Abstract
Microtiter plates with 96 wells are being increasingly used for biofilm studies due to their high throughput, low cost, easy handling, and easy application of several analytical methods to evaluate different biofilm parameters. These methods provide bulk information about the biofilm formed in each well but lack in detail, namely, regarding the spatial location of the biofilms. This location can be obtained by microscopy observation using optical and electron microscopes, but these techniques have lower throughput and higher cost and are subjected to equipment availability. This work describes a differential crystal violet (CV) staining method that enabled the determination of the spatial location of Escherichia coli biofilms formed in the vertical wall of shaking 96-well plates. It was shown that the biofilms were unevenly distributed on the wall with denser cell accumulation near the air-liquid interface. The results were corroborated by scanning electron microscopy and a correlation was found between biofilm accumulation and the wall shear strain rates determined by computational fluid dynamics. The developed method is quicker and less expensive and has a higher throughput than the existing methods available for spatial location of biofilms in microtiter plates.
1. Introduction
Biofilms are defined as structured microbial communities that are attached to a surface and encapsulated within a self-produced matrix [1, 2]. They constitute a serious problem for public health because of the increased resistance of biofilm-associated microorganisms to antimicrobial agents and their potential to cause infections in patients with indwelling medical devices [1, 3].
Intensive studies on the mechanisms of biofilm formation and resistance have encouraged the development of different in vitro platforms, such as microtiter plates (MTPs), which are one of the most widely used biofilm model systems [4, 5]. In these systems, biofilms are formed on the bottom and on the wall [6] of the microtiter plate wells (most commonly a 96-well plate) or they are grown on the surface of a coupon placed in the wells of the MTP (most commonly a 6-, 12-, or 24-well plate). The large number of advantages offered by these straightforward and user-friendly systems explain their widespread use (Table 1). In addition, several standard assays are available for the determination of different parameters related to the biofilm in MTPs [7]. They can be categorized into biofilm biomass assays (quantitation of matrix and both living and dead cells), viability assays (determination of viable cells), and matrix quantitation assays (through specific staining of matrix components) (Table 1). Microtiter plates have been intensively used in clinical research for screening of antimicrobial compounds [8, 9] and for studying biofilm formation [10, 11] and inhibition [12, 13]. The most widely used method for following biofilm formation in MTPs is the crystal violet (CV) staining, derived from the original Christensen et al. [14] method, which only measured biofilm biomass at the bottom of the well. CV is a basic dye that stains both living and dead cells by binding to negatively charged surface molecules and polysaccharides in the extracellular matrix of biofilms [15]. Later, the CV assay was modified to increase its accuracy and to allow for biofilm biomass quantitation in the entire well by the solubilization of the dye [16, 17]. This method can therefore be considered a bulk method which provides information about the total amount of biofilms produced without revealing any information about biofilm localization. It has been shown that the biofilm distribution in the wall of a 96-well MTP may not be uniform when dynamic conditions are used (when the MTP is shaken with an orbital motion) [6]. This biofilm heterogeneity can be analyzed, for instance, by microscopy either using standard optical microscopy [18], confocal laser scanning microscopy (CLSM) [19], or scanning electron microscopy (SEM) [20]. The relative advantages and limitations of these two later techniques are presented in Table 2. It is interesting to point out that, for the most part, these microscopy analyses are made at the bottom of the well, disregarding the biofilm that is formed on the vertical wall. Both techniques enable spatial localization of the biofilms, but are inherently low throughput techniques, with a high cost and are subjected to equipment availability. The aim of this work was to develop a low cost and high throughput method that would enable the quantitation of the total amount of biofilm produced inside a well but providing the same information about the spatial location of the formed biofilm. A differential CV staining method was here developed by combining the high throughput features of the usual CV staining but enabling spatial localization of the biofilm without the use of expensive equipment.
Table 1.
Advantages of MTPs as biofilm reactors and standard methods for quantitation of biofilm parameters in MTPs.
Table 2.
