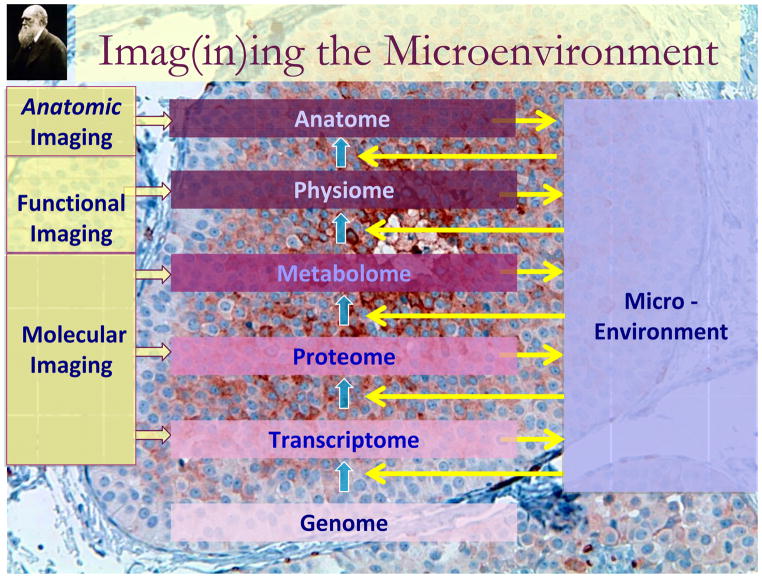
While it is commonly assumed that “cancer is a genetic disease”, it is significantly more complicated than that. Cancers are complex, evolving, multiscale ecosystems that are characterized by profound spatial and temporal heterogeneity. Understanding a cancer requires a detailed understanding of the complex dynamical system that is cancer. The interactions are nonlinear in that small changes in one variable can have large changes on another. These multiple interacting phenotypes and spatial scales can only be integrated and understood with the application of appropriate mathematical and computational models. Imaging is central to this investigation because it can characterize spatial variations in the tumor phenotype and environment in a non-destructive way so that, by re-interrogating at a later date, the system dynamics over time can be captured. Figure 1 illustrates a conceptual basis for the roles of imaging in understanding cancer biology. The effect of an altered genome is manifested by the complement of genes that are expressed, the “transcriptome”. The conversion from gene to transcript is a non-linear process and can be affected by genetic events such as mutations or single nucleotide polymorphisms (SNPs), but also by epigenetic events such as DNA methylation, histone methylation, acetylation, chromatin dynamics and the activity of micro-RNAs (1–8). Even with a full description of these molecular events, the final transcripts are regulated by interaction with the microenvironment surrounding the cell. This figure also shows that these different levels of organization can be interrogated with non-invasive molecular, functional or anatomic imaging. All three types of imaging are interrelated and the delineations are somewhat arbitrary, they each have distinguishing characteristics. Molecular imaging is relegated to measuring the levels or activities of specific macromolecules or metabolic pathways in vivo. Functional imaging is devoted to measuring specific organ functions such as perfusion and cell density. At the highest level of organization, anatomic imaging can be very quantitative and can identify underlying heterogeneity in microenvironmental conditions, gene expression, and metabolic phenotypes.
Figure 1.
Functional, Molecular and Anatomic Imaging of Cancer processes.
