Abstract
Autophagy protects cells from harmful substances such as protein aggregates, damaged mitochondria and intracellular pathogens and has been implicated in a variety of diseases. Selectivity of autophagic processes is mediated by cargo receptors that link cargo to Atg8 family proteins on the developing autophagosomal membrane. To avoid collateral degradation during constitutive autophagic pathways the autophagic machinery must not only select cargo but also exclude non-cargo material. Here we show that cargo directly activates the cargo receptor Atg19 by exposing multiple Atg8 binding sites. Furthermore, Atg19 mediates tight apposition of the cargo and Atg8-coated membranes in a fully reconstituted system. These properties are essential for the function of Atg19 during selective autophagy in vivo. Our results suggest that cargo receptors contribute to tight membrane bending of the isolation membrane around the cargo.
Introduction
Macroautophagy (hereafter autophagy) is the major pathway for the bulk degradation of cellular material. This is achieved by the de novo formation of double membrane-bound vesicles called autophagosomes. During autophagosome formation small membrane structures called isolation membranes (or phagophores) are generated which capture cellular material as they grow1-4. Subsequently, they fuse with the endo-lysosomal system where the inner membrane and the cargo are degraded5. Owing to its ability to degrade cargo selectively it serves as an cellular essential quality control system6. Autophagy has been shown to play a protective role against neurodegeneration, uncontrolled infection by intracellular pathogens and cancer2,6-9.
The selectivity of the autophagic process is determined by cargo receptors that selectively link the cargo to the isolation membrane. They achieve this by selective recognition of the cargo and interaction with proteins of the Atg8 family on the isolation membrane10, 11. Binding to Atg8 family proteins is mediated by so-called LIR (or LRS) motifs12,13-15. During constitutive autophagic processes the autophagic machinery must also ensure that non-cargo material is excluded from autophagosomes in order to avoid constant loss of cellular material into lysosomes. This exclusivity can only be achieved if the isolation membrane is tightly apposed to the cargo as it grows.
The yeast cargo receptor Atg19 is a distant relative of the mammalian p62/SQSTM1 and NBR1 (neighbour of BRCA1 gene 1) cargo receptors16. Atg19 mediates the delivery of the precursor aminopeptidase I (prApe1) and other substances into the vacuole in a constitutive process called cytoplasm-to-vacuole-targeting (Cvt) pathway17-21. The Cvt pathway is mechanistically equivalent to selective autophagy in mammals3, 5, 13-16.
Since the Cvt pathway is constitutive, it is important that other cellular material is not constantly lost into the vacuole. Consequently, the membrane of the autophagosome-like Cvt vesicles is tightly wrapped around the prApe1 oligomers22. In contrast, when yeast cells starve they respond by the formation of autophagosomes that are larger than Cvt vesicles and contain random cytoplasmic material22. During this process the cargo receptors Atg19 and its relative Atg34 tether cargo to the isolation membrane23, 24. They thereby confer some cargo selectivity to autophagosome formation but not exclusivity because bulk cellular material is not excluded from it sequestration within autophagosomes.
Here we find that cargo binding to the Atg19 receptor positively regulates its association with Atg8. Atg8 binding by Atg19 is not only mediated by its known LIR motif but by multiple cryptic Atg8 binding sites. Furthermore, in vitro reconstitution reveals that Atg19 is sufficient for the tight apposition of Atg8 positive membranes with the cargo. Our results suggest that certain cargo receptors contribute to bending of the isolation membrane around the cargo to exclude non-cargo material from its sequestration into autophagosomes.
Results
LIR-independent interaction of Atg19 with Atg8
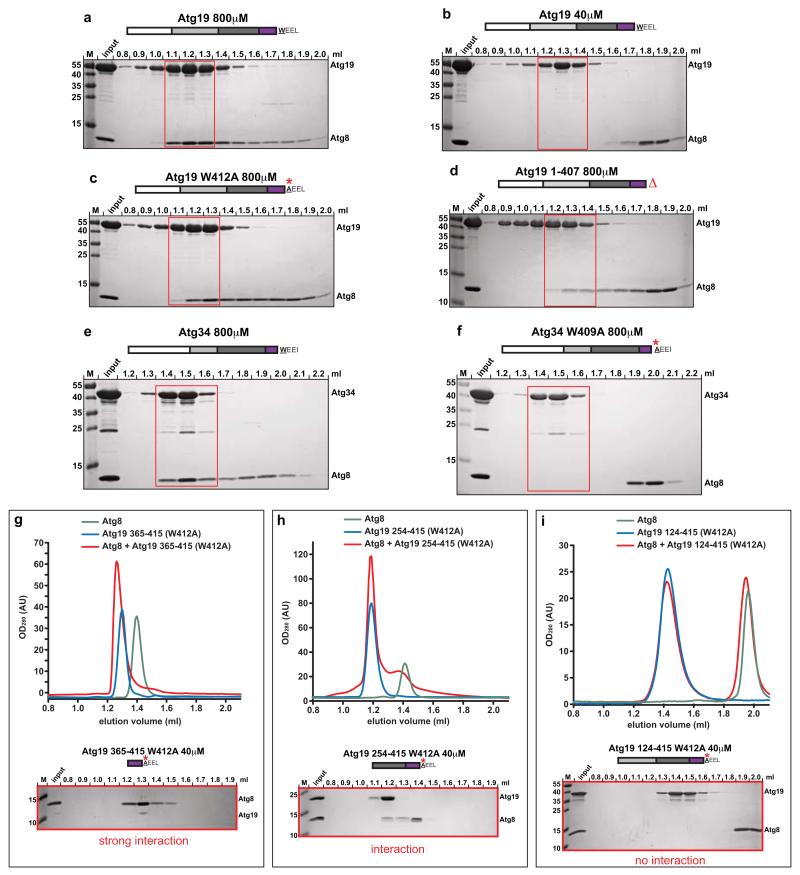
We first studied the interaction between Atg19 and Atg8 in solution using size exclusion chromatography (SEC) (Fig. 1). At high (800μM) but not low (40μM) concentrations, Atg8 and Atg19 co-eluted (Fig. 1a,b). Consistently, we determined the dissociation constant (KD) to be about 35μM by isothermal titration calorimetry (ITC) (Supplementary Fig. 1). This value is about 10-20 fold lower compared to the KD for other cargo receptors interacting with Atg8 family proteins25. The interaction of cargo receptors with Atg8 family proteins has been shown to occur via so-called LC3-interacting regions (LIRs, also called LRS, W412EEL415 in Atg19)10, 12-15. To test the requirement of the known LIR motif for the interaction of Atg19 with Atg8, we mutated W412 to A or deleted entire LIR motif13. Unexpectedly, although these mutations weakened the interaction with Atg8 these Atg19 mutants still bound to Atg8 (Fig. 1c,d). This indicates that the known LIR motif contributes to Atg8 interaction but is not essential. In contrast, the interaction between the Atg34 cargo receptor and Atg8 was entirely dependent on the known LIR motif since the equivalent mutant (W409A) showed no interaction with Atg8 (Fig. 1e,f).