Advantages and limitations of scanning electron microscopy (SEM) and confocal laser scanning microscopy (CLSM) on biofilm analysis.
| Technique | Advantages | Limitations | References |
|---|---|---|---|
| Scanning electron microscopy | High resolution Wide range of magnifications Good comparative information Ability to image complex shapes |
Not real time Requires additional sample preparation Limited quantification |
[45–49] |
|
| |||
| Confocal laser scanning microscopy | Living, fully hydrated samples Non-invasive Quantitative evaluation Reflection and fluorescence mode |
Low resolution Narrow range of magnifications Not applicable to thick biofilms |
[35, 47, 50–52] |
2. Materials and Methods
2.1. Computational Fluid Dynamic (CFD) Simulations
Numerical simulations were made in Ansys Fluent CFD package (version 13.0) as previously described [6]. A cylindrical well (diameter of 6.6 mm and height of 11.7 mm) was built in Design Modeller 13.0 and discretized into a grid of 18,876 hexahedral cells by Meshing 13.0. Simulation was made for a shaking diameter of 50 mm and frequency of 150 rpm. The location of the interface was determined by Ansys Fluent, as well as the magnitude of the shear strain rate. The time averaged shear strain rate was obtained by averaging the steady state shear strain rate of the liquid side during a complete orbit.
2.2. Biofilm Formation in 96-Well Microtiter Plates
Escherichia coli JM109(DE3) from Promega (USA) was used to form biofilms in 96-well microtiter plates because this strain has shown a good biofilm forming ability in both turbulent [21] and laminar [6] flow conditions. Its Its genotype is endA1, recA1, gyrA96, thi, hsdR17 (rk −, mk +), relA1, supE44, λ −, Δ(lac-proAB), (F′,traD36, proAB, lacIqZΔM15), and λ(DE3). Cells from an overnight culture were prepared as previously described [6] and biofilms were produced by pipetting 20 μL of these cells into six wells of sterile 96-well flat-bottomed microtiter plates (Orange Scientific, USA) filled with 180 μL of nutrient media. The media used was identical to the one described in [22], except for glucose. A glucose concentration of 1 g L−1 was used in this work since it has been demonstrated that this concentration originated the maximum biofilm amount at 24 h in different shaking conditions [6]. Microtiter plates were placed for 24 h at 30°C in an orbital shaking incubator with 50 mm of diameter at 150 rpm (CERTOMAT BS-1, Sartorius AG, Germany).
2.3. Biofilm Quantitation by Crystal Violet (CV) Staining
The amount of biofilm formed was measured using the CV dye in a differential form; this means that the well has been divided into four different sections (Figure 1(d)) and the corresponding vertical wall section was stained sequentially up to a maximum volume of 200 μL. Biofilm quantitation on the bottom of the well was performed by using 25 μL of CV for staining (as detailed below). Section 1 corresponded to a volume between 25 and 50 μL (and a height of 0.77 mm). Section 2 corresponded to a volume between 50 and 100 μL, section 3 corresponded to a volume between 100 and 150 μL and section 4 corresponded to a volume between 150 and 200 μL. Sections 2, 3, and 4 had an equivalent height of 1.54 mm.
Figure 1.
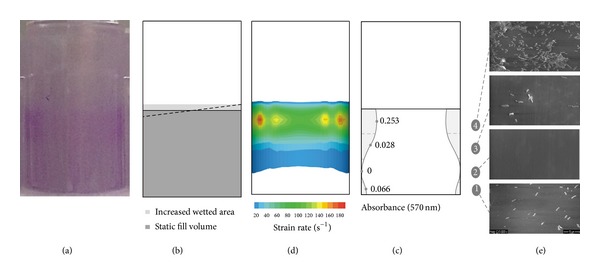
Biofilm localization in shaking 96-well microtiter plates placed in a 50 mm incubator at 150 rpm. (a) Photograph of a well stained with crystal violet; (b) schematic representation of a well where the dark grey area corresponds to the wetted area without shaking, the light grey area represents the area increase upon shaking, and the dotted line depicts the inclination of the air-liquid interface; (c) time averaged shear strain rates (values below 20 s−1 are not represented); (d) illustration of the biofilm distribution on the vertical wall assayed by the differential CV staining; (e) representative scanning electron micrographs of the wall sections defined in image (d); (5000x magnification bar = 10 μm).
