Abstract
The hammerhead ribozyme has long been considered a prototype for understanding RNA catalysis, but discrepancies between the earlier crystal structures of a minimal hammerhead self-cleaving motif and various biochemical investigations frustrated attempt to understand hammerhead ribozyme catalysis in terms of structure. With the discovery that a tertiary contact distal from the ribozyme’s active site greatly enhances its catalytic prowess, and the emergence of new corresponding crystal structures of full-length hammerhead ribozymes, a unified understanding of catalysis in terms of the structure is now possible. A mechanism in which the invariant residue G12 functions as a general base, and the 2′-OH moiety of the invariant G8, itself forming a tertiary base pair with the invariant C3, is the general acid, appears consistent with both the crystal structure and biochemical experimental results. Originally discovered in the context of plant satellite RNA viruses, the hammerhead more recently has been found embedded in the 3′-untranslated region of mature mammalian mRNAs, suggesting additional biological roles in genetic regulation.
1. DISCOVERY AND CONTEXT
The fundamental importance of RNA to all of biology has become increasingly clear within the last 25 years or so. It is within this time frame that catalytic RNAs were discovered and first characterized, beginning with the Group I Intron1 and RNase P,2 closely followed by the hammerhead ribozyme,3 which was the third catalytic RNA to be discovered.
The discovery that RNA in some cases can be an enzyme not only forces us to reexamine our notions of biological catalysis, but suggests that by understanding ribozyme chemistry, we might learn more about how life may have originated from an “RNA World” inhabited by self-replicating ribozymes. The finding that the ribosome is indeed a ribozyme4,5 underscores the relevance of RNA catalysis in today’s protein-dominated world. The more recent discoveries of RNA interference and micro-RNAassociated mechanisms of gene regulation further emphasize the central importance of RNA to understanding gene regulation as well as development of new RNA-based technologies for gene manipulation and silencing.6 The discovery that riboswitches7 and in some cases ribozymes,8 including a variant of the hammerhead ribozyme,9,10 are also involved in regulating gene expression illustrates how intimately RNA structure, function, and catalysis are involved in many aspects of biological control. Quite possibly, the most revolutionary discoveries in RNA molecular biology have yet to be made.
1.1. Biological context
The hammerhead RNA, unlike RNase P, is not a true enzyme in its natural biological context, in that it is a single-folded strand of RNA that undergoes autocatalytic self-cleavage. A trivial modification, separating the hammerhead RNA into an enzyme strand and a substrate strand by removing a nonessential connecting loop, creates a true catalyst capable of multiple turnover. Hence the terms “hammerhead RNA” and “hammerhead ribozyme” tend to be used interchangeably.
The hammerhead RNA was first discovered in the satellite RNA of tobacco ringspot virus,3 a 371 nucleotide single-stranded covalently closed circular genome that is parasitic upon the tobacco ringspot virus and is replicated via a rolling-circle mechanism. Linear concatameric complementary copies of the satellite RNA genome cleave themselves into monomeric fragments; the cleavage points occur highly specifically at regular intervals and are embedded within hammerhead RNA motifs. Hammerhead RNAs have subsequently been discovered in several other RNA sequences involved in rolling-circle replication.11 Most are plant virus or virus-like genomes, but a few occur as transcripts of repetitive DNA in the animal kingdom as well.
1.2. A prototype ribozyme
The hammerhead ribozyme in many respects is a model or prototype ribozyme in the same sense that RNase A, lysozyme, and the serine protease family have served enzymology as prototype protein enzymes for many years. The hammerhead, being a comparatively simple and wellcharacterized ribozyme, is quite possibly the most intensively studied ribozyme, both from the point of view of mechanistic biochemical characterizations and structural investigations.12 After the discovery of RNase P and the Group I intron ribozymes, both of which are comparatively large and complex catalytic RNAs, the discovery of the hammerhead ribozyme offered the first hope that the phenomenon of RNA catalysis might be best understood within the framework of a smaller, more tractable RNA that catalyzes a simple phosphodiester isomerization reaction. The first ribozyme crystal structures were, in fact, those of minimal hammerhead ribozymes,13–15 but they seemed to create more questions than compelling explanations for RNA catalysis.16,17
Within the past 5 years, it has become apparent that acid–base catalysis and electrostatic transition-state stabilization are universal catalytic strategies of such fundamental importance that they appear in all of the structurally characterized small ribozymes and protein enzymes such as RNase A that catalyze RNA reactions.18 Yet, each ribozyme appears to have evolved a unique and different strategy to achieve similar goals. Hence, the need to explain ribozyme catalysis in terms of a unified mechanistic understanding has become even more compelling. The potential relevance of ribozyme catalysis to gene regulation and to the origin of life each further underscores the fundamental importance of the problem.
2. THE MINIMAL SEQUENCE
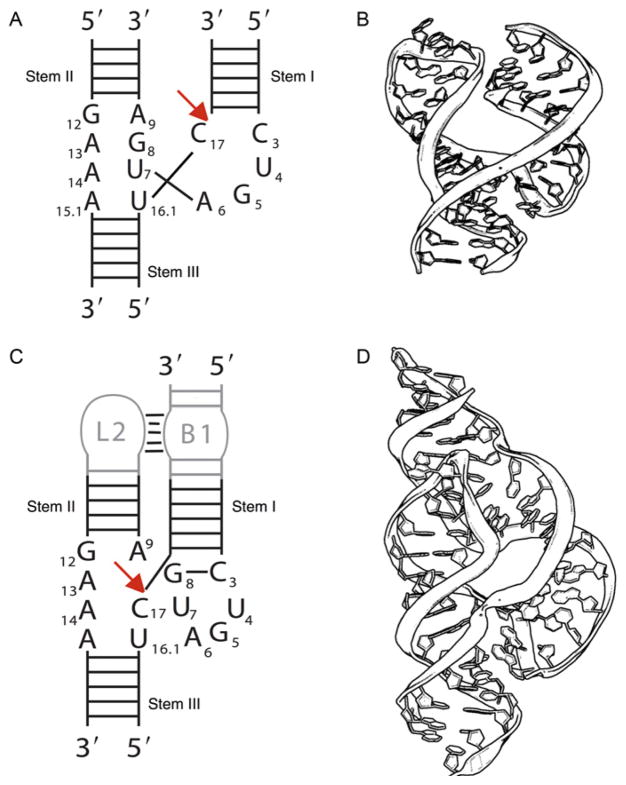
The minimal hammerhead ribozyme (Fig. 1.1A and B) consists of a core region of 15 conserved (mostly invariant) nucleotides flanked by three helical stems. In 2003, it finally became clear that optimal activity required the presence of a tertiary interaction between stem(s) I and II. Although there is little apparent sequence variation, the contact appears to be present in most if not all hammerhead sequences. Although the minimal hammerhead has a turnover rate of approximately 1 min−1, full-length sequences that include a tertiary contact, described in Section 3, are up to 1000-fold more active.19–21
Figure 1.1.
