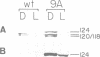Abstract
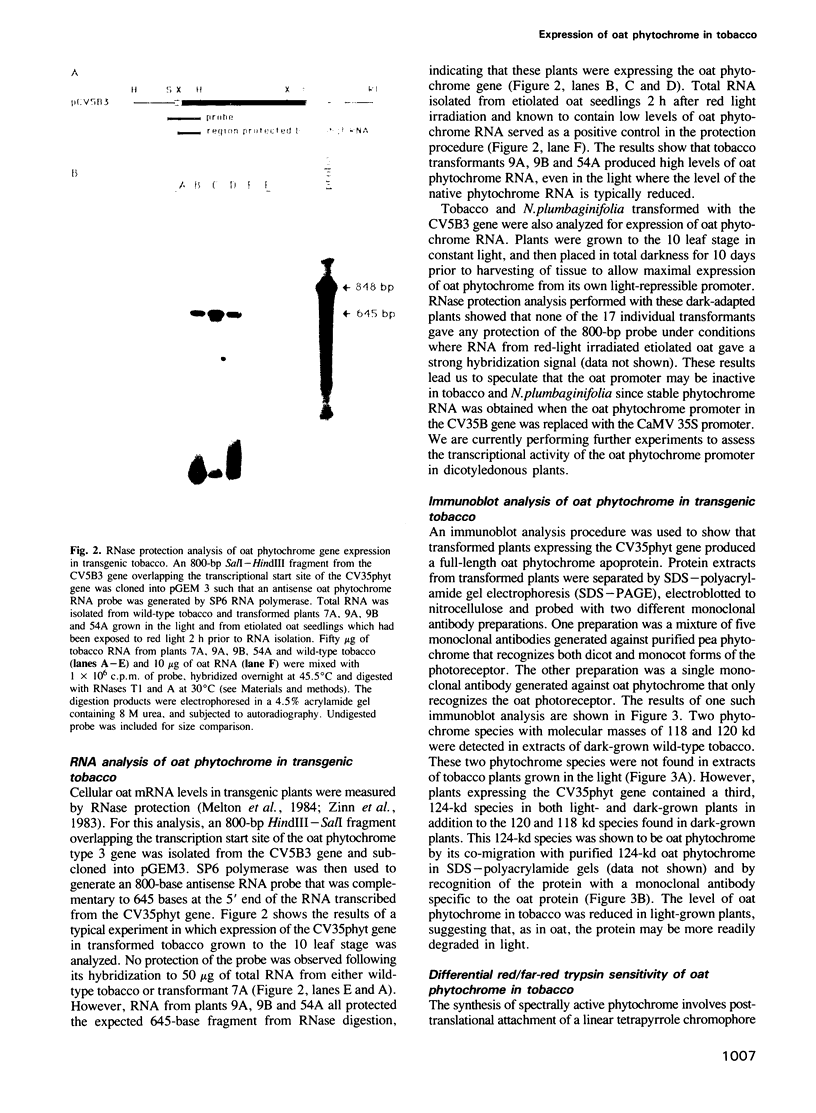
A chimeric oat phytochrome structural gene with an uninterrupted coding region was constructed for expression of the monocot protein in transgenic plants. The structural gene was placed under the transcriptional control of either a light-regulated oat phytochrome promoter or the constitutively active cauliflower mosaic virus 35S promoter. These genes were then introduced into Nicotiana tabacum and N.plumbaginifolia. None of the regenerated plants showed expression of oat phytochrome RNA when transcription was controlled by the oat promoter. In contrast, RNA was obtained in plants when the structural gene was functionally linked to the 35S promoter. Transformants expressing oat phytochrome RNA produced a full length 124-kd polypeptide that was recognized by oat-specific anti-phytochrome monoclonal antibodies. The oat protein was a substrate for chromophore addition in tobacco as judged by its red/far-red photoreversible sensitivity to trypsin degradation. Production of oat phytochrome in transgenic plants gave rise to increased phytochrome spectral activity in both light- and dark-grown plants. This increased phytochrome content resulted in phenotypic changes in transformed plants, including semi-dwarfism, darker green leaves, increased tillering and reduced apical dominance. The possible significance of expressing a biologically active phytochrome in transgenic plants is discussed.