Prior to CV staining, the contents of the microtiter plates were discarded and the wells were washed with 200 μL of sterile water to remove non-adherent bacteria [16]. Then, the biofilms were fixed with 250 μL of 96% ethanol [23]. The first section of the well to be quantified was the bottom (not represented in Figure 1(d)), which corresponds to a volume of 25 μL and a height of 0.77 mm. The cells adhered to this region were stained for 5 min with the correspondent volume of 1% (v/v) crystal violet (Merck, Germany) and the dye bound to this specific region was solubilized with 200 μL of 33% (v/v) acetic acid (VWR, Portugal). The absorbance was measured at 570 nm using a microtiter plate reader (SpectraMax M2E, Molecular Devices, UK). The biofilms formed in the successive sections of the well (sections 1 to 4 of Figure 1(d)) were quantified using the described procedure, specifically using the CV volume equivalent to the maximum level of each section (section 1—50 μL; section 2—100 μL; section 3—150 μL; section 4—200 μL). The absorbance corresponding to each of the defined sections and presented on Figure 1(d) was calculated by subtracting the absorbances from the previous sections and considering the dilution factor. To quantify the biofilm formed in each of the well sections, six replica wells were used per experiment and three independent experiments were performed.
2.4. Scanning Electron Microscopy (SEM)
Microtiter plate wells containing 24 h old biofilms were imaged by SEM as previously described [24]. Briefly, biofilms were fixed in 3% (w/w) glutaraldehyde in cacodylate buffer, dehydrated with ethanol and hexamethyldisilazane (HMDS), and sputter-coated with a palladium-gold thin film. The different sections along the vertical wall of three independent wells were observed with a SEM/EDS system (FEI Quanta 400FEG ESEM/EDAX Genesis X4 M, FEI Company, USA).
3. Results and Discussion
The effect of orbital shaking on biofilm formation in 96-well microtiter plates was firstly assessed by the conventional procedure of crystal violet staining [6, 16]. Through direct observation of the stained wells (prior to solubilization of the dye), it was possible to observe that biofilms were mainly formed on the wall and not on the bottom of the well. Moreover, since the amount of produced biofilms is proportional to the amount of CV adsorbed, it seemed that the biofilm was unevenly distributed in the cylindrical wall and higher amounts were formed closer to the air-liquid interface (Figure 1(a)).
It is widely known that hydrodynamics affects biofilm formation [25–27] as a result of the shear forces that can modulate cell adhesion to a given surface [28–31]. Even though microtiter plates are broadly accepted as biofilm formation reactors by the scientific community, there is still a lack of information on the impact of hydrodynamics on biofilm formation in this system. In this work, the flow inside the wells was simulated using computational fluid dynamics (Figures 1(b) and 1(c)) in order to find out if the hydrodynamic conditions were related to the uneven biofilm distribution that was visible to the naked eye after CV staining (Figure 1(a)). Figure 1(b) represents a scale model of a microwell with the wetted area without orbital motion, the area increase upon shaking, and the inclination of the air-liquid interface. For the shaking conditions chosen for this work (50 mm of orbital diameter and 150 rpm), an area gain of 8.5% and a maximum angle of 7.8° were obtained. Similar to what was observed in the photograph of the stained well (Figure 1(a)), the shear strain rate distribution was not uniform along the wall, being much higher in the liquid side near the interface (Figure 1(c)). In this region, CFD simulations revealed some spots, corresponding to a maximum shear strain rate of 180 s−1, which are related to regions of unstable vortices [6].
Observing the results of the differential CV staining (Figure 1(d)), the amount of biofilm detected along the different sections of the wall was not constant. The highest value of absorbance (corresponding to the highest amount of biofilm biomass) was measured in the region located immediately below the liquid level (section 4). In the intermediate sections of the well, the absorbance values were about 10 times lower than near the interface (section 3) or almost zero (section 2), increasing slightly in the section closer to the bottom of the well (section 1).
In order to validate the results obtained by the differential CV staining, SEM analysis was performed on the vertical wall of the wells. Figure 1(e) shows scanning electron micrographs representative of the four sections defined in the application of the differential CV staining (Figure 1(d)). SEM analysis showed that E. coli adhesion varied across the wall and that higher attachment occurred closer to the air-liquid interface (section 4) compared to the intermediate (sections 2 and 3) and near bottom (section 1) regions of the well. Biofilms consisting of bacterial aggregates were observed near the interface, while in the lower regions of the well there were only few cells randomly distributed on the surface (Figure 1(e)).