The minimal and full-length hammerhead ribozymes. (A) A schematic representation of the secondary structure of the minimal hammerhead ribozyme. (B) The crystal structure of a corresponding minimal hammerhead ribozyme. The longer strand is the enzyme and the shorter strand is the substrate. (C) A schematic representation of the full-length hammerhead ribozyme emphasizing the presence of a tertiary contact between stem(s) I and II. (D) The crystal structure of a corresponding full-length hammerhead ribozyme. Again, the longer strand is the enzyme and the shorter strand is the substrate.
2.1. Enzymology
2.1.1 Rate enhancement
The rate of non-site-specific, spontaneous decay of RNA is highly dependent upon the secondary structural context, but is on average about 10−6 min−1.22 Hence, the rate enhancement provided by an optimized minimal hammerhead is on the order of 106, and for the full-length natural hammerhead can be as much as 109. To achieve this magnitude of rate enhancement, not to mention site specificity, the hammerhead ribozyme must adopt several effective catalytic strategies simultaneously. Each of these is separated perhaps somewhat artificially and analyzed below.
2.1.2 Metal ions and catalysis
Originally, it was believed that all ribozymes, including the hammerhead ribozyme, were obligate metalloenzymes.23,24 Mg2+ ion is assumed to be the biologically relevant divalent cation, although the hammerhead is active in the presence of a variety of divalent cations.25 Proposed roles for Mg2+ ion in catalysis included both acid and base catalysis components24,26 (with Brønsted and Lewis variants of this proposal articulated) as well as direct coordination of the pro-R nonbridging phosphate oxygen of the scissile phosphate for transition-state stabilization. Mg2+ ion has also been implicated in structural roles that facilitate formation of the active ribozyme.27–35
In 1998, it was demonstrated that the hammerhead, along with the hairpin and VS ribozymes (but not the HDV ribozyme) could also function in the absence of divalent metal ions as long as a high enough concentration of positive charge was present (molar quantities of Li+, Na+, or even the nonmetallic NH4+ ion permit cleavage to take place). The study suggested that ribozymes were not strictly metalloenzymes.36,37
Considering the volume of research devoted to understanding the mechanistic roles of divalent metal ions in hammerhead ribozyme catalysis and the fundamental tenet of ribozyme enzymology that all ribozymes are metalloenzymes, it was unexpected to find that at least three of the four small naturally occurring ribozymes can function reasonably efficiently in the absence of divalent metal ions; a fact that was discovered in the course of performing experimental controls for time-resolved crystallographic freezetrapping experiments in crystals of the minimal hammerhead ribozyme.36–38 It now appears that RNA folding and nonspecific electrostatic transitionstate stabilization accounts for much, if not all, of the catalytic enhancement over background rates found with these ribozymes.36,37 For example, hammerhead 16.1, which is considered to be an optimized hammerhead ribozyme sequence for single-turnover reactions, cleaves only threefold faster in the presence of 10 mM MgCl2 and 2 M Li2SO4 than it does in the presence of 2 M Li2SO4 alone.36,37 The rates of hairpin and VS ribozymes in 2 M Li2SO4 actually exceed those measured under “standard” low ionic strength conditions, and the rate of cleavage for the non-optimized hammerhead sequence used for crystallization is fivefold enhanced in 2 M Li2SO4 alone versus standard reaction conditions. The non-optimized sequence used for crystallization tends to form alternative, inactive structures in solution, such as a dimer of the enzyme strands, which dominate at lower ionic strength.
This result implied that any chemical role of Mg2+ ion in the ribozyme reaction was likely to be one of comparatively nonspecific electrostatic stabilization rather than more direct participation in the chemical step of catalysis. Moreover, the result implied that the RNA itself was an active participant in the chemistry of catalysis rather than serving as a passive scaffold for binding metal ions that served the roles of general acid and base catalysts. With the subsequent structural elucidation of the hairpin39,40 and full-length hammerhead41 structures, it was, in fact, revealed that RNA bases and other functional groups were positioned to provide the moieties likely responsible for acid–base catalysis.
2.1.3 Acid–base chemistry
Originally, hydrated Mg2+ and other hydrated divalent metal ions were thought to play the direct chemical role of general base and general acid in ribozyme catalysis, with the RNA itself serving as an ancillary and passive scaffold upon which metal ions would bind and would be positioned in the active site.
With the discovery that the hairpin, hammerhead, and VS ribozymes were not strictly metalloenzymes,36,37 it became apparent that in at least these three cases, the RNA itself must be an active participant in the chemistry of catalysis rather than serving merely as a metal ion-binding scaffold. The crystal structure of the hairpin ribozyme,40 in contrast to the HDV ribozyme42,43 that is in fact a metalloenzyme, soon validated this prediction. However, it was not apparent from the crystal structure of the minimal hammerhead13–15,44 what functional groups might be involved in acid–base catalysis. Consequently, the focus of biochemical mechanistic investigations in the hammerhead turned to this problem.
The invariant core residues G12 and G8 in the hammerhead ribozyme were finally identified in 2005 as likely candidates for participation in acid–base chemistry by careful purine modification studies conducted by John Burke and coworkers.45,46 Substitution of G12 (pKa 9.5) with inosine (pKa 8.7), 2,6-diaminopurine (pKa 5.1) or 2-aminopurine (pKa 3.8) shifts the reaction rate profile in a manner consistent with G12’s suggested role in general base (or acid) catalysis without significantly perturbing ribozyme folding.45 Similar substitutions at G8 also implicated this invariant residue in acid–base catalysis, but in this case, the modifications also partially inhibited ribozyme folding.45 These experiments could not determine specifically whether an individual nucleotide, such as G12, was the general acid or the general base, but clearly implicated G12 and G8 in acid–base catalysis.
2.1.4 Kinetics
The minimal hammerhead ribozyme, under “standard” reaction conditions (10 mM Tris, pH 7.5, 10 mM MgCl2) has a turnover rate on the order of 1 min−1, a Km of about 10 mm, and a log-linear dependence of rate on pH with a slope of 0.7. Above pH 8.5–9.0 (depending upon reaction conditions), the rate becomes pH independent, suggesting an apparent kinetic pKa of about 8.5–9.0.25,47,48 This observation is consistent with both Mg2+ and guanine-mediated acid–base chemistry. The full-length hammerhead ribozyme shows similar pH dependence, but the cleavage rate is up to 1000-fold enhanced (i.e., approximately 15 s−1).49 There exists no compelling evidence that the reaction is sequential rather than concerted, although this remains an issue for debate. It is perplexing that the pH dependence of the rate-limiting step is similar in both the minimal and full-length ribozymes, despite the remarkable reaction rate difference.