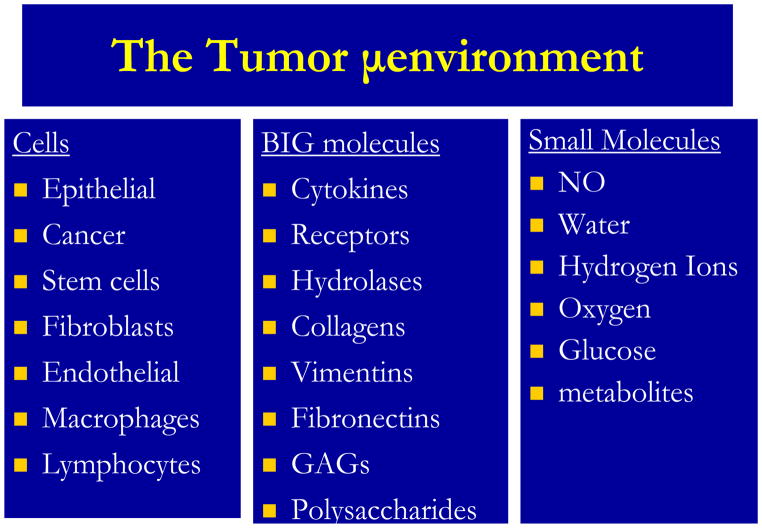
As levels of organization increase in complexity, the interactions between levels remain non-linear and involve complex interactions of the underlying cellular genotypes and phenotypes with the microenvironment. A fundamental outstanding problem in modern biology is how to directly link the current wealth of genetic data, not only with clinical outcome, but crucially with the cellular phenotype. The so called genotype-phenotype mapping is at the heart of our understanding of many of the engines that drive tumor growth and progression; however, it has to some extent been ignored. This is in no small part due to the sheer complexity of the processes involved: thousands of genes result in the regulation of a multitude of proteins, which interact and signal within complex pathways to drive a cellular response in individual cell that modulates its signaling in response microenvironmental cues and signals from other cells. As shown in the figure, the gene expression patterns also, in turn, are exported from the cell to the microenvironment, which provides a conduit for cell-cell communication. Figure 2 lists the complex mixture of components of the tumor microenvironment. The conversion of gene transcripts to protein is also non-linear and is greatly affected by microenvironmental interactions. The microenvironment contains cells, proteins and small molecules, all of which can impact tumor growth and response to therapy. These complex interactions between intra- and extracellular processes give rise to intra-tumoral heterogeneity. Indeed, it can be shown by mathematical modeling that complex heterogeneities can arise even from a limited set of cell-environment interactions(9). As a consequence of these interactions, the microenvironment is spatially and temporally heterogeneous and thus gene expression and cell phenotypes are highly variable, even within the same lesion. The consequences of this cellular heterogeneity are enormous and challenging. For example, phenotypic heterogeneity is the most significant factor underlying evolution rates (10). An emerging concept states that tumors with the most heterogeneity are to be more readily adaptable to perturbations such as chemotherapy and hence, have the worst prognosis. I can be predicted that, as a result of selection pressure (e.g. from treatment), a phenotypic convergence occurs (11). This convergence maybe transient and need not involve a convergence in genotype. In fact, mathematical models suggest that multiple genotypes can produce similar phenotypes (12). Such transient convergence at the phenotype scale is precisely the sort of thing that modern imaging can highlight,
Figure 2.
Components of the Tumor Microenvironment.
Intratumoral variability dramatically confounds our ability to study in-vivo cancers by molecular diagnostics. Typical molecular characterizations such as microarray studies measure the transcriptome in a large number of cells. The relevance of this single, average measurement in a population of high phenotypic and genotypic variance will likely be very limited. In such studies, the impact of heterogeneity can only be appreciated with large data sets from a large study population, i.e. “systems biology”. Consequently, the applicability of these data to individual patients is limited. Modern imaging and, more to the point, modern image analyses, have begun to explore the importance of tumor heterogeneity in progression and response and have the potential to improve diagnosis, prognosis and prediction for individual patients.
Molecular imaging of the metabolome: HIF and the Warburg Effect
An example illustrating the biology and imaging of tumor heterogeneity can be found with the hypoxia-inducible factor, HIF. Since its discovery in 1992 (13), studies of HIF have truly revolutionized understanding of normal and pathological physiology. In that time, there have been over 6,000 primary literature reports on HIF and over 1,000 review articles. The role of this factor in the etiopathogenesis of cancer is indisputable (14, 15). The alpha subunit of this transcription factor is constitutively produced in most cells. In the presence of oxygen, it is acted on by an enzyme, proline hydroxylase, which will add hydroxyl groups to specific proline residues on the protein. These are, in turn, recognized by another enzyme, the von Hippel-Lindau tumor suppressor, which will decorate the protein with small ubiquitin peptides. Ubiquitinated proteins are recognized by a proteosome which will specifically degrade the protein and keep the levels low. In the absence of oxygen, HIF-1α (or HIF-2α) increase and combine with a beta subunit to create an active transcription factor. This induces the transcription of over 60 genes that result in increased glucose metabolism, increased angiogenesis, increased iron metabolism and resistance to cell death (14). These are all associated with a physiological response to hypoxia, such as that seen in wound healing. In many cancers, HIF becomes aberrantly stabilized and this results in spatially and temporally inappropriate angiogenesis, glycolysis and resistance to programmed cell death, PCD (16, 17), which are three hallmarks of cancer (18). The consequences of angiogenesis, glycolysis and PCD resistance can be imaged in patients using dynamic contrast enhanced (DCE) MRI, 18F-deoxyglucose (FDG) PET, and diffusion MRI, respectively and these can be used to measure response to anti-HIF therapy (19–21).