Figure 1.
Atg19 and Atg34 interaction with Atg8
(a-f) Atg8 was co-incubated with Atg19 or Atg34 (wild type and mutants) at concentrations of 40μM and 800μM, respectively and run on a size exclusion column. Aliquots of individual fractions were run on SDS-PAGE gels and stained by Coomassie. The white box indicates the N-terminal domain, the light grey box the coiled-coil domain, the dark grey box the Ams1-binding domain and the purple box indicates the C-terminal domain. (g-f) Atg8 was co-incubated with the indicated Atg19 domains at 40 μM and run on a size exclusion column. Individual elution profiles are shown above; Coomassie stained SDS-PAGE gels below. Columns: (a-f, i: S200 3.2/30; g, h: S75 3.2/30). M: size marker. All experiments shown have been done two times.
To identify the Atg8 binding activity independent of the classical LIR motif in Atg19 we investigated the ability of Atg19 fragments containing the W412A mutation to interact with Atg8. Strikingly, the short C-terminal fragment (365-415 W412A) showed a strong interaction with Atg8 at 40μM (Fig. 1g), stronger than the wild type full-length protein (Fig. 1b), and this was maintained when the fragment was further extended by 111 amino acids (254-415 W412A; Fig. 1h). However, when a further extension to include the coiled-coil domain was introduced (amino acids 124-415 W412A) the interaction became undetectable at low concentration (40μM) (Fig. 1i). These results suggest that the C-terminus of Atg19 (but not Atg34) interacts strongly with Atg8 independent of the known LIR motif and further that the coiled-coil domain inhibits this interaction.
Cargo activates the Atg8-Atg19 interaction
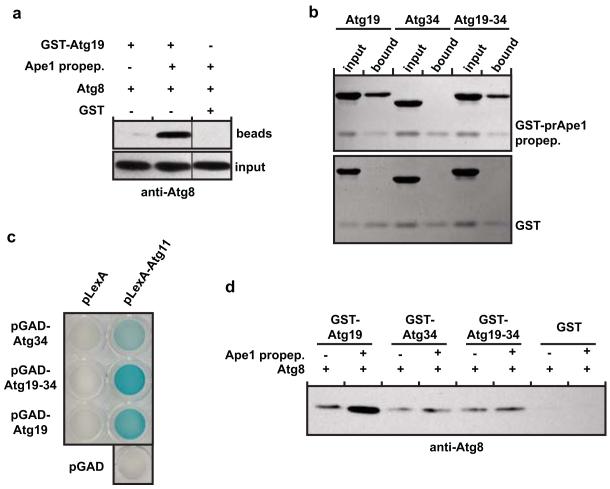
The coiled-coil domain of Atg19 mediates the interaction with the prApe1 cargo23. We therefore reasoned that the coiled-coil domain mediated inhibition of the interaction of the Atg19 C-terminus with Atg8 might be relieved upon addition of the prApe1 cargo. This was indeed the case as the presence of the Atg19 binding prApe1 propeptide23 greatly stimulated the amount of Atg8 pulled down by Atg19 (Fig. 2a and Supplementary Figs 1). To test if the C-terminus of Atg19 that showed strong LIR-independent interaction with Atg8 is specifically required for the prApe1 cargo stimulated interaction with Atg8, we constructed an Atg19-Atg34 chimeric protein by exchanging the C-terminus of Atg19 with the C-terminus of Atg34 (Supplementary Fig. 1). Atg34 showed no interaction with Atg8 independent of the classical LIR motif (Fig. 1e,f). The Atg19-Atg34 chimera efficiently bound the prApe1 propeptide (Fig. 2b and Supplementary Fig. 1) and still interacted with the scaffold protein Atg11 (Fig. 2c)23 but the propeptide did not stimulate Atg8 binding (Fig. 2d).
Figure 2.
The C-terminus of Atg19 interacts in a cargo-dependent manner with Atg8
(a) Anti-Atg8 western blot of the input and pellet fractions of a pull down experiment using GST-Atg19 as bait and Atg8 as prey. The black vertical line indicates that unrelated samples were run on the same gel but in between the two samples on the left and the one on the right (see Supplementary Fig. 7). (b) Coomassie stained gels of pull down experiments using the GST-prApe1 propeptide as bait to pull down Atg19, the Atg19-Atg34 chimera and Atg34, respectively. The Atg19-Atg34 chimera contained residues 1-361 of Atg19 and residues 348-412 of Atg34. (c) Yeast-two-hybrid assay testing for the interaction of Atg11 with Atg19, the Atg19-Atg34 chimera and Atg34, respectively. (d) Anti-Atg8 western blot using the indicated GST fusion proteins as bait to pull down Atg8. The pull downs were conducted in the presence or absence of the prApe1 propeptide. See Supplementary Fig. 1 for input gel. All experiments shown have been done two times. Images of uncropped western blots and gels can be found in Supplementary Figure 7.