2.2. Crystal structure
The crystal structure of a minimal hammerhead ribozyme (Fig. 1.1B) was the first near-atomic resolution structure of a ribozyme to be determined.13–15 However, the minimal hammerhead ribozyme sequence crystallizes in what is now recognized as an “open,” apparently precatalytic conformation50,51 in which four of the invariant residues (C3, U4, G5, and A6) form a uridine turn structure13,52 similar to that found in the anticodon loop of tRNA, and the remaining conserved residues augment or extend stem II via stacked sheered GA pairs.14,15 Together, these residues form a three-strand junction, in which the augmented stem II stacks upon stem II, and stem(s) I branches out via the uridine turn and the cleavage-site nucleotide.
The first hammerhead ribozyme structure, solved by McKay and coworkers in 1994,13 was that of a minimal hammerhead RNA enzyme strand bound to a DNA substrate-analogue inhibitor, and in 1995 a different all-RNA hammerhead construct having a 2′-OMe inhibitory substitution of the nucleophilic 2′-OH of C17 appeared.14,15 Subsequently, structures of minimal hammerheads without modified nucleophiles appeared in various precatalytic conformational states,44 and finally a structure of the cleavage product appeared53 in 2000, providing the opportunity to construct the first “molecular movie” of ribozyme catalysis.
2.3. Experimental discord
It was immediately apparent from the first hammerhead crystal structures13 that a conformational change would need to take place to position the attacking nucleophile in line for activation of the cleavage reaction. The requirement for this conformational change motivated subsequent crystallographic freeze-trapping experiments.36,37
Meanwhile, a growing list of discrepancies between the minimal hammerhead ribozyme structure and mechanistic biochemical experiments designed to probe transition-state interactions began to accumulate.17 The observed hydrogen-bonding patterns within the minimal hammerhead crystal structures could not explain the immutability of G8, G12, G5, C3, and a number of other core residues.16 Even more concerning was evidence that the phosphate of A9 and the scissile phosphate, separated by 18 Å in the minimal hammerhead crystal structures, might bind a single metal ion in the transition state of the reaction.54 Such an interaction would require the two phosphates to approach each other within about 4.4 Å, but this requirement could be demonstrated to be incompatible with the minimal hammerhead crystal structure unless significant unwinding or base-unpairing were to take place in one or more of the helices.55
3. THE FULL-LENGTH SEQUENCE
When the hammerhead RNA was first discovered, it was observed to be embedded within an $370 nucleotide single-stranded genomic satellite RNA, most of which could be deleted while preserving the RNA’s catalytic properties.3 Eventually, it was found that about 13 core nucleotides and a minimal number of flanking helical nucleotides were all that was required for a respectable catalytic turnover rate of 1 to 10 min−1, and this “minimal” hammerhead construct became the focus of attention.56,57 It thus came as a great surprise to most in the field when in 2003 it was finally discovered that for optimal activity the hammerhead ribozyme in actuality requires the presence of sequences in stem(s) I and II. These sequences interact to form tertiary contacts (Fig. 1.1C), but were removed in the process of eliminating seemingly superfluous sequences from the hammerhead ribozyme; the standard reductionist approach often employed in molecular biology.58 Once the full ramifications of this revelation became apparent, that is, that the entire field had been studying the residual catalytic activity of an overzealously truncated version of the full-length ribozyme, attention shifted away from the minimal constructs. It also quickly became apparent that a crystal structure of the full-length hammerhead ribozyme, in which these distal tertiary contacts were present, might be of considerable interest.
3.1. Biological context
Apparently, all naturally occurring, biologically active hammerhead RNA sequences possess a tertiary contact that enhances their ability to fold into a catalytically competent structure. That this was always overlooked is testimony to the lack of any clear sequence conservation pattern.
3.2. Enzymology
Many of the biochemical experiments designed to probe the nature of catalysis in the minimal hammerhead ribozyme structure attempted to measure the effects of structural alterations upon the rate-limiting step (presumed to be the chemical step) of the self-cleavage reaction. In general, the observations made in the context of the minimal hammerhead ribozyme are also relevant to the full-length hammerhead.50,51 The most straightforward explanation of this fact is that both the minimal and full-length hammerhead structures are believed to pass through what is essentially the same transition state.50,51 The full-length hammerhead is thus believed to accelerate the self-cleavage reaction primarily by stabilizing the precatalytic structure in a manner that is unavailable to the minimal hammerhead due to a lack of the tertiary contact between stem(s) I and II.
The hammerhead ribozyme sequence derived from Schistosoma Sma1 is arguably the most extensively characterized of full-length hammerhead sequences. The cleavage kinetics and internal equilibrium have been thoroughly investigated, revealing significant surprises. The apparent cleavage rate at pH 8.5 in 200 mM Mg2+ is at least 870 min−1, which in actuality is a lower bound as there is also a significant rate of ligation under these conditions. In contrast to minimal hammerheads that show a log-linear dependence of rate on pH up to about pH 8.5, the Sma1 hammerhead has a lower apparent pKa that is dependent upon Mg2+ concentration. At 100 mM Mg2+, the apparent pKa is about 7.5–8.49 The Sma1 hammerhead is also a rather efficient ligase,59 revealing internal equilibrium constants (Kint1/4[EP]/[ES]) as small as 0.5 in the presence of high concentrations of Mg2+, and as small as 1.3 under physiological concentrations of Mg2+. Cleavage and ligation reaction rates are also highly dependent upon the identity of the divalent cation present, with Mn2+ accelerating the reaction almost two orders of magnitude relative to Mg2+. This suggests that the ability to coordinate soft ligands (perhaps including the N7 of G10.1) optimizes catalysis, whereas simply folding the RNA is only weakly dependent upon the identity of the divalent cation present.60
3.3. Crystal structure
The full-length hammerhead structure (Fig. 1.1D) reveals how tertiary interactions occurring remotely from the active site prime the ribozyme for catalysis. G12 and G8, two invariant residues previously identified in biochemical studies to be potential acid–base catalysts, are in fact positioned in a way that is consistent with their suggested roles. In contrast to the minimal hammerhead structure, the nucleophile in the full-length structure is aligned with the scissile phosphate which in turn is positioned proximal to the A9 phosphate, and previously unexplained roles of other conserved nucleotides become apparent within the context of a distinctly new fold that nonetheless accommodates the previous structural studies. These interactions allow us to explain the previously irreconcilable sets of experimental results in a unified, consistent, and unambiguous manner.41
Figure 1.2A is a close-up of the active site. The light blue dotted lines are conventional hydrogen-bonding interactions. The other dotted lines represent interactions that may be relevant to the catalytic mechanism. The structure includes an introduced modification, a 2′-OMeC at the cleavage site, to prevent abstraction of the 2′-H from the nucleophilic oxygen. G12 is positioned in a manner consistent with a role as the general base in the reaction. A transiently deprotonated G12 might then be able to abstract a proton from the 2′-OH, generating the required attacking nucleophile for the cleavage reaction. The 2′-O is pre-positioned for in-line attack, and a second hydrogen-bonding interaction between the 2′OH of G8 and the leaving group 5′-O of C1.1 may represent a general acid catalytic mechanism. The invariant G8 forms a Watson–Crick base pair with C3, another invariant residue. Mutation of either one of these abrogates ribozyme activity completely, but a double mutation (i.e., C8/G3) that restores the base pair restores activity to the hammerhead ribozyme. Thus, it appears that the ribose of G8 rather than the nucleobase provides the relevant acidic moiety for catalysis, although other factors are no doubt involved.