Most likely the spatial location of the biofilms (assayed by differential staining and SEM analysis) is conditioned by the non-homogeneous distribution of shear strain calculated by CFD during shaking. The higher cell density of the biofilms formed closer to the interface is probably due to the higher oxygen and substrate mass transfer from the bulk solution to the biofilm [32], resulting from a more efficient liquid mixing in this region. These results are in agreement with previous studies [29, 33, 34] showing that higher shear forces promote the formation of denser biofilms.
Both methods used for biofilm analysis in this work (differential CV staining and SEM) enabled a higher detail in analysis of biofilms grown in 96-well microtiter plates, which is not achieved by the spectrophotometric methods commonly used in MTPs (Table 1). Indeed, methods such as the traditional CV assay provide bulk data from a biofilm and are classified as macroscale methods, whereas SEM is a microscale technique [24]. It is interesting to notice that the differential CV staining presented in this work corresponds to an intermediate scale between the traditional CV assay and SEM analysis. The traditional CV assay quantifies the biofilm formed on the wall and bottom of each well of a microtiter plate, which corresponds to an area of about 146 mm2 (for a 96-well plate), while the method proposed here evaluates wall sections of about 14 and 28 mm2. In comparison, the area covered by a SEM image taken at 5000x magnification was of 2 × 10−3 mm2, and this technique enables detailed analysis of individual cells within the biofilm rather than simply determining their localization. SEM offers higher magnification (ranging from 20x to approximately 30,000x) and resolution (from 50 to 100 nm), together with the ability of imaging complex shapes (Table 2). It is also highly recommended for the visualization of cellular morphology, cell-to-cell interactions, and matrix components within biofilms [35]. However, one must bear in mind that most laboratories are not equipped with an electron microscope and this technique has a considerably lower throughput than the methods described in Table 1.
Observation of biofilms formed on the bottom of the wells of microtiter plates is very common using optical microscopes at magnifications of 100 to 200x, which typically cover areas of 0.3 mm2 [36–39]. These observations have provided some information about the architecture of the biofilms formed on the bottom of the wells (particularly when CLSM is used) but usually disregard the biofilm that forms on the vertical wall. It has been shown that in dynamic conditions the amount of biofilm formed on the vertical wall can be higher than the one formed on the bottom of the well [6], and therefore a method was developed in this work to determine the spatial localization of these biofilms in a high throughput manner, using common laboratory equipment. The area under analysis in each of the four sections defined for the differential staining is equivalent to the area analyzed by optical microscopy using a 15x magnification. This area could be further reduced (thus increasing the precision of the method) by dividing the well in sections of smaller height.
The novel approach presented in this work demonstrates that the CV dye can be extremely useful in locating the adherent cells in microtiter plates when used in a differential way. The method is slightly more laborious and slower than the traditional staining procedure, but it requires fewer resources and has higher throughput than other techniques that are used to determine the spatial location of biofilms.
