Abstract
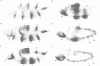
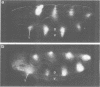
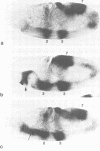
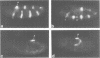
The pair-rule gene even-skipped (eve) plays a key role in the regulatory hierarchy governing segmentation in Drosophila. Here we describe the use of P-transformation and eve promoter fusions to identify cis elements that regulate the periodic seven-stripe eve pattern. A distal region of the eve promoter, located between -5.9 and -5.2 kb, controls autoregulation. Sequences from this region will induce striped expression of a heterologous hsp70 basal promoter in the presence, but not absence, of endogenous eve+ products. Autoregulatory activity was localized to a 200-bp region of the distal eve promoter. We also provide evidence that individual eve expression stripes are regulated by separate cis sequences. eve promoter sequences located between -4.7 and -3 kb upstream of the transcription start site are important for the initiation of stripe 3, whereas sequences between -1.7 and -0.4 kb are needed for stripes 2 and 7. It is possible that these latter regions are directly regulated by the products of gap genes.
Full text
PDF


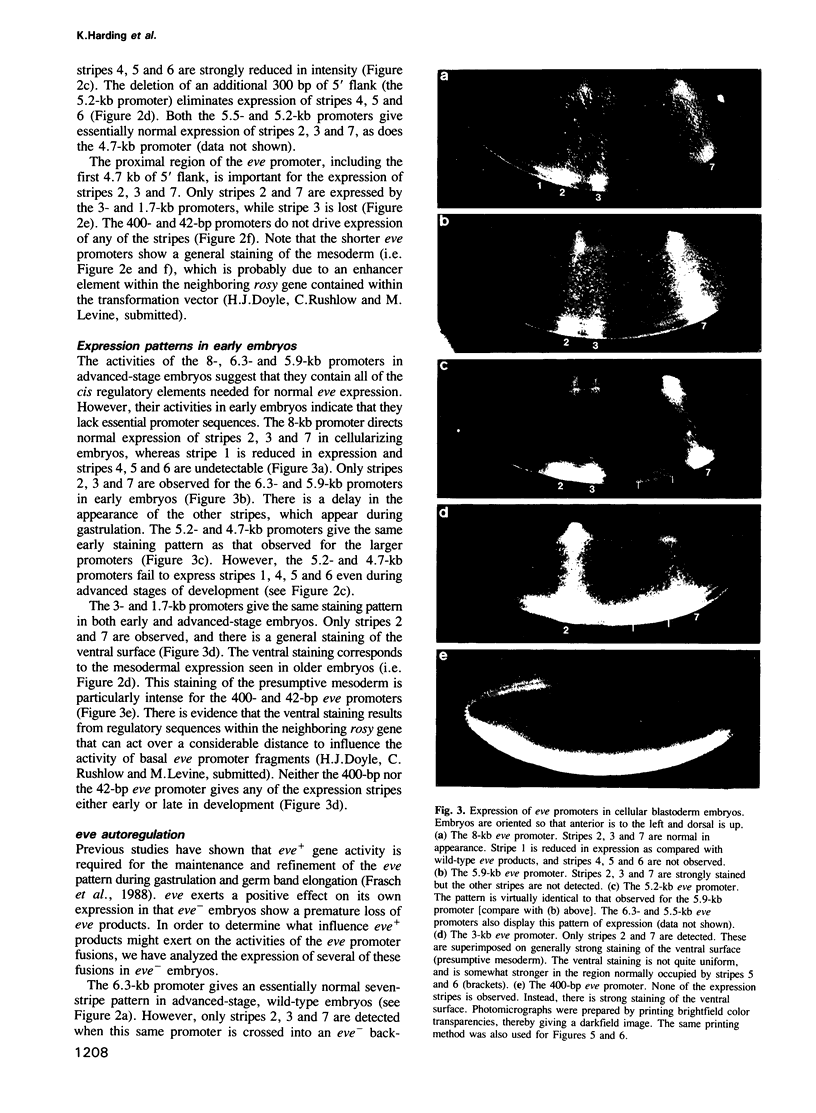

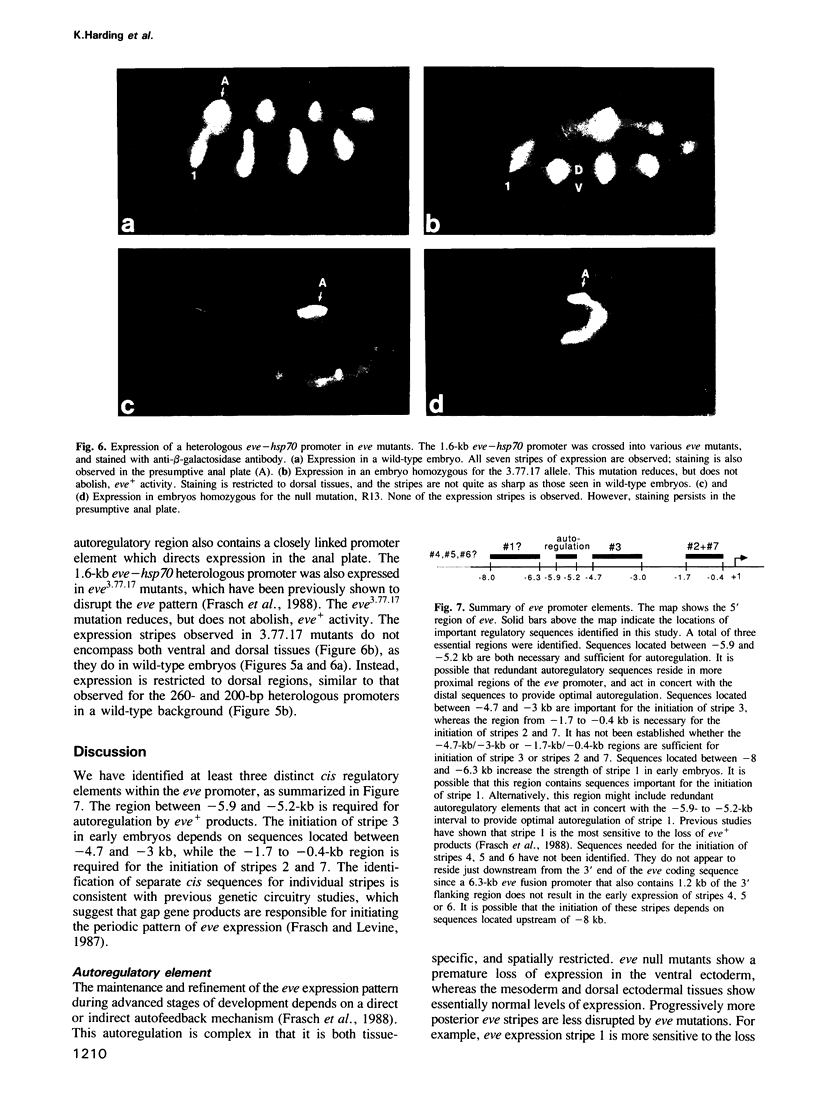


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987 Sep;101(1):1–22. [PubMed] [Google Scholar]
- Carroll S. B., Laughon A., Thalley B. S. Expression, function, and regulation of the hairy segmentation protein in the Drosophila embryo. Genes Dev. 1988 Jul;2(7):883–890. doi: 10.1101/gad.2.7.883. [DOI] [PubMed] [Google Scholar]
- Carroll S. B., Scott M. P. Zygotically active genes that affect the spatial expression of the fushi tarazu segmentation gene during early Drosophila embryogenesis. Cell. 1986 Apr 11;45(1):113–126. doi: 10.1016/0092-8674(86)90543-x. [DOI] [PubMed] [Google Scholar]
- DiNardo S., Kuner J. M., Theis J., O'Farrell P. H. Development of embryonic pattern in D. melanogaster as revealed by accumulation of the nuclear engrailed protein. Cell. 1985 Nov;43(1):59–69. doi: 10.1016/0092-8674(85)90012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S., O'Farrell P. H. Establishment and refinement of segmental pattern in the Drosophila embryo: spatial control of engrailed expression by pair-rule genes. Genes Dev. 1987 Dec;1(10):1212–1225. doi: 10.1101/gad.1.10.1212. [DOI] [PubMed] [Google Scholar]
- Fjose A., McGinnis W. J., Gehring W. J. Isolation of a homoeo box-containing gene from the engrailed region of Drosophila and the spatial distribution of its transcripts. Nature. 1985 Jan 24;313(6000):284–289. doi: 10.1038/313284a0. [DOI] [PubMed] [Google Scholar]