Atg19 contains multiple Atg8 binding sites
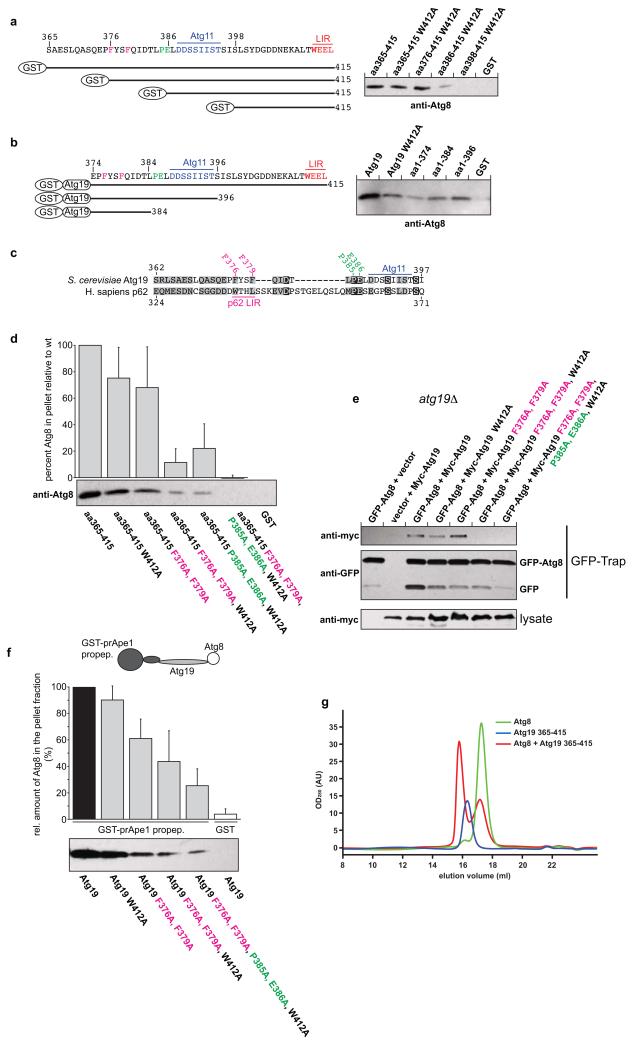
To identify the Atg8 binding sites independent of the classical LIR motif within the C-terminus of Atg19 we fused a series of shortened versions of the C-terminal domain (residues 365-415) of Atg19 to GST and tested its interaction with Atg8 in pull down experiments (Fig. 3a and Supplementary Fig. 1). Consistent with our SEC results (Fig 1c,d) the W412A mutation did not abolish the interaction with Atg8 (Fig. 3a and Supplementary Figs 1 and 2). Residues 398-415 containing the W412A mutation showed no interaction with Atg8 suggesting that the interaction of the extreme C-terminal region of Atg19 is dependent on the known LIR motif (Fig. 3a and Supplementary Fig. 1). Extension of the C-terminal domain (residues 386-415, 376-415 and 365-415, respectively) in context of the W412A mutation enhanced the interaction with Atg8 (Fig. 3a).
Figure 3.
The C-terminus of Atg19 contains multiple Atg8 binding sites
(a, b) Anti-Atg8 western blots using the indicated GST fusion proteins shown on the left as bait to pull down Atg8. See Supplementary Fig. 1 for input gels. (c) Alignment of the Atg19 C-terminus lacking the LIR motif with the region around the LIR motif of human p62 (NP_003891.1). The amino acids shown in magenta and green indicate the residues mutated in the experiments shown in (d-f). (d) Anti-Atg8 western blots with the indicated GST-fusion proteins as bait to pull down Atg8. Quantification of the amount of Atg8 pulled down by the indicated GST fusion protein. The graph is based on 3 independent experiments (N = 3), one of which is shown below the graph. Shown are the averages and standard deviations. (e) GFP-Trap pull down experiment using yeast cells with integrated GFP-Atg8 transformed with the indicated Myc-Atg19 constructs. (f) Pull down experiment using the GST-prApe1 propeptide to pull down Atg8 via the indicated Atg19 proteins. The graph is based on 3 experiments (N = 3), one of which is shown below the graph. Shown are the averages and s.d. See Supplementary Fig. 2 for input gel. (g) Representative profile of a size exclusion chromatography run using a 10/300 S200 column. Atg8 was run at 400μM and the Atg19 C-terminus at 40μM. For the complex run both proteins were pre-incubated at the concentrations mentioned. The average number of Atg8 molecules shifted into the complex peak per 1 Atg19 C-terminus was 4.14 (average of 3 experiments). The experiments shown in (a, b, d, f, g) have been conducted three times; the experiment shown in (e) has been conducted two times. Images of uncropped western blots and gels can be found in Supplementary Figure 7.
In a complementary approach we tested a series of C-terminally truncated fragments of GST-Atg19 for their ability to pull down Atg8 (Fig. 3b and Supplementary Fig. 1). Deletion of residues 375-415 (amino acids, aa1-374) drastically decreased but did not fully abolish its interaction with Atg8 (Fig. 3b). Residues 1-384 showed slightly increased binding to Atg8 compared to GST-Atg19 aa1-374 while GST-Atg19 aa1-396 showed a further enhanced interaction (Fig. 3b). Thus the C-terminus of Atg19 contains multiple Atg8 binding sites, one site localizes to the known LIR motif, one site is close to the Atg11 binding site and at least one site localizes N-terminally of the Atg11 binding site.
In order to identify these Atg8 binding sites we aligned the sequence of the C-terminal domain of Atg19 lacking the known, C-terminal LIR motif to the LIR-containing regions of other cargo adaptors. Phenylalanine residues F376 and F379 aligned with the two hydrophobic residues of the LIR motif of the human cargo receptor p62 (W338 and L341)12, 15 (Fig. 3c). To test if F376 and F379 contribute to Atg8 binding we mutated these residues to alanine. Indeed, the F376A, F379A double mutant showed slightly reduced binding to Atg8 (Fig. 3d and Supplementary Fig. 2). When F379A was co-introduced with W412A the resulting double mutant showed severely reduced binding (Supplementary Fig. 2) as did the F376A, F379A, W412A triple mutant (Fig. 3d and Supplementary Fig. 2). We conclude that one additional Atg8 binding site localizes to the region around F376 and F379.