Keywords: chimeric constructions, phenotypic changes, phytochrome expression, transgenic plants
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- A simple and general method for transferring genes into plants. Science. 1985 Mar 8;227(4691):1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey H. P., Barker R. F., Idler K. B., Lissemore J. L., Quail P. H. Analysis of cloned cDNA and genomic sequences for phytochrome: complete amino acid sequences for two gene products expressed in etiolated Avena. Nucleic Acids Res. 1985 Dec 9;13(23):8543–8559. doi: 10.1093/nar/13.23.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey H. P., Barker R. F., Idler K. B., Murray M. G., Quail P. H. Nucleotide sequence and characterization of a gene encoding the phytochrome polypeptide from Avena. Gene. 1987;61(3):339–348. doi: 10.1016/0378-1119(87)90197-1. [DOI] [PubMed] [Google Scholar]
- Hershey H. P., Colbert J. T., Lissemore J. L., Barker R. F., Quail P. H. Molecular cloning of cDNA for Avena phytochrome. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2332–2336. doi: 10.1073/pnas.81.8.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith B., Chua N. H. Monocot and dicot pre-mRNAs are processed with different efficiencies in transgenic tobacco. EMBO J. 1986 Oct;5(10):2419–2425. doi: 10.1002/j.1460-2075.1986.tb04516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarias J. C., Mercurio F. M. Structure function studies on phytochrome. Identification of light-induced conformational changes in 124-kDa Avena phytochrome in vitro. J Biol Chem. 1985 Feb 25;260(4):2415–2423. [PubMed] [Google Scholar]
- Litts J. C., Kelly J. M., Lagarias J. C. Structure-function studies on phytochrome. Preliminary characterization of highly purified phytochrome from Avena sativa enriched in the 124-kilodalton species. J Biol Chem. 1983 Sep 25;258(18):11025–11031. [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio F. M., Houghten R. A., Lagarias J. C. Site-directed antisera to the chromophore binding site of phytochrome: characterization and cross-reactivity. Arch Biochem Biophys. 1986 Jul;248(1):35–42. doi: 10.1016/0003-9861(86)90398-x. [DOI] [PubMed] [Google Scholar]
- Nagy F., Kay S. A., Chua N. H. Gene regulation by phytochrome. Trends Genet. 1988 Feb;4(2):37–42. doi: 10.1016/0168-9525(88)90064-9. [DOI] [PubMed] [Google Scholar]
- Odell J. T., Nagy F., Chua N. H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. 1985 Feb 28-Mar 6Nature. 313(6005):810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- Pratt L. H., Kidd G. H., Coleman R. A. An immunochemical characterization of the phytochrome destruction reaction. Biochim Biophys Acta. 1974 Sep 13;365(1):93–107. doi: 10.1016/0005-2795(74)90253-0. [DOI] [PubMed] [Google Scholar]
- Quail P. H., Colbert J. T., Peters N. K., Christensen A. H., Sharrock R. A., Lissemore J. L. Phytochrome and the regulation of the expression of its genes. Philos Trans R Soc Lond B Biol Sci. 1986 Nov 17;314(1166):469–480. doi: 10.1098/rstb.1986.0066. [DOI] [PubMed] [Google Scholar]
- Shanklin J., Jabben M., Vierstra R. D. Red light-induced formation of ubiquitin-phytochrome conjugates: Identification of possible intermediates of phytochrome degradation. Proc Natl Acad Sci U S A. 1987 Jan;84(2):359–363. doi: 10.1073/pnas.84.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock R. A., Lissemore J. L., Quail P. H. Nucleotide and amino acid sequence of a Cucurbita phytochrome cDNA clone: identification of conserved features by comparison with Avena phytochrome. Gene. 1986;47(2-3):287–295. doi: 10.1016/0378-1119(86)90072-7. [DOI] [PubMed] [Google Scholar]
- Vierstra R. D., Langan S. M., Haas A. L. Purification and initial characterization of ubiquitin from the higher plant, Avena sativa. J Biol Chem. 1985 Oct 5;260(22):12015–12021. [PubMed] [Google Scholar]
- Vierstra R. D., Quail P. H. Photochemistry of 124 kilodalton Avena phytochrome in vitro. Plant Physiol. 1983 May;72(1):264–267. doi: 10.1104/pp.72.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski P., Joos H., Genetello C., Leemans J., Montagu M. V., Schell J. Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J. 1983;2(12):2143–2150. doi: 10.1002/j.1460-2075.1983.tb01715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn K., DiMaio D., Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983 Oct;34(3):865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]