Figure 1.2.
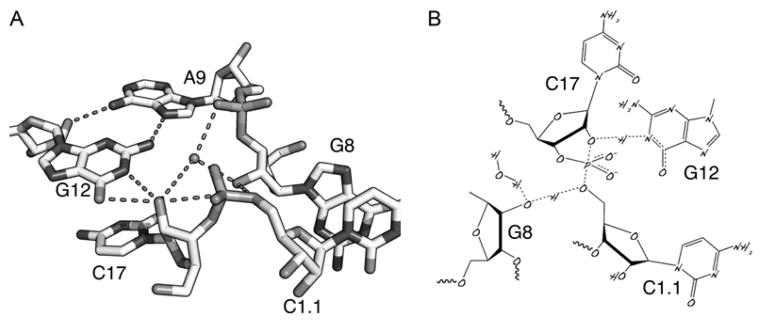
The active site of the full-length hammerhead ribozyme permits a mechanism to be proposed. (A) Closeup of the crystal structure of the full-length hammerhead ribozyme showing G12 positioned for general base catalysis, the 2′-OH of G8 poised for acid catalysis, and the attacking nucleophile, the 2′-O of C17, positioned for an in-line attack upon the adjacent scissile phosphate of C1.1. A9 helps to position G12 and may also engage in transition-state stabilization of the pentacoordinate oxyphosphorane transition state. (B) A mechanistic diagram illustrating partial proton dissociation and transfer in a putative transition state.
3.4. Resolution of experimental discord
Many of the biochemical experiments designed to probe transition-state interactions and the chemistry of catalysis appeared to be irreconcilable with the minimal hammerhead crystal structures. For example, the invariant core residues G5, G8, G12, and C3 in the minimal hammerhead ribozyme were each observed to be so fragile that changing even a single exocyclic functional group on any one of these nucleotides resulted in abolition of catalytic activity, yet few of these residues appeared to form hydrogen bonds involving the Watson–Crick faces of the nucleobases. A particularly striking and only recently observed example consisted of G8 and G12, which had been identified as possible participants in acid–base catalysis. After we demonstrated that the hammerhead ribozyme does not require divalent metal ions for catalysis, it gradually became apparent that the RNA itself, rather than passively bound divalent metal ions, must play a direct chemical role in any acid–base chemistry within the hammerhead active site. However, it was completely unclear how G12 and G8 could accomplish this, given the original structures of the minimal hammerhead ribozyme. In addition, the attacking nucleophile in the original structures, that is, the 2′-OH of C17, was not in a position amenable to in-line attack upon the adjacent scissile phosphate.16 Perhaps most worrisome were experiments that suggested the A9 and scissile phosphates must come within about 4 Å of one another in the transition state based upon double phosphorothioate substitution and soft metal ion rescue experiments.54 The distance between these phosphates in the crystal structure was about 18 Å, with no clear mechanism for close approach if the stem IIand stem(s) I A-form helices were treated as rigid bodies. Taken together, these results appeared to suggest that a fairly large-scale conformational change must take place to reach the transition state within the minimal hammerhead ribozyme structure. For these reasons, results from the two sets of experiments (biochemical vs. crystallographic) appeared not only to be at odds, but completely and hopelessly irreconcilable, and they generated a substantial amount of discord in the field. No compelling evidence for dismissing either set of experimental results was ever successfully made, although some claims to the contrary were made in favor of each.
The resolution of this vexing conundrum came only with the crystal structure of the full-length hammerhead ribozyme in which C17 is positioned for in-line attack, and the invariant residues C3, G5, G8, and G12 all appear involved in vital interactions relevant to catalysis. Moreover, the A9 and scissile phosphates are observed to be 4.3 Å apart, which is consistent with the idea that these phosphates when modified could bind a single thiophilic metal ion. The structure also reveals how two invariant residues, G12 and G8, are positioned within the active site in a manner consistent with their previously proposed roles in acid–base catalysis. G12 is within hydrogen-bonding distance to the 2′-O of C17, the nucleophile in the cleavage reaction, and the ribose of G8 hydrogen bonds to the leaving group 5′-O, while the nucleobase of G8 forms a Watson–Crick pair with the invariant C3. The crystal structure of the full-length hammerhead ribozyme thus clearly addressed the major concerns that appeared irreconcilable with earlier minimal hammerhead structures.
3.5. Mechanistic proposals
Based upon the arrangement of invariant nucleotides in the hammerhead active site, as well as the solvent structure in a combined crystallographic and molecular dynamics investigation, we have formulated a testable hypothesis for how the chemical mechanism of cleavage works. Our proposal is that a specifically bound water molecule accepts a proton from G12. G12 must ionize to function as the general base, and the proton is replaced by that from the 2′-OH of C17. The original G12 proton can then be relayed directly to the 2′-OH of G8 to replace a proton that must be donated to the 5′-O leaving group of C1.1 as the phosphodiester backbone is cleaved. This mechanism (Fig. 1.2B) conserves the number of protons during the phosphodiester isomerization. It is testable in that it predicts that altering the pKa of either the purine base at position 12 or the 2′-OH at position 8 will alter the cleavage rate without inducing gross structural perturbations. There are also opportunities for transition-state stabilization of the accumulating negative charges in the pentacoordinate oxyphosphorane. We propose that either the exocyclic amine of A9 or a divalent cation can perform this function.