The alignment of Atg19 with p62 also revealed homology around the Atg11 binding site (Fig. 3c). In particular, P385 and E386 are conserved. Interestingly, P385 and E386 are absent from Atg34. Since our pull down experiments suggested that one Atg8 binding site localizes around the Atg11 binding site of Atg19 (Fig. 3a,b) we mutated P385 and E386 to alanine. When P385A, E386A was combined with W412A the resulting triple mutant showed reduced Atg8 binding compared to the W412A mutant C-terminus (Fig. 3d). Additional introduction of F376A, F379A to generate a F376A, F379A, P385A, E386A, W412A mutant resulted in complete loss of Atg8 binding under the tested conditions (Fig. 3d).
We corroborated these findings firstly by preforming in vivo pull down experiments (Fig. 3e). These results show that the previously reported LIR motif around W412 is indeed not the only Atg8 site in vivo while the F376A, F379A, W412A triple mutant lost detectable Atg8 binding activity, at least under these conditions.
Using an in vitro experimental setup that resembles the situation in vivo (Fig. 3f and Supplementary Fig. 2) we found that the W412A mutation and the F376A, F379A double mutation each reduced binding to some extent but that simultaneous mutation of all three residues severely decreased Atg8 interaction. Additional mutation of P385A, E386A further weakened the interaction with Atg8 (Fig. 2f).
In order to test if one Atg19 C-terminus molecule can indeed simultaneously interact with multiple Atg8 molecules we incubated the Atg19 C-terminus with a 10-fold excess of Atg8 and run the mixture on a size exclusion column (Fig. 3g). By calculating the fraction of the Atg8 peak that was shifted into the Atg8-Atg19 C-terminus complex peak we determined the ratio of Atg19 C-terminus/Atg8 to be about 1/4 suggesting that about 4 molecules of Atg8 can simultaneously interact with one Atg19 C-terminus.
In summary, we have identified two regions within the C-terminus of Atg19 that contribute to Atg8 binding (apart from the known LIR motif around W412). The first region localizes to F376 and F379 and the second region localizes to P385 and E386. There may be an additional Atg8 binding site N-terminally of F376 (Fig. 3b) but if present it is too weak to be detectable in a reliable manner.
Reconstitution of Atg8 mediated recruitment of Atg19 to the membrane
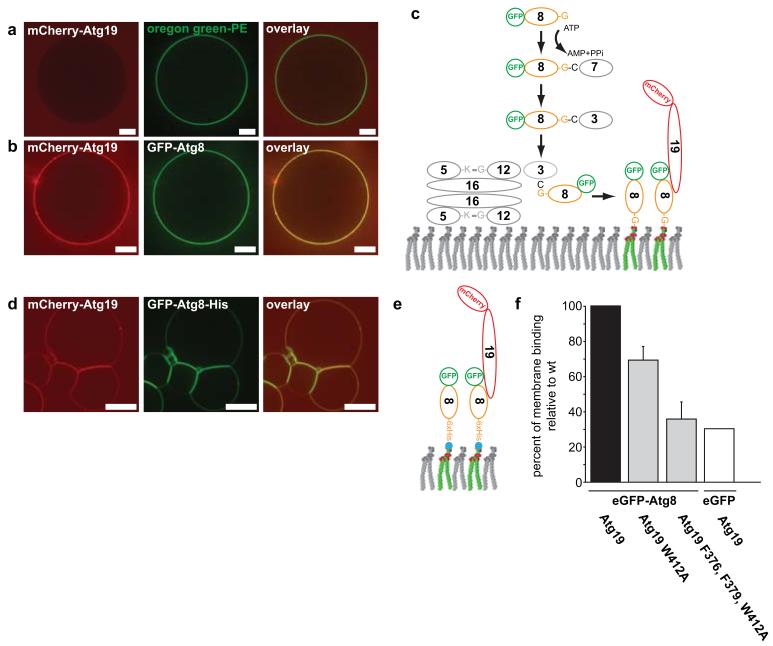
In the cell, the concentrations of Atg19 and Atg8 are in the nM range and thus very low26, 27. However, the Atg8-Atg19 interactions occur at a membrane where the local concentrations are increased. We therefore analysed the effect of Atg8 membrane localization on Atg19 recruitment in a reconstituted system using giant unilamellar vesicles (GUVs) (Fig. 4). In the absence of Atg8 on the GUV membrane no Atg19 recruitment was detectable (Fig. 4a). When we reconstituted Atg8 conjugation using recombinant Atg3, Atg7 and the Atg12–Atg5-Atg16 complex28 we detected efficient recruitment of Atg19 to the GUV membrane (Fig. 4b,c). To more accurately control the amount of Atg8 on the membrane we added a C-terminal 6xHis-tagged version of Atg8 to GUVs containing Ni-lipids. Under these conditions Atg8 detectably recruited Atg19 and Atg34 at low nM concentrations (Supplementary Fig. 3). Thus, the local concentration of Atg8 on the membrane was sufficient for recruitment of Atg19 and Atg34 to the membrane at concentrations that are close to the concentrations found in vivo26, 27. Consistent with these findings we observed a dramatically increased interaction in pull down assays when we used GST-Atg8 concentrated on beads to pull down Atg19 (Supplementary Fig. 4) (compared to the reversed situation shown in Fig. 3b and Supplementary Figs 1 and 2). Correlating with reduced Atg8 interaction of the Atg19 W412A and F376A, F379A, W412A mutants in the pull down assays (Fig. 3 and Supplementary Fig. 2) the recruitment of these mutant Atg19 proteins to Atg8 harbouring GUVs was dramatically reduced (Fig. 4d-f).
Figure 4.
