acetonitrile (ACN; Thermo-Fisher Scientific).
Equilibration/wash solution: 0.1% TFA.
Elution solution: 50% ACN, 0.1% TFA.
3 Methods
3.1 Label Nanoparticles with Coomassie Brilliant Blue R250
Combine nanoparticles (see Note 1) with 0.01% (weight/volume) of CBB (see Note 2) in sterile PBS. Mix on a micro-centrifuge tube tumbler rotator at 15 rpm for 12 h at room temperature (see Note 5).
Centrifuge the mixture at 20,000 × g for 5 min to pellet the particles and then remove the supernatant with a pipette. Add sterile PBS to the tube and briefly resuspend the particles by sonicating them at 20% amplitude for 1–3 s with a sonicator probe (see Note 6). This will remove the excess dye. Repeat the PBS wash and sonication cycle five times or until no more dye is visible in the supernatant (Fig. 1a).
Store labeled particles in sterile PBS at 4°C until ready for use.
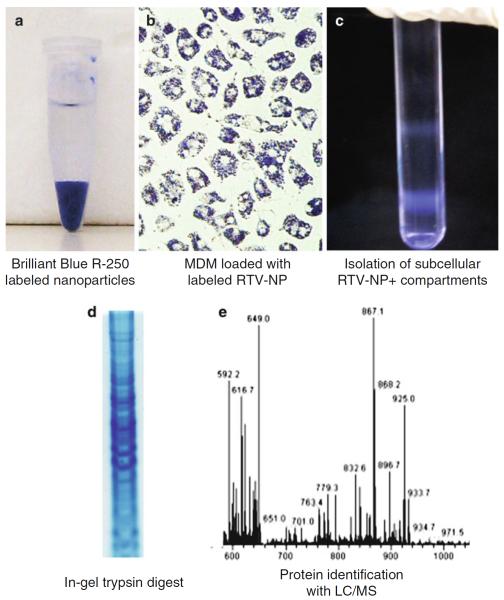
Fig. 1.
Representative images of processes for nanoparticle staining, cell treatment, endosome enrichment and protein processing and identification. Crystalline nanoparticles that have been labeled with CBB with all unbound dye washed away (a). Human monocyte-derived macrophages after being treated with CBB-labeled nanoparticles. Note the cells have developed a purple color after ingesting the labeled nanoparticles (b). Enriched endosomes containing proteins stained by CBB-labeled nanoparticles are seen as bands on a sucrose gradient after being centrifuged at 100,000 × g for 1 h at 4°C (c). One lane of a gel showing labeled proteins separated by molecular weight (d). Chromatogram of protein fractionation followed by identification using LC/MS-MS (e)
3.2 Treat Cells with CBB-Labeled Nanoparticles
Wash cells three times for 10 min with 37°C sterile PBS to remove residual serum protein.
Add nanoparticles in sterile PBS to cells at desired concentration.
Incubate at 37°C to allow cells to take up nanoparticles. The cells will visibly become blue as they take up the labeled nanoparticles (Fig. 1b) (see Note 7).
Once the cells have taken up the nanoparticles, wash the cells three times with 37°C sterile PBS to remove any residual non-internalized nanoparticles. Keep cells in PBS and immediately begin homogenization.
3.3 Homogenization of Nanoparticle-Loaded Cells
Remove PBS and add 6 mL of homogenization buffer to each flask if working in a T75 culture flask. Adjust the buffer volume to the minimum necessary for covering the dish surface if working with other culture systems.
Detach cells from bottom of flask using a cell lifter.
Add entire volume to Dounce homogenizer and grind cells with 15 strokes (see Note 8).
Add entire volume to a 15 mL centrifuge tube and remove cellular debris and nuclei by centrifuging at 500 × g for 10 min at 4°C.
Remove supernatant and centrifuge for 1 h at 100,000 × g at 4°C. Resuspend the pellet in 3 mL of 10% sucrose, 10 mM imidazole, pH 7.4, solution and store on ice.