Functional and Metabolic Imaging of tumor heterogeneity: DCE-MRI, FDG-PET and Diffusion
Dynamic Contrast Enhanced MRI monitors the time-activity uptake curves of small molecular weight contrast agents with T1-weighted imaging. These data can be modeled in a pixel-wise fashion to yield functional maps of vascular parameters such as blood volume, volume of extravascular extracellular space and the blood vessel transfer function, Ktrans, which is related to permeability (22, 23). Jackson et al (24)have recently and authoritatively reviewed DCE-MRI data from a variety of cancer patients and have put forth the hypothesis that the heterogeneity of enhancement is of more prognostic value than is the average value taken over the entire region-of-interest. In order to assess heterogeneity, the signal measured over time must come from the same volume of tissue for each sample. In practice this is difficult to accomplish below the neck, due to physiologic motion (respiration and cardiac). While motion artifacts can be reduced by cardiac gating or acquisitions during breath hold, this invariably reduces accuracy. More modern approaches collect data with free breathing and a navigator echo, and utilize post acquisition processing (25). Pixelwise analysis of data has shown to have higher predictive value than single ROI analyses. For example, compared to the mean Ktrans values from a single ROI, the SD of Ktrans values was shown to more accurately delineate benign from malignant breast lesions (26). As described in Jackson et al, histogram analyses are proving to be very powerful for tumor diagnosis, prognosis and therapy monitoring(24). More sophisticated analytical paradigms are being applied, with greater accuracy and success (27). In cervical cancers, heterogeneities of enhancement kinetics have been quantified using masked wavelet decomposition coefficients. These have been modeled and shown to be highly predictive (>95%) of both recurrence and survival (28).
As described elsewhere in this volume, PET imaging can be used to monitor the distribution of a number of position emitting tracers, and data can be kinetically modeled to yield metabolic transfer coefficients. In cancer, no tracer has had a larger impact than 19F-labeled deoxyglucose (FDG), which measures the uptake and trapping of a glucose analog. Since it was first used for tumor detection in 1980 (29), more than 2 million patients have been imaged worldwide for the purpose of cancer staging and diagnosis. For many cancers, the diagnostic accuracy of FDG PET is over 90% (30). The two major FDG metabolizing enzymes: the glucose transporter, GLUT-1 and the hexokinase, HK-2, are both HIF client proteins and both correlate highly to FDG uptake (31). Thus their heterogeneous distribution mirrors that of heterogeneous perfusion measured by DCE-MRI. There is abundant evidence that this heterogeneity of FDG uptake is prognostic. For example, Eary et al (32) have developed a new heterogeneity-analysis algorithm that was a strong independent predictor of patient outcome in 238 sarcoma patients. Statistical analyses show that heterogeneity analysis is a strong independent predictor of patient outcome. Additional predictive power can be obtained if tumor shape functions are incorporated into the analysis(33). In cervical cancer, the inter-tumoral maximum value of FDG uptake (SUVMax) has been shown to be predictive of outcome. However, more recent studies have shown that SUV heterogeneity was more predictive of lymph node metastasis at diagnosis and radiation response and progression-free survival (34). In this study, volumes were derived using adaptive image segmentation and using the pretreatment FDG-PET/CT images. Intratumoral heterogeneity was expressed as the derivative (dV/dT) of the volume-threshold function. Notably, there the correlations of dV/dT to histology or SUVMax were insignificant suggesting that it is an independent predictor. In non-small cell lung cancer (NSCLC), heterogeneity of FDG uptake is used to identify “hot spots” for radiotherapy and to predict recurrence (35).