Acknowledgments
The authors acknowledge the financial support provided by Operational Programme for Competitiveness Factors (COMPETE), European Fund for Regional Development-FEDER, and the Portuguese Foundation for Science and Technology (FCT), through Projects PTDC/EBB-BIO/102863/2008 and PTDC/EBB-BIO/104940/2008. Luciana Gomes acknowledges the receipt of a Ph.D. Grant from FCT (SFRH/BD/80400/2011). João Miranda and José Araújo (Transport Phenomena Research Center, FEUP) are acknowledged for the numerical simulations.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annual Review of Microbiology. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 3.Hancock V, Ferrières L, Klemm P. Biofilm formation by asymptomatic and virulent urinary tract infectious Escherichia coli strains. FEMS Microbiology Letters. 2007;267(1):30–37. doi: 10.1111/j.1574-6968.2006.00507.x. [DOI] [PubMed] [Google Scholar]
- 4.Coenye T, Nelis HJ. In vitro and in vivo model systems to study microbial biofilm formation. Journal of Microbiological Methods. 2010;83(2):89–105. doi: 10.1016/j.mimet.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 5.Teodósio JS, Simões M, Melo LF, Mergulhão FJ. Platforms for in vitro biofilm studies. In: Simões M, Mergulhão F, editors. Biofilms in Bioengineering. New York. NY, USA: Nova Science; 2013. pp. 45–62. [Google Scholar]
- 6.Moreira JMR, Gomes LC, Araújo JDP, et al. The effect of glucose concentration and shaking conditions on Escherichia coli biofilm formation in microtiter plates. Chemical Engineering Science. 2013;94:192–199. [Google Scholar]
- 7.Peeters E, Nelis HJ, Coenye T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. Journal of Microbiological Methods. 2008;72(2):157–165. doi: 10.1016/j.mimet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Erriu M, Pili FMG, Tuveri E, et al. Oil essential mouthwashes antibacterial activity against Aggregatibacter actinomycetemcomitans: a comparison between antibiofilm and antiplanktonic effects. International Journal of Dentistry. 2013;2013:5 pages. doi: 10.1155/2013/164267.164267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minardi D, Cirioni O, Ghiselli R, et al. Efficacy of tigecycline and rifampin alone and in combination against Enterococcus faecalis biofilm infection in a rat model of ureteral stent. Journal of Surgical Research. 2012;176(1):1–6. doi: 10.1016/j.jss.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez C, Mende K, Beckius M, et al. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infectious Diseases. 2013;13, article 47 doi: 10.1186/1471-2334-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hell E, Giske C, Hultenby K, et al. Attachment and biofilm forming capabilities of Staphylococcus epidermidis strains isolated from preterm infants. Current Microbiology. 2013;67(6):712–717. doi: 10.1007/s00284-013-0425-3. [DOI] [PubMed] [Google Scholar]
- 12.Lin M-H, Chang F-R, Hua M-Y, Wu Y-C, Liu S-T. Inhibitory effects of 1,2,3,4,6-penta-O-galloyl-β-D-glucopyranose on biofilm formation by Staphylococcus aureus . Antimicrobial Agents and Chemotherapy. 2011;55(3):1021–1027. doi: 10.1128/AAC.00843-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De La Fuente-Núñez C, Korolik V, Bains M, et al. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrobial Agents and Chemotherapy. 2012;56(5):2696–2704. doi: 10.1128/AAC.00064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen GD, Simpson WA, Younger JJ. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. Journal of Clinical Microbiology. 1985;22(6):996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Yan Z, Xu J. Quantitative variation of biofilms among strains in natural populations of Candida albicans . Microbiology. 2003;149(2):353–362. doi: 10.1099/mic.0.25932-0. [DOI] [PubMed] [Google Scholar]