Reconstitution of Atg8-dependent Atg19 recruitment to the membrane
(a) GUVs incubated with mCherry-Atg19. The membrane was marked by incorporation of an Oregon green labelled lipid. (b) GUV coated with GFP-Atg8 incubated with mCherry-Atg19. (c) Scheme showing the proteins and reaction resulting in GFP-Atg8 conjugation to the GUV shown in (b). (d) GUVs containing nickel lipids incubated with GFP-Atg8-6xHis and wild type mCherry-Atg19. (e) Experimental setup of the experiment shown in (d). (f) Quantification of membrane binding by the indicated mCherry-Atg19 proteins. The experimental setup is shown in (e). The graph is based on 3 experiments. Shown are the averages and s.d. of these 3 experiments (N = 3, based on 318 GUVs for Atg19, 171 GUVs for Atg19 W412A, 166 GUVs for Atg19 F376A, F379A, W412A, 188 GUVs for the eGFP control). Scale bars: 5μm. Abbreviations: 3: Atg3; 5: Atg5; 7: Atg7; 8: Atg8; 12: Atg12; 16: Atg16; 19: Atg19. Experiments shown in (a, b) have been conducted two times; the experiment in (d) three times.
Reconstitution of membrane bending during selective autophagy
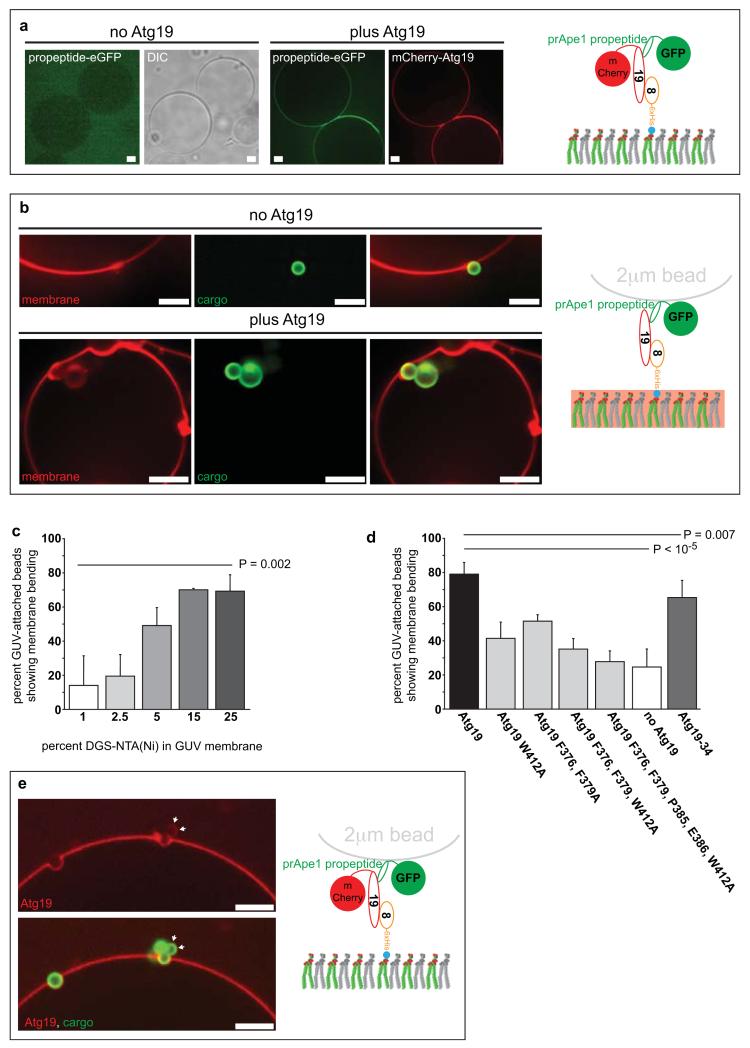
Next we asked if Atg19 could recruit cargo to Atg8-coated membranes. To this end we added the propeptide of the prApe1 cargo to Atg8-coated GUVs (Fig. 5a). This was indeed the case since only upon addition of Atg19 did we detect recruitment of the Ape1 propeptide to the GUV membrane (Fig. 5a). To mimic the in vivo situation where the prApe1 propeptide is concentrated on the surface of the prApe1 oligomer we coated 2μm diameter polystyrene beads with the prApe1 propeptide (Fig. 5b-e and Supplementary Fig. 3). In the absence of Atg19 only a few beads were found peripherally attached to the Atg8-coated GUV membrane. In contrast, in the presence of Atg19, the beads attached much more frequently and we also noticed that Atg19 bound directly to the prApe1 propeptide-coated beads (Fig. 5e and Supplementary Fig. 3). Strikingly, in the presence of Atg19 the GUV membrane was closely wrapped around parts of the prApe1 propeptide beads (Fig. 5b,e and Supplementary Fig. 3). Frequently, the beads were almost completely invaginated into the lumen of the GUVs (Fig. 5b). Atg19 mediated membrane bending positively correlated with the density of Atg8 on the membrane (Fig. 5c). Furthermore, the ability of the Atg19 mutants to bend the membrane around the beads correlated well with their Atg8 binding activity (Figs 5d and 3f). In the absence of Atg19 few membrane bending events were observed (Fig. 5d), and these were also far less pronounced. Thus, the tripartite interaction between prApe1, Atg19 and Atg8 is sufficient for close membrane-cargo apposition.
Figure 5.
Reconstitution of Atg8 and Atg19 dependent cargo recruitment to the membrane
(a) prApe1 propeptide recruitment to Atg8 harbouring GUVs in the presence and absence of Atg19. The experimental setup is shown on the right. (b) Atg19 dependent envelopment of prApe1-coated beads with Atg8 positive membranes. The experimental setup is shown on the right. The membrane was labelled by incorporation of a rhodamine labelled lipid. (c) Quantification of the percentage of GUV-associated eGFP-Ape1 coated beads showing detectable membrane bending. (N = 3; total GUVs per condition = 40). The experimental setup is shown in (b). The density of Atg8-6xHis on the membrane was titrated by different amounts of Ni-lipids in the GUV membrane. The graph is based on at least 3 independent experiments. Shown are the averages and standard deviations. (d) Quantification of the percentage of GUV-associated eGFP-Ape1 coated beads showing detectable membrane bending. The quantification is based on 3 independent experiments for the mutant Atg19 versions and 16 independent experiments for wild type Atg19 and the no Atg19 control (Atg19: N = 16, total GUVs = 382; Atg19 W412A: N = 3, total GUVs = 55; Atg19 F376A, F379A: N = 3, total 66 GUVs; Atg19 F376A, F379A, W412A: N = 3, total GUVs = 84; Atg19 F376A, F379A, P385A, E386E, W412A: N = 3, total GUVs = 60; Atg19-34: N = 4, total GUVs = 110; no Atg19: N = 16, total GUVs = 382). Shown are the averages and standard deviations. The experimental setup is as shown in (b) the p values in (c) and (d) were calculated using a 2-tailed Student’s T-test. (e) Visualization of Atg19 during the envelopment of prApe1-coated beads with Atg8 containing membranes. Note that Atg19 directly binds the prApe1-coated beads (arrows). The experimental setup is shown on the right. Scale bars: 5μM. Abbreviations: 8: Atg8; 19: Atg19; DIC: Differential interference contrast. All experiments have been conducted at least three times.