3.4 Enrichment of Nanoparticle-Laden Compartments
Take sucrose solutions and set up sucrose gradient in 12 mL thin-walled ultracentrifuge tube. Take 3 mL of 60% sucrose solution and place it in the bottom of the tube followed by layering 3 mL each of 35 and 20% sucrose solutions one on top of the next in the order given (see Note 9).
Carefully add the supernatant from the last step of Subheading 3.3 to the top of the sucrose gradient and centrifuge at 100,000 × g at 4°C for 1 h.
Using a 3 mL syringe with an 18-gauge needle, carefully perforate the tube at the end of the blue sucrose band and aspirate the solution until the color disappears (Fig. 1c). Transfer each band to a separate ultracentrifuge tube (see Note 10).
Pellet the nanoparticle-enriched subcellular compartments by centrifuging at 100,000 × g at 4°C for 1 h. Remove supernatant and wash pellet with PBS and subsequent centrifugation at 100,000 × g at 4°C for 1 h to remove residual sucrose.
3.5 Sample Processing for 1D Electrophoresis and Mass Spectrometry Analysis
Solubilize enriched subcellular compartments in lysis buffer by resuspending the pellet and pipetting five times (see Note 11).
Precipitate proteins using a ReadyPrep™ 2D Cleanup Kit (GE Healthcare) per manufacturer's instructions.
Quantify protein using a 2D Quant kit (GE Healthcare) per the manufacturer's instructions.
Run samples on Bis–Tris 4–12% and 7% Tris-Glycine gels (Invitrogen) to separate low and high molecular weight proteins.
Incubate gels in fixation buffer for 1 h at room temperature followed by staining with colloidal coomassie for 24 h at room temperature (Fig. 1d).
Destain gels by rinsing with destaining solution until solution is light blue or clear.
Manually excise the bands using a razor blade and place each band in a separate glass autosampler vial. Excise each gel band in several pieces to increase surface contact (see Note 12).
Perform in-gel tryptic digestion using the In-Gel Tryptic Digestion Kit per the manufacturer's instructions.
3.6 Peptide Purification and Concentration for Mass Spectrometry Analysis and Protein Identification
Resuspend the extracted peptides from the in-gel tryptic digestion procedure in 30 μL of resuspension buffer and vortex vigorously for 5 min.
Wet Zip-Tip pipette tip by aspirating and releasing wetting solution three times.
Equilibrate tip by pipetting and discarding equilibration/wash solution three times.
Bind peptides to the zip tip by pipetting 15 times inside the sample tube then discard sample.
Wash tip three times (and discard fluid) in equilibration/wash solution.
Elute the peptides into a vial insert by pipetting 10 μL at a time of the elution solution (100 μL). Keep vials on ice until all samples are zip tipped.
Freeze samples brie fly at −80°C then SpeedVac to dryness.
Resuspend peptides with the appropriate volume of 0.1% formic acid and analyze by LC-MS/MS (see Note 13).
4 Notes
This protocol has been used only for crystalline antiretroviral nanoparticles coated in lipophilic surfactants such as poloxamer-188 (P188), 1,2-distearoyl-phosphatidyl ethanolaminemethyl-polyethyleneglycol-2000 (mPEG2000-DSPE), and 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) (3). We suggest using rigid nanoparticles that have a lipophilic coating because the dye will easily label the particle without disrupting its structure. If other types of nanocarriers are used, it is highly encouraged that the nanoparticles are re-characterized after labeling to ensure that they have not been altered.
This protocol has been used only with Brilliant Blue R250. However, the goal is to coat the particles with a visible dye that will label the proteins that the nanoparticles come into contact with. Thus, other dyes that label proteins and are readily seen in the visible light spectrum, such as bromophenol blue, could also be used. It is unknown how using Brilliant Blue R250 (or other such dyes) to label different types of nanoparticles will affect their physical properties. Thus, it is highly encouraged that the nanoparticles be re-characterized after labeling to ensure that they have not been altered.