The roles of G12 and G8 in general base and general acid catalysis, respectively, have been examined using chemical modification strategies in a hammerhead RNA sequence closely resembling that of the crystal structure.
To test the hypothesis that G12 is the general base, an affinity label was synthesized to identify the relevant functionality. The full-length hammerhead ribozyme was titrated with a substrate analogue possessing a 2′bromoacetamide group at C17. The electrophilic 2′-bromoacetamide group alkylated the general base, which was then identified as N1 of G12 by footprinting analysis. In addition, the experiment provided evidence that the pKa of G12 is perturbed downward to about 8.5 in the context of the hammerhead active site structure relative to unstructured RNA.61
To test the hypothesis that the 2′-OH of G8 participates in general acid catalysis, either by itself or accompanied by a divalent metal ion, a bridging phosphorothioate substrate analogue, in which the leaving group oxygen atom is replaced by a sulfur atom, was synthesized and characterized in a full-length hammerhead ribozyme self-cleavage reaction.62 Cleavage of the unmodified substrate, unlike the modified leaving group, was inhibited by modification of the G8 2′-OH, and evidence for involvement of a divalent metal ion assisting in pKa perturbation of the general acid was also obtained. Hence, it appears that the functional groups identified in the crystal structure as the main participants in acid–base catalysis indeed do so.
4. HAMMERHEAD RIBOZYMES IN MAMMALIAN GENE REGULATION
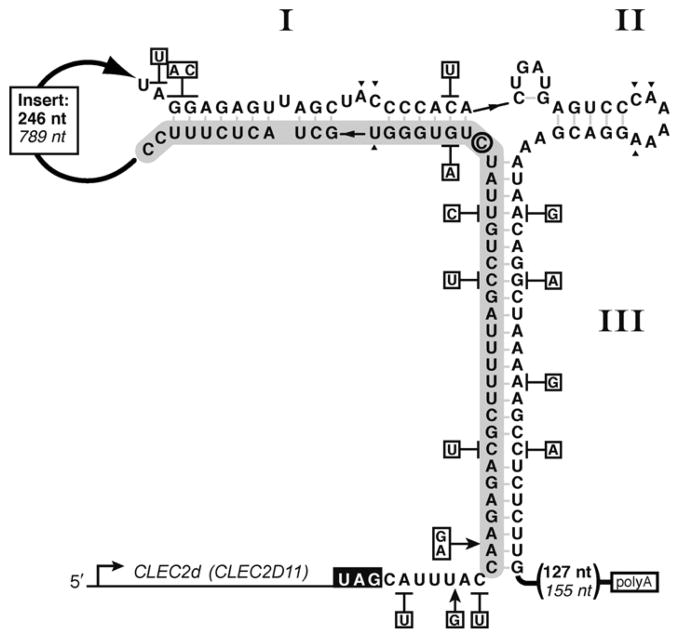
Discovery of conserved mammalian hammerhead ribozymes suggests that the hammerhead motif’s biological role extends beyond processing of satellite RNA and viroid replication products, and into the dominion of cellular functions.9,10 Uncovered by bioinformatic searches of available genomes, the new class of hammerhead ribozymes is found in 3′ untranslated regions (UTRs) of several mammalian C-type lectin type II (CLEC2) genes.63 The formation of active hammerhead ribozymes between the stop codon and the polyadenylation (polyA) signal sequence (Fig. 1.3) leads to cis-cleavage of the 3′ UTR and reduction of associated gene expression.9,10 Significantly, these sequences represent full-length ribozymes including tertiary interactions necessary for physiologically relevant catalytic rates. Here, we provide an overview of the mammalian mRNA-associated CLEC2 ribozymes.
Figure 1.3.
Sequence arrangement and secondary structure model of rodent CLEC2d-associated hammerhead ribozymes. Secondary structure of the mouse ribozyme sequence is shown. The rat ribozyme single nucleotide- and base pair-differences are indicated in boxes adjacent to the mouse sequence. The stop codon is denoted in white. The “substrate” sequence is shown on a gray background. The insertion sequence separating the two ribozyme segments is abridged with a thick arrow, and helices are identified by roman numerals. Rat insertion length and distance to polyA site are in italics. The predicted cleavage site is 3′ to the active site cytosine (circled).
To date, 12 CLEC2 hammerhead ribozymes have been identified in 9 mammalian species (Fig. 1.4). Two structures are found in mouse, three in rat, and one in each of the following mammalian genomes: tree shrew, hedgehog, horse, elephant, cow, dog, and platypus.9,10 All 12 are located immediately downstream of genomic sequences that share varying degrees of homology with the CLEC2 gene family. Two hammerhead ribozymes in mouse CLEC genes (mCLEC2dand mCLEC2e) and one in rat (rCLEC2D11) reside within the 3′ UTRs of known protein coding genes. The incomplete proteome annotation of the other seven species prevents verification that their hammerhead ribozymes are embedded in mature transcripts. However, the horse and platypus ribozymes are located within the approximated 3′ UTRs of predicted CLEC2-like genes. The bestcharacterized CLEC2 family member resides in mCLEC2d. This gene encodes a cell surface ligand (CLRB) that is recognized by natural killer (NK) cells through an inhibitory NKR-P1 receptor.64 Engagement of the NK cell-associated receptor with the CLRB ligand initiates an inhibitory signal such that the loss of CLRB expression increases NK cell-mediated cytotoxicity.
Figure 1.4.

Comparison of the CLEC2 hammerhead ribozyme sequences. Alignments of verified and predicted CLEC2 hammerhead ribozyme sequences. The sequences of the substrate and enzyme segments were aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The remainders of 3′-UTRs are denoted as length of sequence in parentheses to the predicted stop codon and polyA signal. For reference, the active site cytosine is indicated with an arrowhead, and other conserved catalytic core nucleotides are boxed. Residues predicted to form base pairs in the double helices (including GU pairs) are highlighted in gray and correspond to stems indicated in the labels below the alignment. Asterisks mark nucleotides that are identical in all sequences.