- 16.Stepanović S, Vuković D, Dakić I, Savić B, Švabić-Vlahović M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. Journal of Microbiological Methods. 2000;40(2):175–179. doi: 10.1016/s0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 17.Crémet L, Corvec S, Batard E, et al. Comparison of three methods to study biofilm formation by clinical strains of Escherichia coli . Diagnostic Microbiology and Infectious Disease. 2013;75(3):252–255. doi: 10.1016/j.diagmicrobio.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Martinez LR, Casadevall A. Specific antibody can prevent fungal biofilm formation and this effect correlates with protective efficacy. Infection and Immunity. 2005;73(10):6350–6362. doi: 10.1128/IAI.73.10.6350-6362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bridier A, Le Coq D, Dubois-Brissonnet F, Thomas V, Aymerich S, Briandet R. The spatial architecture of Bacillus subtilis biofilms deciphered using a surface-associated model and in situ imaging. PLoS ONE. 2011;6(1) doi: 10.1371/journal.pone.0016177.e16177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kostenko V, Salek MM, Sattari P, Martinuzzi RJ. Staphylococcus aureus biofilm formation and tolerance to antibiotics in response to oscillatory shear stresses of physiological levels. FEMS Immunology and Medical Microbiology. 2010;59(3):421–431. doi: 10.1111/j.1574-695X.2010.00694.x. [DOI] [PubMed] [Google Scholar]
- 21.Teodósio JS, Simões M, Mergulhão FJ. The influence of nonconjugative Escherichia coli plasmids on biofilm formation and resistance. Journal of Applied Microbiology. 2012;113(2):373–382. doi: 10.1111/j.1365-2672.2012.05332.x. [DOI] [PubMed] [Google Scholar]
- 22.Teodósio JS, Simões M, Melo LF, Mergulhão FJ. Flow cell hydrodynamics and their effects on E. coli biofilm formation under different nutrient conditions and turbulent flow. Biofouling. 2011;27(1):1–11. doi: 10.1080/08927014.2010.535206. [DOI] [PubMed] [Google Scholar]
- 23.Shakeri S, Kermanshahi RK, Moghaddam MM, Emtiazi G. Assessment of biofilm cell removal and killing and biocide efficacy using the microtiter plate test. Biofouling. 2007;23(2):79–86. doi: 10.1080/08927010701190011. [DOI] [PubMed] [Google Scholar]
- 24.Gomes L, Moreira JMR, Miranda JM, et al. Macroscale versus microscale methods for physiological analysis of biofilms formed in 96-well microtiter plates. Journal of Microbiological Methods. 2013;95(3):342–349. doi: 10.1016/j.mimet.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Tay J-H. The essential role of hydrodynamic shear force in the formation of biofilm and granular sludge. Water Research. 2002;36(7):1653–1665. doi: 10.1016/s0043-1354(01)00379-7. [DOI] [PubMed] [Google Scholar]
- 26.Stoodley P, Wilson S, Hall-Stoodley L, Boyle JD, Lappin-Scott HM, Costerton JW. Growth and detachment of cell clusters from mature mixed-species biofilms. Applied and Environmental Microbiology. 2001;67(12):5608–5613. doi: 10.1128/AEM.67.12.5608-5613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wäsche S, Horn H, Hempel DC. Influence of growth conditions on biofilm development and mass transfer at the bulk/biofilm interface. Water Research. 2002;36(19):4775–4784. doi: 10.1016/s0043-1354(02)00215-4. [DOI] [PubMed] [Google Scholar]
- 28.Busscher HJ, van der Mei HC. Microbial adhesion in flow displacement systems. Clinical Microbiology Reviews. 2006;19(1):127–141. doi: 10.1128/CMR.19.1.127-141.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simões M, Pereira MO, Sillankorva S, Azeredo J, Vieira MJ. The effect of hydrodynamic conditions on the phenotype of Pseudomonas fluorescens biofilms. Biofouling. 2007;23(4):249–258. doi: 10.1080/08927010701368476. [DOI] [PubMed] [Google Scholar]
- 30.Teodósio JS, Silva FC, Moreira JMR, et al. Flow cells as quasi ideal systems for biofouling simulation of industrial piping systems. Biofouling. 2013;29(8):953–966. doi: 10.1080/08927014.2013.821467. [DOI] [PubMed] [Google Scholar]
- 31.Van Loosdrecht MCM, Eikelboom D, Gjaltema A, Mulder A, Tijhuis L, Heijnen JJ. Biofilm structures. Water Science and Technology. 1995;32(8):35–43. [Google Scholar]
- 32.Moreira JMR, Teodósio JS, Silva FC, et al. Influence of flow rate variation on the development of Escherichia coli biofilms. Bioprocess and Biosystems Engineering. 2013;36(11):1787–1796. doi: 10.1007/s00449-013-0954-y. [DOI] [PubMed] [Google Scholar]