Multiple Atg8 interaction sites are required for Atg19-dependent selective autophagy
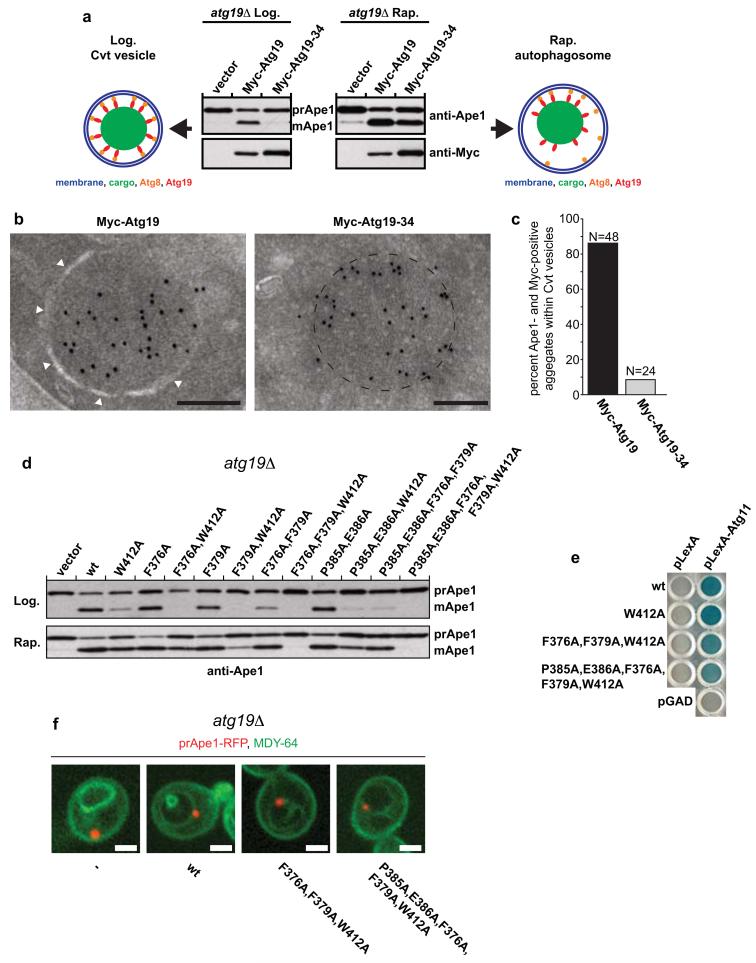
To test the requirements for the function of Atg19 during selective autophagy in vivo, we monitored prApe1 processing under nutrient rich conditions, where only the selective Cvt pathway is active, and in cells treated with rapamycin, where bulk autophagy mediates transport of prApe1 into the vacuole (Fig. 6a)22. Intriguingly, the Atg19-Atg34 chimeric protein was unable to support the Cvt pathway (Fig. 6a). Consistent with the ability of the Atg19-Atg34 chimera to bind the prApe1 propeptide (Fig. 2b), immuno-electron microscopy showed that the protein localized to the prApe1 oligomers (Fig. 6b and Supplementary Fig. 5). However, in contrast to Atg19 the Atg19-34 chimera did not support the growth of an isolation membrane around the prApe1 particles (Fig. 6b,c). Upon rapamycin treatment Atg19-Atg34 was almost fully functional (Fig. 6a) suggesting that multiple Atg8 binding sites are required for transport of the prApe1 cargo during selective (Cvt) but not bulk autophagy. This rapamycin effect was totally dependent on the presence of Atg17, a scaffold protein essential for autophagosome formation but not the Cvt pathway29 (Supplementary Fig. 6).
Figure 6.
Requirements for the Atg19-Atg8 interaction for cargo delivery during selective and bulk autophagy.
(a) Anti-Ape1 western blot of atg19Δ cells transformed with the indicated expression constructs. The lower Ape1 band indicates prApe1 processing and thus its delivery into the vacuole. (b) Representative electron micrographs of ypt7Δ, atg19Δ yeast cells grown under Cvt conditions expressing the indicated proteins labelled with an anti-Myc antibody. The white arrowheads indicate the isolation membrane. The dashed line indicates the circumference of the prApe1 oligomer. Gold particles: 10nm. Scale bars: 200nm. See Supplementary Fig. 5 for full images. (c) Quantification of electron micrographs of yeast cells expressing either Myc-Atg19 or Myc-Atg19-Atg34 co-labelled with anti-Myc (10nm gold) and anti-prApe1 (5nm gold). See also Supplementary Fig. 5. (3 independent experiments, Myc-Atg19: N = 48, Myc-Atg19-34: N = 24). (d) Anti-Ape1 western blot of atg19Δ cells transformed with the indicated expression constructs. The lower Ape1 band indicates prApe1 processing and thus its delivery into the vacuole. Supplementary Fig. 6 shows the expression of the Myc-tagged proteins and a quantification of the assay. (e) Yeast-two-hybrid assay testing for the interaction of Atg11 with Atg19 and the indicated Atg19 mutants. (f) Atg19Δ yeast cells expressing the indicated Atg19 proteins and prApe1-RFP were labelled with the vacuolar membrane dye MDY-64. Scale bars: 2μM. The experiments in (a-d, f) have been conducted 3 times; the experiment in (e) two times. Images of uncropped western blots and gels can be found in Supplementary Figure 7.