There are a number of methods for measuring nanoparticle size and charge. Companies such as Horiba, Malvern, TSI, and many others offer equipment that will simultaneously measure both the size and charge of nanoparticles. No one method or machine is preferred for this protocol.
This protocol has been used primarily with primary human monocytes and human monocyte-derived macrophages. However, any cells that will take up the nanoparticle being tested could be used. Adjust the protocol appropriately to accommodate for the cell type being used. It is suggested to use at least 100 × 106 cells in order to purify enough protein for proteomic analysis.
Typically the labeling procedure can be carried out in a 0.5 mL microcentrifuge tube. When using this small of a volume, using just a few grains (about 1–3 grains) of Brilliant Blue R250 dye will be enough to sufficiently label the particles without altering their physical characteristics. If too much dye is used, there is a risk of nanoparticle aggregation. It does not take much dye to label the particles so use less dye rather than more.
The purpose of sonication is to resuspend the particles so that they can be efficiently washed. However, it is possible to over-heat or dissolve the particles with too much sonication. Therefore, a brief sonication (10 s) to resuspend the particles is all that is necessary.
The investigators will need to adjust the cell-nanoparticle exposure time based on endocytic activity of the targeted cells and the size and coating of their nanoformulations. It is suggested to allow for maximal nanoparticle uptake since this will allow for a better identification of nanoparticle-laden compartments.
When using the Dounce homogenizer, press and pull the piston hard enough to keep the solution flowing past at a steady rate. Excess force is not necessary and could break the homogenizer.
When forming the sucrose gradient, carefully add each subsequent layer of sucrose by gently pouring it down the side of the tube. This will help to prevent mixing of the layers and will increase the intensity of the protein bands that form during centrifugation.
Do not disturb the contents of the ultracentrifuge tube when removing it from the centrifuge. The fractions need to be collected within 15 min after centrifugation; otherwise the bands will diffuse. When collecting the fractions (2–4 blue bands representing nanoparticle-laden compartments), use separate needles and syringes for each band. Start from the top band. Insert the needle-syringe at the bottom of band line facing up and aspirate until very little blue is left. Do not remove the syringe after band aspiration. Insert a new needle-syringe at the level of the next lower band and repeat.
Use nitrile gloves when handling the gel and the in-gel tryptic digestion solutions. Latex can interfere with downstream mass spectrometry analysis. Use a clean surface and equipment and avoid contact of gloves with skin or hair during the processing of gel bands. Dust and shedding epithelial cells can be a major contaminant of the samples, and it compromises the mass spectrometry analysis.
HPLC-grade reagents and HPLC-grade water should be used to make solutions used for in-gel tryptic digestion and peptide extraction (zip-tipping).
The volume necessary for resuspension of samples depends on the method used for mass spectrometry analysis and the instrument configuration (nanospray or electrospray). Typically, peptides from in-gel tryptic digestion are resuspended in 4–8 μL of 0.1% formic acid and are run in a nanospray configuration.
Acknowledgments
The work was supported by the National Institutes of Health grants 1P01 DA028555, 2R01 NS034239, 2R37 NS36126, P01 NS31492, P20RR 15635, P01MH64570, and P01 NS43985 (to H.E.G.) and a research grant from Baxter Healthcare. The authors thank Ms. Robin Taylor for critical reading of the manuscript and outstanding graphic and literary support.
References
- 1.Nowacek A, Gendelman HE. NanoART, neuroAIDS and CNS drug delivery. Nanomedicine (Lond) 2009;4:557–574. doi: 10.2217/nnm.09.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nowacek A, Kosloski LM, Gendelman HE. Neurodegenerative disorders and nanoformulated drug development. Nanomedicine (Lond) 2009;4:541–555. doi: 10.2217/nnm.09.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kadiu I, Nowacek A, McMillan J, Gendelman HE. Macrophage endocytic trafficking of anti-retroviral nanoparticles. Nanomedicine (Lond) 2011;6:975–994. doi: 10.2217/nnm.11.27. [DOI] [PMC free article] [PubMed] [Google Scholar]