- 33.Kwok WK, Picioreanu C, Ong SL, et al. Influence of biomass production and detachment forces on biofilm structures in a biofilm airlift suspension reactor. Biotechnology and Bioengineering. 1998;58(4):400–407. doi: 10.1002/(sici)1097-0290(19980520)58:4<400::aid-bit7>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 34.Rochex A, Godon J-J, Bernet N, Escudié R. Role of shear stress on composition, diversity and dynamics of biofilm bacterial communities. Water Research. 2008;42(20):4915–4922. doi: 10.1016/j.watres.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 35.Bridier A, Meylheuc T, Briandet R. Realistic representation of Bacillus subtilis biofilms architecture using combined microscopy (CLSM, ESEM and FESEM) Micron. 2013;48:65–69. doi: 10.1016/j.micron.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 36.White MG, Piccirillo S, Dusevich V, Law DJ, Kapros T, Honigberg SM. Flo11p adhesin required for meiotic differentiation in Saccharomyces cerevisiae minicolonies grown on plastic surfaces. FEMS Yeast Research. 2011;11(2):223–232. doi: 10.1111/j.1567-1364.2010.00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quave CL, Estévez-Carmona M, Compadre CM, et al. Ellagic acid derivatives from Rubus ulmifolius inhibit Staphylococcus aureus biofilm formation and improve response to antibiotics. PLoS ONE. 2012;7(1) doi: 10.1371/journal.pone.0028737.e28737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lizcano A, Chin T, Sauer K, Tuomanen EI, Orihuela CJ. Early biofilm formation on microtiter plates is not correlated with the invasive disease potential of Streptococcus pneumoniae . Microbial Pathogenesis. 2010;48(3-4):124–130. doi: 10.1016/j.micpath.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melo AS, Colombo AL, Arthington-Skaggs BA. Paradoxical growth effect of caspofungin observed on biofilms and planktonic cells of five different Candida species. Antimicrobial Agents and Chemotherapy. 2007;51(9):3081–3088. doi: 10.1128/AAC.00676-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duetz WA. Microtiter plates as mini-bioreactors: miniaturization of fermentation methods. Trends in Microbiology. 2007;15(10):469–475. doi: 10.1016/j.tim.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Honraet K, Goetghebeur E, Nelis HJ. Comparison of three assays for the quantification of Candida biomass in suspension and CDC reactor grown biofilms. Journal of Microbiological Methods. 2005;63(3):287–295. doi: 10.1016/j.mimet.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 42.Kumar S, Wittmann C, Heinzle E. Review: minibioreactors. Biotechnology Letters. 2004;26(1):1–10. doi: 10.1023/b:bile.0000009469.69116.03. [DOI] [PubMed] [Google Scholar]
- 43.Azevedo NF, Lopes SP, Keevil CW, Pereira MO, Vieira MJ. Time to “go large” on biofilm research: advantages of an omics approach. Biotechnology Letters. 2009;31(4):477–485. doi: 10.1007/s10529-008-9901-4. [DOI] [PubMed] [Google Scholar]
- 44.Toté K, Berghe DV, Maes L, Cos P. A new colorimetric microtitre model for the detection of Staphylococcus aureus biofilms. Letters in Applied Microbiology. 2008;46(2):249–254. doi: 10.1111/j.1472-765X.2007.02298.x. [DOI] [PubMed] [Google Scholar]
- 45.Norton TA, Thompson RC, Pope J, et al. Using confocal laser scanning microscopy, scanning electron microscopy and phase contrast light microscopy to examine marine biofilms. Aquatic Microbial Ecology. 1998;16(2):199–204. [Google Scholar]
- 46.Serra DO, Richter AM, Klauck G, et al. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio. 2013;4(2):e100103–e100113. doi: 10.1128/mBio.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart PS, Murga R, Srinivasan R, De Beer D. Biofilm structural heterogeneity visualized by three microscopic methods. Water Research. 1995;29(8):2006–2009. [Google Scholar]
- 48.Surman SB, Walker JT, Goddard DT, et al. Comparison of microscope techniques for the examination of biofilms. Journal of Microbiological Methods. 1996;25(1):57–70. [Google Scholar]
- 49.Hannig C, Follo M, Hellwig E, Al-Ahmad A. Visualization of adherent micro-organisms using different techniques. Journal of Medical Microbiology. 2010;59(1):1–7. doi: 10.1099/jmm.0.015420-0. [DOI] [PubMed] [Google Scholar]
- 50.Bridier A, Dubois-Brissonnet F, Boubetra A, Thomas V, Briandet R. The biofilm architecture of sixty opportunistic pathogens deciphered using a high throughput CLSM method. Journal of Microbiological Methods. 2010;82(1):64–70. doi: 10.1016/j.mimet.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 51.Fedel M, Caciagli P, Chistè V, et al. Microbial biofilm imaging ESEM vs. HVSEM. Imaging & Microscopy. 2007;9(2):44–47. [Google Scholar]
- 52.Palmer Jr. RJ, Haagensen JJ, Neu T, Sternberg C. Confocal microscopy of biofilms—spatiotemporal approaches. In: Pawley JB, editor. Handbook of Biological Confocal Microscopy. New York, NY, USA: Springer; 2006. pp. 870–888. [Google Scholar]