Diffusion MRI measures the motion of water during a time interval between two pulsed magnetic field gradients. Since the probability of motion is reduced in the presence of diffusion barriers, such as cell membranes, diffusion MRI has been shown to be sensitive to cell density (20). Since successful therapy often leads to reductions in cell density, diffusion MRI changes have been used as a biomarker for therapy response (36–38). Quantitative diffusion MRI requires comparisons of multiple scans at different gradient strengths, and is readily applicable in non-moving organs such as the brain. Heterogeneity measures have been shown to be predictive of outcome. In brain cancer, for example, an index has been derived, which is a robust measure of lesion heterogeneity (39). This alpha index is predictive of stage and progression (40). In visceral organs, quantitative diffusion is made difficult because of motion (41). Thus diffusion images are typically collected at only a single diffusion weighting. Although quantitative values of diffusion are not derived, the data are nonetheless very amenable to pixelwise analysis of heterogeneity.
Anatomic Imaging and gene expression patterns: Radiomics
Referring again to figure 1, the physiology and anatomy of organs and tumors is driven by gene expression patterns which are a product of cellular genetics interfacing with the microenvironment. Over the last few years, it has become clear that distinct sub-regions of tumors, identifiable by MR imaging, have distinct gene expression patterns (31, 42–44). This indicates that underlying molecular biology can affect the “anatome”. Recently, there have been attempts to determine if quantitative analysis of the anatome can be used to infer an underlying molecular gene expression pattern. This involves “radiomics” which is the extraction of quantitative features from radiographic images. Relating these to gene expression patterns using sophisticated bioinformatic approaches is sometimes termed “radiogenomics”. The central hypothesis of cancer radiomics is that tumor imaging features reflect underlying gene expression patterns. In the simplest cases, changes in expression of specific genes can affect specific imageable parameters, such as vascular-endothelial growth factor (VEGF) effects on perfusion or survival gene effects on tumor density, both of which are measurable by MRI (45, 46).
The idea that imaging features reflect underlying differences in gene expression is the basis for image-guided biopsy, which has clearly shown that tumors exhibit distinct regional variations in gene expression that are correlated with image features, such as perfusion (42, 43, 47). Image features also underlie the tremendous power of CAD1 systems (48–50). The use of image features has been elevated to a new level through the work of Kuo and colleagues, who have compared extractable features from MRI or CT to predict global gene expression patterns in gliomas, GBM, and hepatocellular carcinoma, HCC, respectively (51, 52). In GBM, there are clear correlations between histopathology grade and MR imaging features (53). However, the diversity of MR phenotypes is greater than that of histology, e.g. tumors of similar histopathology can exhibit distinctly different MR imaging patterns (54). MR-visible contrast enhancement and tissue density can predict specific hypoxia- and proliferation-associated gene expression patterns (51). Furthermore, overexpression of the epidermal growth factor receptor, EGFR, and an infiltrative phenotype could be inferred from image features, which correlated negatively to survival. Interestingly, intratumoral heterogeneity was one of the most predictive of imaging features.
Conclusion
Cancers are typically complex dynamical systems that respond and evolve rapidly to perturbations such as chemotherapy. The multiscale interactions of the genome, transcriptome, proteome, metabolome, physiome, and anatome are non-linear and will likely be understood only through development and application of appropriate mathematical tools. The resulting spatial and temporal heterogeneity of cancers render investigation and measurement of the underlying genetic, phenotypic, and environmental characteristics extraordinarily difficult. Molecular imaging is uniquely suited to investigate this dynamical cancer ecosystem. New methods to identify and quantify the cellular and environmental heterogeneity of the system may prove to be molecular imaging’s most valuable contribution.