Next we addressed the relevance of the additional regions in Atg19 that contribute to Atg8 binding (Fig. 3). Consistent with the biological relevance of these sites the W412A mutation in the LIR motif of Atg19 was not essential for prApe1 processing, both during Cvt and autophagy (Fig. 6d and Supplementary Fig. 6). Next, we tested the effect of the F376A and F379A mutations on prApe1 processing. In isolation the two mutations had weak if any effects on prApe1 processing during the Cvt pathway (Fig. 6d and Supplementary Fig. 6). However, when either of the two single mutations was combined with the W412A mutation the Cvt pathway was non-functional while prApe1 processing was still apparent under rapamycin-induced autophagic conditions (Fig. 6d and Supplementary Fig. 6). We observed an almost identical effect for the F376A, F379A double mutant (Fig. 6d and Supplementary Fig. 6) but when the F376A, F379A double mutant was combined with the W412A mutation Ape1 processing was lost under both Cvt and autophagic conditions (Fig. 6d and Supplementary Fig. 6).
An identical phenomenon was observed for P385 and E386 (Fig. 6d and Supplementary Fig. 6). Introduction of P385A, E386A only slightly decreased prApe1 processing under rich conditions. When combined with W412A or F376A, F379A the P385A, E386A mutation drastically reduced prApe1 processing during the selective Cvt pathway but much less under autophagic conditions (Fig. 6d and Supplementary Fig. 6). None of the mutations affected Atg11 binding (Fig. 6e) or the localization of the prApe1 cargo to the vacuole (Fig. 6f).
Discussion
Here we have provided a number of fundamental insights into the action of cargo receptors during selective autophagy.
In the absence of cargo Atg19 binds only weakly to Atg8. Binding of the prApe1 cargo to Atg19 enhances interaction of Atg19 with Atg8. Thus the cargo has an active role during its sequestration into autophagosomes. This is consistent with the requirement of the prApe1 cargo for assembly of the pre-autophagosomal structure (PAS) for selective autophagy during nutrient rich conditions in yeast30. We thus provide a molecular mechanism for the requirement of the cargo and the Atg19 receptor for PAS formation during nutrient rich conditions23, 30. The inhibition of cargo receptors in the absence of cargo may be a general feature as it prevents cargo receptors from associating with the growing isolation membrane in the absence of their respective cargo.
We found that Atg19 contains multiple Atg8 binding sites. One of these sites localizes to two phenylalanine residues (F376 and F379) that align to the LIR motif of human p62. Given the hydrophobic nature and the spacing between these two residues it is likely that they bind to Atg8 in a manner that is similar to the binding of other LIR motifs to Atg8 family proteins13-15, 31. In fact it is becoming increasingly clear that LIR motifs can deviate from the canonical W/F/YxxL/I/V sequence31-34. In addition, we found that mutation of P385 and E386 to alanine negatively affected Atg8 binding by Atg19, in particular in the context of other Atg8 binding mutations. This site shows no resemblance to previously identified LIR motifs. Interestingly P385 and E386 are also found in the p62 (Fig. 3c). Future studies will have to elucidate how P385 and E386 contribute to Atg8 binding. Atg19 may contain another Atg8 binding site N-terminally of F376 (Fig. 3b). In combination the multiple Atg8 binding sites result in high avidity interactions with Atg8 family proteins. This in turn favours the interaction with Atg8 family proteins when they are locally concentrated on the isolation membrane.
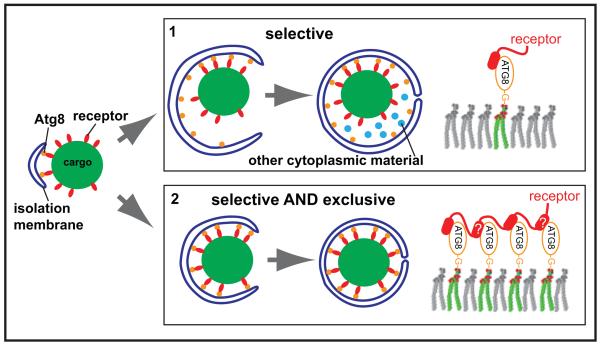
In case of Atg19, multiple sites per se are not required for the interaction of Atg19 with Atg8 as the C-terminus of Atg34 that lacks detectable Atg8-binding sites beside the known C-terminal LIR motif is able to functionally replace the C-terminus of Atg19 during bulk autophagy. Instead, the multiple Atg8 interaction sites in the C-terminus of Atg19 may provide sufficient energy to tightly wrap the membrane around the cargo and therefore allow the exclusion of non-cargo material during selective autophagy (Fig. 7). Consistently, Atg19 was sufficient for tight membrane-cargo apposition in a fully reconstituted system and this activity correlated with the number of Atg8-Atg19 interactions (Fig. 5).
Figure 7.
Model for the differential actions of cargo receptors during selective and bulk autophagy. In order to promote exclusion of non-cargo material cargo receptors interact with Atg8 via multiple sites to wrap the membrane around the cargo material.
Recently it has been proposed that membrane localized Atg8 assembles together with the Atg12–Atg5-Atg16 complex into a membrane scaffold on the isolation membrane and that the cargo receptor Atg32 disrupts this scaffold due to its LIR-dependent interaction with Atg835. Thus tight, LIR-dependent interactions of cargo receptors with Atg8 family proteins on the membrane may more efficiently disrupt this putative scaffold.
The Atg19-Atg34 chimera was rather effective in the in vitro reconstitution assay (Fig. 5). This may be due the fact that this assay is less stringent then the in vivo situation where the Cvt particles are around 150nm in diameter as opposed to 2μm for the cargo mimetic beads in the reconstituted assay.