- Frasch M., Hoey T., Rushlow C., Doyle H., Levine M. Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 1987 Mar;6(3):749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M., Levine M. Complementary patterns of even-skipped and fushi tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes Dev. 1987 Nov;1(9):981–995. doi: 10.1101/gad.1.9.981. [DOI] [PubMed] [Google Scholar]
- Frasch M., Warrior R., Tugwood J., Levine M. Molecular analysis of even-skipped mutants in Drosophila development. Genes Dev. 1988 Dec;2(12B):1824–1838. doi: 10.1101/gad.2.12b.1824. [DOI] [PubMed] [Google Scholar]
- Gergen J. P., Butler B. A. Isolation of the Drosophila segmentation gene runt and analysis of its expression during embryogenesis. Genes Dev. 1988 Sep;2(9):1179–1193. doi: 10.1101/gad.2.9.1179. [DOI] [PubMed] [Google Scholar]
- Harding K., Rushlow C., Doyle H. J., Hoey T., Levine M. Cross-regulatory interactions among pair-rule genes in Drosophila. Science. 1986 Aug 29;233(4767):953–959. doi: 10.1126/science.3755551. [DOI] [PubMed] [Google Scholar]
- Hiromi Y., Gehring W. J. Regulation and function of the Drosophila segmentation gene fushi tarazu. Cell. 1987 Sep 11;50(6):963–974. doi: 10.1016/0092-8674(87)90523-x. [DOI] [PubMed] [Google Scholar]
- Hiromi Y., Kuroiwa A., Gehring W. J. Control elements of the Drosophila segmentation gene fushi tarazu. Cell. 1985 Dec;43(3 Pt 2):603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- Hoey T., Levine M. Divergent homeo box proteins recognize similar DNA sequences in Drosophila. Nature. 1988 Apr 28;332(6167):858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- Howard K., Ingham P. Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell. 1986 Mar 28;44(6):949–957. doi: 10.1016/0092-8674(86)90018-8. [DOI] [PubMed] [Google Scholar]
- Howard K., Ingham P., Rushlow C. Region-specific alleles of the Drosophila segmentation gene hairy. Genes Dev. 1988 Aug;2(8):1037–1046. doi: 10.1101/gad.2.8.1037. [DOI] [PubMed] [Google Scholar]
- Ingham P. W., Baker N. E., Martinez-Arias A. Regulation of segment polarity genes in the Drosophila blastoderm by fushi tarazu and even skipped. Nature. 1988 Jan 7;331(6151):73–75. doi: 10.1038/331073a0. [DOI] [PubMed] [Google Scholar]
- Ingham P. W., Ish-Horowicz D., Howard K. R. Correlative changes in homoeotic and segmentation gene expression in Krüppel mutant embryos of Drosophila. EMBO J. 1986 Jul;5(7):1659–1665. doi: 10.1002/j.1460-2075.1986.tb04409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988 Sep 1;335(6185):25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Karess R. E., Rubin G. M. Analysis of P transposable element functions in Drosophila. Cell. 1984 Aug;38(1):135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- Kornberg T. Engrailed: a gene controlling compartment and segment formation in Drosophila. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1095–1099. doi: 10.1073/pnas.78.2.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg T., Sidén I., O'Farrell P., Simon M. The engrailed locus of Drosophila: in situ localization of transcripts reveals compartment-specific expression. Cell. 1985 Jan;40(1):45–53. doi: 10.1016/0092-8674(85)90307-1. [DOI] [PubMed] [Google Scholar]
- Kuroiwa A., Hafen E., Gehring W. J. Cloning and transcriptional analysis of the segmentation gene fushi tarazu of Drosophila. Cell. 1984 Jul;37(3):825–831. doi: 10.1016/0092-8674(84)90417-3. [DOI] [PubMed] [Google Scholar]
- Lawrence P. A., Johnston P., Macdonald P., Struhl G. Borders of parasegments in Drosophila embryos are delimited by the fushi tarazu and even-skipped genes. 1987 Jul 30-Aug 5Nature. 328(6129):440–442. doi: 10.1038/328440a0. [DOI] [PubMed] [Google Scholar]
- Lawrence P. A., Morata G. Compartments in the wing of Drosophila: a study of the engrailed gene. Dev Biol. 1976 Jun;50(2):321–337. doi: 10.1016/0012-1606(76)90155-x. [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Ingham P., Struhl G. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell. 1986 Dec 5;47(5):721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Levine M. S., Hafen E., Kuroiwa A., Gehring W. J. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. 1984 Mar 29-Apr 4Nature. 308(5958):428–433. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. Hierarchical inductions of cell states: a model for segmentation in Drosophila. J Cell Sci Suppl. 1986;4:357–381. doi: 10.1242/jcs.1986.supplement_4.20. [DOI] [PubMed] [Google Scholar]
- Mismer D., Rubin G. M. Analysis of the promoter of the ninaE opsin gene in Drosophila melanogaster. Genetics. 1987 Aug;116(4):565–578. doi: 10.1093/genetics/116.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Kluding H., Jürgens G. Genes affecting the segmental subdivision of the Drosophila embryo. Cold Spring Harb Symp Quant Biol. 1985;50:145–154. doi: 10.1101/sqb.1985.050.01.020. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980 Oct 30;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982 Oct 22;218(4570):348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C. Vectors for P element-mediated gene transfer in Drosophila. Nucleic Acids Res. 1983 Sep 24;11(18):6341–6351. doi: 10.1093/nar/11.18.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4115–4119. doi: 10.1073/pnas.81.13.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]