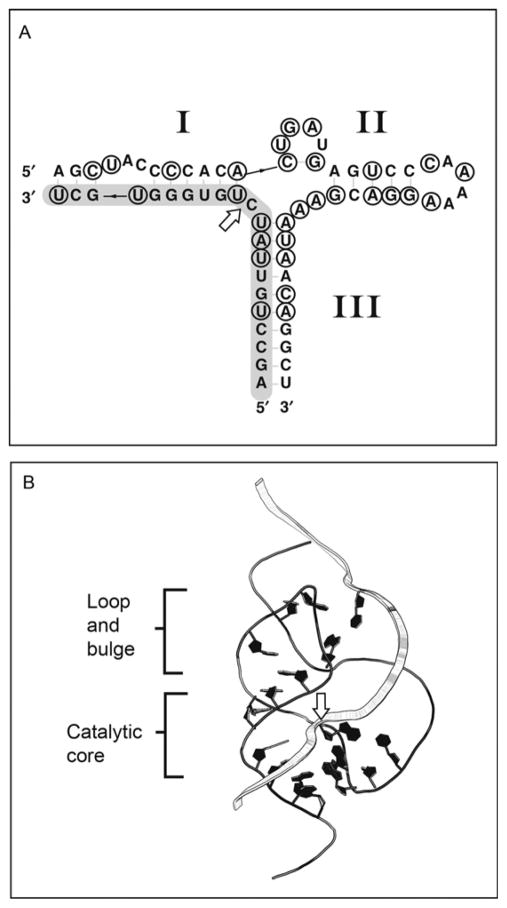
All 12 ribozymes have a similar global arrangement: They are type III hammerheads that contain large, non-conserved, intervening sequences in place of Loop I. Sequence alignment revealed remarkable conservation of the hammerhead motif’s catalytic core including nucleotides necessary to establish the catalytically important tertiary interactions (Fig. 1.5). Additional conservation is observable in the secondary structure in the form of compensatory mutations that maintain the hammerhead’s secondary structure (Fig. 1.4). Structural similarity and specific association with orthologous genes, including CLEC-like sequence in the platypus genome,65 imply that all 12 CLEC2 ribozymes share a common ancestor that arose before monotreme divergence from other therian lineages about 2’0 million years ago.
Figure 1.5.
MappingofCLEC2ribozymeinvariantnucleotidesonthetertiarystructureof the full-length hammerhead ribozyme. (A) Secondary structure of mCLEC2d hammer-head ribozyme. Positions conserved in all CLEC2 ribozyme sequences are circled. The cleavage site is indicated with a white arrow. (B) Positions analogous to invariant CLEC2 ribozyme nucleotides are drawn in black on the string representation of the Schistosome hammerhead ribozyme tertiary structure (PDB ID: 3ZP8).41 The substrate strand is represented as a wide ribbon and the site of bond cleavage is indicated with a white arrow.
A large insertion between substrate and enzyme strands distinguishes the CLEC2 ribozymes not only from other hammerhead ribozymes, but also from most known self-cleaving sequences. In rodent CLEC2 hammerheads, the length of the sequence separating the two critical ribozyme domains ranges from 246 to 789 nt. Nevertheless, the substrate and enzyme segments pair to form a structurally and catalytically accurate hammerhead ribozyme. More specifically, CLEC2 ribozymes contain the signature core of 15 invariant nucleotides flanked by three helices with distal rate-enhancing interactions, and they produce two strands by self-cleaving (Fig. 1.4). Even though compact sequences dominate the collection of all known ribozymes, the discontinuous format of the CLEC2 ribozymes is not without precedent. The widespread nonanimal self-splicing group I and II introns span hundreds to thousands of nucleotides. The CLEC2 ribozyme demonstrates that interruption of functional domains with long sequences does not impede hammerhead ribozyme activity. Further investigation will determine whether these sequence intervals play a role in the regulation of ribozyme function and gene expression.
The mCLEC2d-associated ribozyme is very similar to the well-studied full-length Schistosome ribozyme that is active in physiological conditions.49 The catalytic core is strictly conserved with a single change in position 7, a well-documented variable nucleotide, while the lengths and base pairing of stem(s) I and II preserve the overall architecture of the secondary structure observed in full-length hammerhead ribozymes. More remarkable, the 16 nucleotides of stem II are also identical between the Schistosome and the mCLEC2d-associated ribozymes except for a single-base deviation in the loop region (Fig. 1.3). In full-length hammerhead ribozymes, interaction between the stem II loop and stem(s) I bulge confers a catalytic rate enhancement to the active site through tertiary structure changes and enables activity in physiologically relevant conditions. Considering that nucleotides in the stem(s) I bulge are also conserved between these two hammerhead ribozymes from phylogenetically distant organisms, it is reasonable to expect that CLEC2 ribozyme tertiary structure corresponds to that of the Schistosome hammerhead ribozyme and functions at physiological conditions with similar kinetics.20,41,66 Consistent with these comparisons, cell-based assays using reporter gene constructs conjugated to CLEC2 3′ UTRs demonstrated that the embedded ribozymes significantly reduce expression of the upstream gene by effectively cleaving and destabilizing the mRNA.9,10
Additional features of the CLEC2 hammerhead ribozyme are universally conserved among all hammerhead ribozymes from diverse origins highlighting the importance of these elements to ribozyme activity in vivo. Naturally occurring hammerheads as well as artificially selected motifs have rather variable sequences in the loop and bulge regions with specific combinations particular to different ribozyme sources.67–69 One exception to this is the widely conserved adenosine in the sixth position of Loop II of type I and III ribozymes. In the full-length ribozyme tertiary structure, this base is involved in a noncanonical interaction with the conserved uridine from the substrate strand.41,70 Preservation of this specific combination in the CLEC2 ribozymes emphasizes its importance to activity within the cellular environment. A less appreciated structural element, the CG base pair adjacent to Loop II, is conserved in 11 out of 12 CLEC2 ribozymes. This interaction exists in identical orientation in all but one natural hammerhead ribozyme analyzed to date and in most artificially selected species.67 Preferential CG base pairing emphasizes the need for a reinforced helix at this position possibly due to the nature of the adjacent loop–bulge interaction. However underappreciated, this interaction may play an important role in fine-tuning ribozyme function.
Homology between CLEC2 ribozymes and hammerheads used in structural and biochemical studies can explain mechanistic roles of most nucleotides that are conserved within the CLEC2 ribozyme group (Fig. 1.5). However, other features that are unique to CLEC2 ribozymes suggest roles intrinsic to function in their specific genomic contexts. For example, CLEC2 ribozymes characteristically possess a long $25 bp stem III adjoining the motif to the remainder of the 3′ UTR. Such elongated stems are uncommon in other naturally occurring hammerhead ribozymes even though they can stabilize catalytic structures in vivo.71 Instead, the long stem III may reflect a function specific to the location or particular mode of regulation of CLEC2 ribozyme.