Multiple Atg8 binding sites in Atg19 were specifically needed for the selective Cvt pathway but not for Atg19 dependent delivery of prApe1 to the vacuole during bulk autophagy. Consistently, for Cvt vesicles the membrane is tightly apposing the prApe1 particle22. During bulk autophagy where the prApe1 particle is tethered to the membrane of larger autophagosome22 the faster growth of the isolation membrane may outpace the membrane bending activity of Atg19. Alternatively, during selective autophagy the isolation membrane may be formed directly around the cargo, while during bulk autophagy the cargo may be tethered to a pre-existing isolation membrane not allowing the receptor to mediate tight membrane cargo apposition. Furthermore, a lower density of Atg8 on the membrane during bulk autophagy or posttranslational modification of the Atg19 C-terminus may reduce the membrane bending capacity of Atg19.
Interestingly, the mammalian cargo receptor NBR1 also contains two LIR motifs36. Furthermore, p62 may utilize oligomerization in order to increase the density of Atg8 interaction site to mediate tight apposition of the isolation membrane and the cargo10. Thus, many autophagic cargo receptors may contribute to the bending of the isolation membrane around cellular cargo to ensure exclusivity of the pathway and thereby prevent loss of non-cargo material into the lysosomal system.
Supplementary Material
Acknowledgements
We thank Claudine Kraft and Thorsten Brach for providing reagents and Graham Warren for critical comments on the manuscript. Funding by the University of Vienna is gratefully acknowledged. The research leading to these results has received funding from the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement No. 260304, from the FWF Austrian Science Fund (grant number P25546-B20) and by the EMBO Young Investigator Program to S.M.. We also acknowledge funding by the VIPS Program of the Austrian Federal Ministry of Science and Research and the City of Vienna to J. S.-M. and funding by the Austrian FFG to C.A. The authors declare no conflict of interests.
References
- 1.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 2.Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 3.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 4.Orsi A, Polson HEJ, Tooze SA. Membrane trafficking events that partake in autophagy. Current Opinion in Cell Biology. 2010;22:150–156. doi: 10.1016/j.ceb.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 5.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 6.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-Activated Protein Kinase Connects Energy Sensing to Mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine B, Kroemer G. Autophagy in the Pathogenesis of Disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Youle RJ, van der Bliek AM. Mitochondrial Fission, Fusion, and Stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirkin V, McEwan DG, Novak I, Dikic I. A Role for Ubiquitin in Selective Autophagy. Molecular Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 12.Pankiv S, et al. p62/SQSTM1 Binds Directly to Atg8/LC3 to Facilitate Degradation of Ubiquitinated Protein Aggregates by Autophagy. Journal of Biological Chemistry. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 13.Noda NN, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 14.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Letters. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 15.Ichimura Y, et al. Structural Basis for Sorting Mechanism of p62 in Selective Autophagy. Journal of Biological Chemistry. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 16.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 17.Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 Is a Receptor for the Cytoplasm-to-Vacuole Targeting Pathway. Molecular Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leber R, Silles E, Sandoval IV, Mazón M.a.J. Yol082p, a Novel CVT Protein Involved in the Selective Targeting of Aminopeptidase I to the Yeast Vacuole. Journal of Biological Chemistry. 2001;276:29210–29217. doi: 10.1074/jbc.M101438200. [DOI] [PubMed] [Google Scholar]
- 19.Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric alpha-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. The Journal of biological chemistry. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuga M, Gomi K, Klionsky DJ, Shintani T. Aspartyl aminopeptidase is imported from the cytoplasm to the vacuole by selective autophagy in Saccharomyces cerevisiae. The Journal of biological chemistry. 2011;286:13704–13713. doi: 10.1074/jbc.M110.173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki K, Morimoto M, Kondo C, Ohsumi Y. Selective autophagy regulates insertional mutagenesis by the Ty1 retrotransposon in Saccharomyces cerevisiae. Developmental Cell. 2011;21:358–365. doi: 10.1016/j.devcel.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 22.Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two Distinct Pathways for Targeting Proteins from the Cytoplasm to the Vacuole/Lysosome. The Journal of Cell Biology. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Developmental Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki K, Kondo C, Morimoto M, Ohsumi Y. Selective transport of alpha-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. The Journal of biological chemistry. 2010;285:30019–30025. doi: 10.1074/jbc.M110.143511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rozenknop A, et al. Characterization of the Interaction of GABARAPL-1 with the LIR Motif of NBR1. Journal of Molecular Biology. 2011;410:477–487. doi: 10.1016/j.jmb.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 27.Geng J, Baba M, Nair U, Klionsky DJ. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. The Journal of Cell Biology. 2008;182:129–140. doi: 10.1083/jcb.200711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romanov J, et al. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamada Y, et al. Tor-Mediated Induction of Autophagy via an Apg1 Protein Kinase Complex. The Journal of Cell Biology. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shintani T, Klionsky DJ. Cargo Proteins Facilitate the Formation of Transport Vesicles in the Cytoplasm to Vacuole Targeting Pathway. Journal of Biological Chemistry. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alemu EA, et al. ATG8 Family Proteins Act as Scaffolds for Assembly of the ULK Complex: SEQUENCE REQUIREMENTS FOR LC3-INTERACTING REGION (LIR) MOTIFS. Journal of Biological Chemistry. 2012;287:39275–39290. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Muhlinen N, et al. LC3C, Bound Selectively by a Noncanonical LIR Motif in NDP52, Is Required for Antibacterial Autophagy. Molecular Cell. 2012;48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farre JC, Burkenroad A, Burnett SF, Subramani S. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO reports. 2013;14:441–449. doi: 10.1038/embor.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki H, et al. Structural Basis of the Autophagy-Related LC3/Atg13 LIR Complex: Recognition and Interaction Mechanism. Structure. 2014;22:47–58. doi: 10.1016/j.str.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 35.Kaufmann A, Beier V, Franquelim Henri G., Wollert T. Molecular Mechanism of Autophagic Membrane-Scaffold Assembly and Disassembly. Cell. 2014;156:469–481. doi: 10.1016/j.cell.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 36.Kirkin V, et al. A Role for NBR1 in Autophagosomal Degradation of Ubiquitinated Substrates. Molecular Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.