Footnotes
Computer aided diagnosis
References
- 1.Croce CM. Causes and consequences of micro RNA dysregulation in cancer. Nature reviews. 2009;10(10):704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hager GL, McNally JG, Misteli T. Transcription dynamics. Molecular cell. 2009;35(6):741–53. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munshi A, Shafi G, Aliya N, Jyothy A. Histone modifications dictate specific biological readouts. Journal of genetics and genomics = Yi chuan xue bao. 2009;36(2):75–88. doi: 10.1016/S1673-8527(08)60094-6. [DOI] [PubMed] [Google Scholar]
- 4.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461(7261):193–8. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 5.Zheng YG, Wu J, Chen Z, Goodman M. Chemical regulation of epigenetic modifications: opportunities for new cancer therapy. Medicinal research reviews. 2008;28(5):645–87. doi: 10.1002/med.20120. [DOI] [PubMed] [Google Scholar]
- 6.Carninci P, Yasuda J, Hayashizaki Y. Multifaceted mammalian transcriptome. Current opinion in cell biology. 2008;20(3):274–80. doi: 10.1016/j.ceb.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nature reviews. 2007;8(11):829–33. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 8.Ponder BA. Cancer genetics. Nature. 2001;411(6835):336–41. doi: 10.1038/35077207. [DOI] [PubMed] [Google Scholar]
- 9.Anderson AR, Rejniak KA, Gerlee P, Quaranta V. Microenvironment driven invasion: a multiscale multimodel investigation. Journal of mathematical biology. 2009;58(4–5):579–624. doi: 10.1007/s00285-008-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatenby RA, Vincent TL. Application of quantitative models from population biology and evolutionary game theory to tumor therapeutic strategies. Molecular cancer therapeutics. 2003;2(9):919–27. [PubMed] [Google Scholar]
- 11.Anderson AR, Weaver AM, Cummings PT, Quaranta V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell. 2006;127(5):905–15. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 12.Gerlee P, Anderson AR. Modelling evolutionary cell behaviour using neural networks: application to tumour growth. Bio Systems. 2009;95(2):166–74. doi: 10.1016/j.biosystems.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Molecular and cellular biology. 1992;12(12):5447–54. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semenza GL. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin Cancer Biol. 2009;19(1):12–6. doi: 10.1016/j.semcancer.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 15.Kaelin WG., Jr The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8(11):865–73. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- 16.Gillies RJ, Robey I, Gatenby RA. Causes and consequences of increased glucose metabolism of cancers. J Nucl Med. 2008;49 (Suppl 2):24S–42S. doi: 10.2967/jnumed.107.047258. [DOI] [PubMed] [Google Scholar]
- 17.Gillies RJ, Schornack PA, Secomb TW, Raghunand N. Causes and effects of heterogeneous perfusion in tumors. Neoplasia. 1999;1(3):197–207. doi: 10.1038/sj.neo.7900037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 19.Jordan BF, Runquist M, Raghunand N, et al. Dynamic contrast-enhanced and diffusion MRI show rapid and dramatic changes in tumor microenvironment in response to inhibition of HIF-1alpha using PX-478. Neoplasia. 2005;7(5):475–85. doi: 10.1593/neo.04628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephen RM, Gillies RJ. Promise and progress for functional and molecular imaging of response to targeted therapies. Pharmaceutical research. 2007;24(6):1172–85. doi: 10.1007/s11095-007-9250-3. [DOI] [PubMed] [Google Scholar]