Although ribozyme sequences have been found throughout divergent genomes,72,73 few are known to change levels of gene expression. A single example of ribozyme-mediated gene regulation via a UTR is found in prokaryotes.8 The bacterial GlmS ribozyme represents one class of riboswitches that responds to a variety of small molecule cues. The ribozyme is encoded within the 5′ UTR of the polycistronic mRNA, and by cleaving within this region abrogates expression of downstream genes. In contrast, CLEC2 hammerhead ribozymes are encoded in 3′ UTRs that are eukaryotic hotspots for motifs that posttranscriptionally regulate gene expression. miRNAs and a variety of RNA-binding proteins target the 3′ UTR and cause changes in transcript processing, abundance, or localization.74 Moreover, several causes of aberrant regulation of messenger RNA can be traced to the 3′ UTR.75 All things considered, the hammerhead ribozyme appears to reside in an important regulatory region that provides the possibility that the ribozyme itself can be regulated. More recent discoveries have demonstrated that the hammerhead ribozyme sequence is found to be ubiquitous throughout the tree of life76 and is possibly the most common ribozyme sequence77 apart from RNase P and the peptidyl transferase of the ribosome.
References
- 1.Kruger K, Grabowski PJ, Zaug AJ, et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 2.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 3.Prody GA, Bakos JT, Buzayan JM, et al. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 4.Noller HF, Hoffarth V, Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 5.Nissen P, Hansen J, Ban N, et al. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 6.Zamore PD. RNA interference: big applause for silencing in Stockholm. Cell. 2006;127:1083–1086. doi: 10.1016/j.cell.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 8.Winkler WC, Nahvi A, Roth A, et al. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 9.Martick M, Horan LH, Noller HF, et al. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martick M, Lee TS, York DM, et al. Solvent structure and hammerhead ribozyme catalysis. Chem Biol. 2008;15:332–342. doi: 10.1016/j.chembiol.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Symons RH. Plant pathogenic RNAs and RNA catalysis. Nucleic Acids Res. 1997;25:2683–2689. doi: 10.1093/nar/25.14.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott WG. Ribozymes. Curr Opin Struct Biol. 2007;17:280–286. doi: 10.1016/j.sbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Pley HW, Flaherty KM, McKay DB. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 14.Scott WG, Murray JB, Stoddard BL, Klug A. Capturing the Structure of a Catalytic RNA Intermediate: The Hammerhead Ribozyme. Science. 1996;274:2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- 15.Scott WG, Finch JT, Klug A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- 16.McKay DB. Structure and function of the hammerhead ribozyme: an unfinished story. RNA. 1996;2:395–403. [PMC free article] [PubMed] [Google Scholar]
- 17.Blount KF, Uhlenbeck OC. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- 18.Fedor MJ. Comparative enzymology and structural biology of RNA self-cleavage. Annu Rev Biophys. 2009;38:271–299. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- 19.Scott WG. BiophysicalandbiochemicalinvestigationsofRNAcatalysisinthehammerhead ribozyme. Q Rev Biophys. 1999;32:241–284. doi: 10.1017/s003358350000353x. [DOI] [PubMed] [Google Scholar]
- 20.de la Peña M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khvorova A, Lescoute A, Westhof E, et al. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 22.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pyle AM. Ribozymes: a distinct class of metalloenzymes. Science. 1993;261:709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]
- 24.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dahm SC, Uhlenbeck OC. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry. 1991;30:9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- 26.Dahm SC, Derrick WB, Uhlenbeck OC. Evidence for the role of solvated metal hydroxide in the hammerhead cleavage mechanism. Biochemistry. 1993;32:13040–13045. doi: 10.1021/bi00211a013. [DOI] [PubMed] [Google Scholar]
- 27.Bassi GS, Mollegaard NE, Murchie AI, et al. RNA folding and misfolding of the hammerhead ribozyme. Biochemistry. 1999;38:3345–3354. doi: 10.1021/bi982985r. [DOI] [PubMed] [Google Scholar]
- 28.Bassi GS, Mollegaard NE, Murchie AI, et al. Ionic interactions and the global conformations of the hammerhead ribozyme. Nat Struct Biol. 1995;2:45–55. doi: 10.1038/nsb0195-45. [DOI] [PubMed] [Google Scholar]
- 29.Bassi GS, Murchie AI, Lilley DM. The ion-induced folding of the hammerhead ribozyme: core sequence changes that perturb folding into the active conformation. RNA. 1996;2:756–768. [PMC free article] [PubMed] [Google Scholar]
- 30.Bassi GS, Murchie AI, Walter F, et al. Ion-induced folding of the hammerhead ribozyme: a fluorescence resonance energy transfer study. EMBO J. 1997;16:7481–7489. doi: 10.1093/emboj/16.24.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammann C, Cooper A, Lilley DM. Thermodynamics of ion-induced RNA folding in the hammerhead ribozyme: an isothermal titration calorimetric study. Biochemistry. 2001;40:1423–1429. doi: 10.1021/bi002231o. [DOI] [PubMed] [Google Scholar]
- 32.Hammann C, Lilley DM. Folding and activity of the hammerhead ribozyme. Chembiochem. 2002;3:690–700. doi: 10.1002/1439-7633(20020802)3:8<690::AID-CBIC690>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 33.Hammann C, Norman DG, Lilley DM. Dissection of the ion-induced folding of the hammerhead ribozyme using 19F NMR. Proc Natl Acad Sci USA. 2001;98:5503–5508. doi: 10.1073/pnas.091097498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Penedo JC, Wilson TJ, Jayasena SD, et al. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA. 2004;10:880–888. doi: 10.1261/rna.5268404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou JM, Zhou DM, Takagi Y, et al. Existence of efficient divalent metal ion-catalyzed and inefficient divalent metal ion-independent channels in reactions catalyzed by a hammerhead ribozyme. Nucleic Acids Res. 2002;30:2374–2382. doi: 10.1093/nar/30.11.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray JB, Seyhan AA, Walter NG, et al. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 37.Murray JB, Terwey DP, Maloney L, et al. The structural basis of hammerhead ribozyme self-cleavage. Cell. 1998;92:665–673. doi: 10.1016/s0092-8674(00)81134-4. [DOI] [PubMed] [Google Scholar]
- 38.Murray JB, Dunham CM, Scott WG. A pH-dependent conformational change, rather than the chemical step, appears to be rate-limiting in the hammerhead ribozyme cleavage reaction. J Mol Biol. 2002;315:121–130. doi: 10.1006/jmbi.2001.5145. [DOI] [PubMed] [Google Scholar]