- 21.van Baardwijk A, Dooms C, van Suylen RJ, et al. The maximum uptake of (18)F-deoxyglucose on positron emission tomography scan correlates with survival, hypoxia inducible factor-1alpha and GLUT-1 in non-small cell lung cancer. Eur J Cancer. 2007;43(9):1392–8. doi: 10.1016/j.ejca.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 22.Taylor JS, Tofts PS, Port R, et al. MR imaging of tumor microcirculation: promise for the new millennium. J Magn Reson Imaging. 1999;10(6):903–7. doi: 10.1002/(sici)1522-2586(199912)10:6<903::aid-jmri1>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 23.Leach MO, Brindle KM, Evelhoch JL, et al. The assessment of antiangiogenic and antivascular therapies in early-stage clinical trials using magnetic resonance imaging: issues and recommendations. British journal of cancer. 2005;92(9):1599–610. doi: 10.1038/sj.bjc.6602550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson A, O’Connor JP, Parker GJ, Jayson GC. Imaging tumor vascular heterogeneity and angiogenesis using dynamic contrast-enhanced magnetic resonance imaging. Clin Cancer Res. 2007;13(12):3449–59. doi: 10.1158/1078-0432.CCR-07-0238. [DOI] [PubMed] [Google Scholar]
- 25.Buonaccorsi GA, O’Connor JP, Caunce A, et al. Tracer kinetic model-driven registration for dynamic contrast-enhanced MRI time-series data. Magn Reson Med. 2007;58(5):1010–9. doi: 10.1002/mrm.21405. [DOI] [PubMed] [Google Scholar]
- 26.Issa B, Buckley DL, Turnbull LW. Heterogeneity analysis of Gd-DTPA uptake: improvement in breast lesion differentiation. Journal of computer assisted tomography. 1999;23(4):615–21. doi: 10.1097/00004728-199907000-00024. [DOI] [PubMed] [Google Scholar]
- 27.Rose CJ, Mills SJ, O’Connor JP, et al. Quantifying spatial heterogeneity in dynamic contrast-enhanced MRI parameter maps. Magn Reson Med. 2009;62(2):488–99. doi: 10.1002/mrm.22003. [DOI] [PubMed] [Google Scholar]
- 28.Prescott JW, Zhang D, Wang JZ, et al. Temporal Analysis of Tumor Heterogeneity and Volume for Cervical Cancer Treatment Outcome Prediction: Preliminary Evaluation. J Digit Imaging. 2009 doi: 10.1007/s10278-009-9179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Som P, Atkins HL, Bandoypadhyay D, et al. A fluorinated glucose analog, 2-fluoro-2-deoxy-D-glucose (F-18): nontoxic tracer for rapid tumor detection. J Nucl Med. 1980;21(7):670–5. [PubMed] [Google Scholar]
- 30.Gambhir SS. Molecular imaging of cancer with positron emission tomography. Nature Reviews Cancer. 2002;2(9):683–93. doi: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]
- 31.Zhao S, Kuge Y, Mochizuki T, et al. Biologic correlates of intratumoral heterogeneity in 18F-FDG distribution with regional expression of glucose transporters and hexokinase-II in experimental tumor. J Nucl Med. 2005;46(4):675–82. [PubMed] [Google Scholar]
- 32.Eary JF, O’Sullivan F, O’Sullivan J, Conrad EU. Spatial heterogeneity in sarcoma 18F-FDG uptake as a predictor of patient outcome. J Nucl Med. 2008;49(12):1973–9. doi: 10.2967/jnumed.108.053397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Sullivan F, Roy S, O’Sullivan J, Vernon C, Eary J. Incorporation of tumor shape into an assessment of spatial heterogeneity for human sarcomas imaged with FDG-PET. Biostatistics (Oxford, England) 2005;6(2):293–301. doi: 10.1093/biostatistics/kxi010. [DOI] [PubMed] [Google Scholar]
- 34.Kidd EA, Grigsby PW. Intratumoral metabolic heterogeneity of cervical cancer. Clin Cancer Res. 2008;14(16):5236–41. doi: 10.1158/1078-0432.CCR-07-5252. [DOI] [PubMed] [Google Scholar]
- 35.Petit SF, Aerts HJ, van Loon JG, et al. Metabolic control probability in tumour subvolumes or how to guide tumour dose redistribution in non-small cell lung cancer (NSCLC): an exploratory clinical study. Radiother Oncol. 2009;91(3):393–8. doi: 10.1016/j.radonc.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 36.Hamstra DA, Rehemtulla A, Ross BD. Diffusion magnetic resonance imaging: a biomarker for treatment response in oncology. J Clin Oncol. 2007;25(26):4104–9. doi: 10.1200/JCO.2007.11.9610. [DOI] [PubMed] [Google Scholar]
- 37.Chenevert TL, Meyer CR, Moffat BA, et al. Diffusion MRI: a new strategy for assessment of cancer therapeutic efficacy. Mol Imaging. 2002;1(4):336–43. doi: 10.1162/15353500200221482. [DOI] [PubMed] [Google Scholar]
- 38.Evelhoch JL, Gillies RJ, Karczmar GS, et al. Applications of magnetic resonance in model systems: cancer therapeutics. Neoplasia. 2000;2(1–2):152–65. doi: 10.1038/sj.neo.7900078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennett KM, Hyde JS, Schmainda KM. Water diffusion heterogeneity index in the human brain is insensitive to the orientation of applied magnetic field gradients. Magn Reson Med. 2006;56(2):235–9. doi: 10.1002/mrm.20960. [DOI] [PubMed] [Google Scholar]
- 40.Kwee TC, Galban CJ, Tsien C, et al. Intravoxel water diffusion heterogeneity imaging of human high-grade gliomas. NMR in biomedicine. 2009 doi: 10.1002/nbm.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Bihan D, Poupon C, Amadon A, Lethimonnier F. Artifacts and pitfalls in diffusion MRI. J Magn Reson Imaging. 2006;24(3):478–88. doi: 10.1002/jmri.20683. [DOI] [PubMed] [Google Scholar]
- 42.Hobbs SK, Shi G, Homer R, Harsh G, Atlas SW, Bednarski MD. Magnetic resonance image-guided proteomics of human glioblastoma multiforme. J Magn Reson Imaging. 2003;18(5):530–6. doi: 10.1002/jmri.10395. [DOI] [PubMed] [Google Scholar]
- 43.Van Meter T, Dumur C, Hafez N, Garrett C, Fillmore H, Broaddus WC. Microarray analysis of MRI-defined tissue samples in glioblastoma reveals differences in regional expression of therapeutic targets. Diagn Mol Pathol. 2006;15(4):195–205. doi: 10.1097/01.pdm.0000213464.06387.36. [DOI] [PubMed] [Google Scholar]
- 44.Tebbit CL, Zhai J, Untch BR, et al. Novel tumor sampling strategies to enable microarray gene expression signatures in breast cancer: a study to determine feasibility and reproducibility in the context of clinical care. Breast cancer research and treatment. 2009;118(3):635–43. doi: 10.1007/s10549-008-0301-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee TY, Purdie TG, Stewart E. CT imaging of angiogenesis. Q J Nucl Med. 2003;47(3):171–87. [PubMed] [Google Scholar]
- 46.Mut M, Turba UC, Botella AC, Baskurt E, Lopes MB, Shaffrey ME. Neuroimaging characteristics in subgroup of GBMs with p53 overexpression. J Neuroimaging. 2007;17(2):168–74. doi: 10.1111/j.1552-6569.2007.00112.x. [DOI] [PubMed] [Google Scholar]
- 47.Sullivan DC. Challenges and opportunities for in vivo imaging in oncology. Technology in cancer research & treatment. 2002;1(6):419–22. doi: 10.1177/153303460200100602. [DOI] [PubMed] [Google Scholar]
- 48.Giger ML, Chan HP, Boone J. Anniversary paper: History and status of CAD and quantitative image analysis: the role of Medical Physics and AAPM. Medical physics. 2008;35(12):5799–820. doi: 10.1118/1.3013555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan Y, Giger ML, Li H, Sennett C. Correlative feature analysis on FFDM. Medical physics. 2008;35(12):5490–500. doi: 10.1118/1.3005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Giger ML, Yuan Y, et al. Evaluation of computer-aided diagnosis on a large clinical full-field digital mammographic dataset. Academic radiology. 2008;15(11):1437–45. doi: 10.1016/j.acra.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(13):5213–8. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nature biotechnology. 2007;25(6):675–80. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 53.Henson JW, Gaviani P, Gonzalez RG. MRI in treatment of adult gliomas. The lancet oncology. 2005;6(3):167–75. doi: 10.1016/S1470-2045(05)01767-5. [DOI] [PubMed] [Google Scholar]
- 54.Rees JH, Smirniotopoulos JG, Jones RV, Wong K. Glioblastoma multiforme: radiologic-pathologic correlation. Radiographics. 1996;16(6):1413–38. doi: 10.1148/radiographics.16.6.8946545. quiz 62–3. [DOI] [PubMed] [Google Scholar]