- 39.Rupert PB, Massey AP, Sigurdsson ST, et al. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 40.Rupert PB, Ferre-D’Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 41.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferre-D’Amare AR, Zhou K, Doudna JA. A general module for RNA crystallization. J Mol Biol. 1998;279:621–631. doi: 10.1006/jmbi.1998.1789. [DOI] [PubMed] [Google Scholar]
- 43.Ke A, Zhou K, Ding F, et al. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 44.Scott WG, Murray JB, Arnold JR, et al. Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science. 1996;274:2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- 45.Han J, Burke JM. Model for general acid–base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site. Biochemistry. 2005;44:7864–7870. doi: 10.1021/bi047941z. [DOI] [PubMed] [Google Scholar]
- 46.Heckman JE, Lambert D, Burke JM. Photo-crosslinking detects a compact, active structure of the hammerhead ribozyme. Biochemistry. 2005;44:4148–4156. doi: 10.1021/bi047858b. [DOI] [PubMed] [Google Scholar]
- 47.Hertel KJ, Herschlag D, Uhlenbeck OC. A kinetic and thermodynamic framework for the hammerhead ribozyme reaction. Biochemistry. 1994;33:3374–3385. doi: 10.1021/bi00177a031. [DOI] [PubMed] [Google Scholar]
- 48.Stage-Zimmermann TK, Uhlenbeck OC. Hammerhead ribozyme kinetics. RNA. 1998;4:875–889. doi: 10.1017/s1355838298980876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Canny MD, Jucker FM, Kellogg E, et al. Fast cleavage kinetics of a natural hammerhead ribozyme. J Am Chem Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- 50.Nelson JA, Uhlenbeck OC. Hammerhead redux: does the new structure fit the old biochemical data? RNA. 2008;14:605–615. doi: 10.1261/rna.912608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nelson JA, Uhlenbeck OC. Minimal and extended hammerheads utilize a similar dynamic reaction mechanism for catalysis. RNA. 2008;14:43–54. doi: 10.1261/rna.717908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doudna JA. Hammerhead ribozyme structure: U-turn for RNA structural biology. Structure. 1995;3:747–750. doi: 10.1016/s0969-2126(01)00208-8. [DOI] [PubMed] [Google Scholar]
- 53.Ruffner DE, Stormo GD, Uhlenbeck OC. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990;29:10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- 54.Wang S, Karbstein K, Peracchi A, et al. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry. 1999;38:14363–14378. doi: 10.1021/bi9913202. [DOI] [PubMed] [Google Scholar]
- 55.Murray JB, Szöke H, Szöke A, et al. Capture and visualization of a catalytic RNA enzyme-product complex using crystal lattice trapping and X-ray holographic reconstruction. Mol Cell. 2000;5:279–287. doi: 10.1016/s1097-2765(00)80423-2. [DOI] [PubMed] [Google Scholar]
- 56.Ruffner DE, Uhlenbeck OC. Thiophosphate interference experiments locate phosphates important for the hammerhead RNA self-cleavage reaction. Nucleic Acids Res. 1990;18:6025–6029. doi: 10.1093/nar/18.20.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uhlenbeck OC. A small catalytic oligoribonucleotide. Nature. 1987;328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- 58.Lilley DM. Ribozymes—a snip too far? Nat Struct Biol. 2003;10:672–673. doi: 10.1038/nsb0903-672. [DOI] [PubMed] [Google Scholar]
- 59.Canny MD, Jucker FM, Pardi A. Efficientl igation of the Schistosoma Hammerhead ribozyme. Biochemistry. 2007;46:3826–3834. doi: 10.1021/bi062077r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boots JL, Canny MD, Azimi E, et al. Metal ion specificities for folding and cleavage activity in the Schistosoma hammerhead ribozyme. RNA. 2008;14:1–11. doi: 10.1261/rna.1010808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomas JM, Perrin DM. Probing general base catalysis in the hammerhead ribozyme. J Am Chem Soc. 2008;130:15467–15475. doi: 10.1021/ja804496z. [DOI] [PubMed] [Google Scholar]
- 62.Thomas JM, Perrin DM. Probing general acid catalysis in the hammerhead ribozyme. J Am Chem Soc. 2009;131:1135–1143. doi: 10.1021/ja807790e. [DOI] [PubMed] [Google Scholar]
- 63.Hao L, Klein J, Nei M. Heterogeneous but conserved natural killer receptor gene complexes in four major orders of mammals. Proc Natl Acad Sci USA. 2006;103:3192–3197. doi: 10.1073/pnas.0511280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carlyle JR, Jamieson AM, Gasser S, et al. Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc Natl Acad Sci USA. 2004;101:3527–3532. doi: 10.1073/pnas.0308304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong ES, Sanderson CE, Deakin JE, et al. Identification of natural killer cell receptor clusters in the platypus genome reveals an expansion of C-type lectin genes. Immunogenetics. 2009;61:565–579. doi: 10.1007/s00251-009-0386-7. [DOI] [PubMed] [Google Scholar]
- 66.Ferbeyre G, Smith JM, Cedergren R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol. 1998;18:3880–3888. doi: 10.1128/mcb.18.7.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen X, Denison L, Levy M, et al. Direct selection for ribozyme cleavage activity in cells. RNA. 2009;15:2035–2045. doi: 10.1261/rna.1635209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de la Peña M, Flores R. An extra nucleotide in the consensus catalytic core of a viroid hammerhead ribozyme: implications for the design of more efficient ribozymes. J Biol Chem. 2001;276:34586–34593. doi: 10.1074/jbc.M103867200. [DOI] [PubMed] [Google Scholar]
- 69.Saksmerprome V, Roychowdhury-Saha M, Jayasena S, et al. Artificial tertiary motifs stabilize trans-cleaving hammerhead ribozymes under conditions of sub-millimolar divalentions and high temperatures. RNA. 2004;10:1916–1924. doi: 10.1261/rna.7159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chi YI, Martick M, Lares M, et al. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol. 2008;6:e234. doi: 10.1371/journal.pbio.0060234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Donahue CP, Yadava RS, Nesbitt SM, et al. The kinetic mechanism of the hairpin ribozyme in vivo: influence of RNA helix stability on intracellular cleavage kinetics. J Mol Biol. 2000;295:693–707. doi: 10.1006/jmbi.1999.3380. [DOI] [PubMed] [Google Scholar]
- 72.Salehi-Ashtiani K, Luptak A, Litovchick A, et al. A genome-wide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 73.Webb CH, Riccitelli NJ, Ruminski DJ, et al. Widespread occurrence of self-cleaving ribozymes. Science. 2009;326:953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3:REVIEWS0004. doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lopez de Silanes I, Quesada MP, Esteller M. Aberrant regulation of messenger RNA 3′-untranslated region in human cancer. Cell Oncol. 2007;29:1–17. doi: 10.1155/2007/586139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.delaPeña M, Garcia-Robles I. Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA. 2010;16:1943–1950. doi: 10.1261/rna.2130310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perreault J, Weinberg Z, Roth A, et al. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput Biol. 2011;7:e1002031. doi: 10.1371/journal.pcbi.1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]