Abstract
A new model of cancer progression has been put forward that predicts existence of tumor stem cells (TSCs) in the heterogeneous bulk tumor mass that self-renew, are resistant to chemo- and radiotherapies, and sustain tumor growth during the course of its progression or relapse ( Ailles and Weissman, Curr Opin Biotechnol 18:460–466, 2007; Chan et al., Proc Natl Acad Sci U S A 106:14016–14021, 2009; D’Angelo and Wicha, Prog Mol Biol Transl Sci 95:113–158, 2010; O’Brien, Semin Radiat Oncol 19:71–77, 2009; Park et al., Mol Ther 17:219–230, 2009). Using most advanced methods of cell purification and transplantation, our laboratory and another independent study identified melanoma stem cells as CD271(NFGR/p75)+ cells from surgical human specimens (Boiko et al., Nature 466:133–137, 2010; Civenni et al., Cancer Res 71:3098–3109, 2011). Here we describe in great detail an approach for isolating tumor-initiating cells from freshly resected melanomas.
Keywords: Melanoma stem cell isolation, FACS, Antibody marker, Tissue digestions, Cell transplantation
1. Introduction
In the recent years there have been a number of reports describing identification of melanoma stem cell surface markers and attempts to isolate melanoma tumor-initiating population based on their expression (1–4). Reported evidence lacked consistency in cell surface marker analysis, methods of tissue digestion into single cell suspension, cell separation, and cell transplantation. As a result frequency of melanoma tumor stem cells (MTSCs) varied considerably ranging from 1 in 106 to 1 in 4. Which of the above studies is more representative of human melanomas? Why is there such a marked difference in the incidence of MTSCs and what are the markers that distinguish MTSCs?
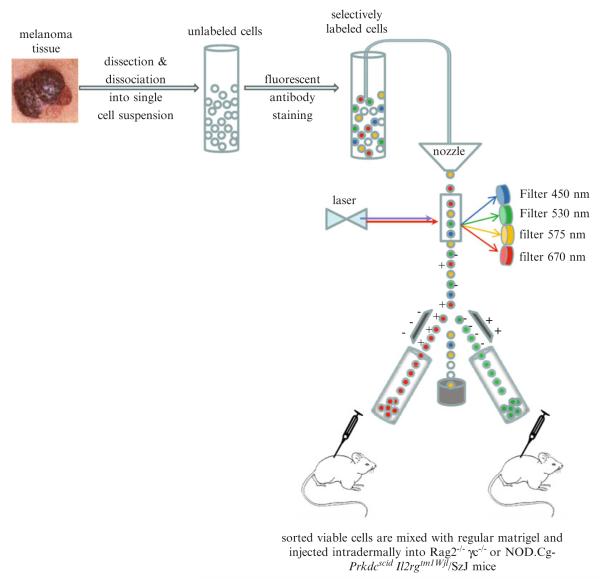
These questions can be addressed by considering several key technical aspects of tumor stem cells (TSCs) identification that are outlined in Fig. 1: (1) Preparation of viable cell suspension from surgical sample by methods that preserve cell surface marker expression. This step has to be approached with extreme care. Protocol(s) of tissue digestion that induce partial or complete cleavage of the marker from the cell surface will generate cells which will cross-contaminate truly negative population with positive cells and ultimately produce unreliable results during transplantation of this mixture in vivo. (2) Cell separation based on their cell surface immunophenotype. While many affinity based column assays have been developed in the past all of them lack ability to separate cells based on multiple parameters simultaneously such as cell shape, live vs. dead cells, singlets vs. doublets, and stromal vs. tumor cells, eventually affecting their purity and usability for transplantation experiments. Therefore, the only method suitable for stem cell isolation from normal and tumorigenic tissues remains the fluorescence-activated cell sorting (FACS) performed under stringent conditions of specific gating strategies that can separate with at least one log of difference for all necessary parameters to distinguish live single cells of different immunophenotypes as revealed by antibody staining (5–8). (3) Cell transplantation is another critical step for assaying tumorigenicity of each candidate cell population. The choice of the in vivo host: the fact is that the more immunocompromised the model, the higher the frequency of engrafting cells will be (3, 9). However, this will only reflect the permissiveness of each particular mouse strain and the ability of the transplanted cells to adjust to the new microenvironment, but the true frequency of human tumorigenic cells can never be found due to the fact that human isogenic tumor cells transplantation are not possible. With the only alternative being the mouse strains humanized with human skin and/or patient specific hematopoietic system. Another vital component is the matrigel that is widely used during tumor cell transplantation assays. Profound differences exist between different types of matrigels in terms of concentration of growth factors that can dramatically affect intrinsic properties of transplanted cells. For example high concentration growth factors type of matrigel was found to promote tumor formation by the cells that have otherwise no tumor forming potential if compared to other types of matrigels (Boiko AD et al. unpublished observations).
Fig. 1.
General outline of melanoma tumor stem cell (MTSC) isolation.
In addition, one must look carefully at the source of melanoma cells that are being used for tumorigenic assays; in many cases they are derived from either xenopassaged tumors or cell lines. The disadvantage of such approaches is that prolonged cell passaging in nonphysiological microenvironment (either in mouse or as a sphere/cell line culture) is likely to select for cell subsets whose functional and expression profiles are the results of adaptation to these conditions and have little to do with original patient tumor. Because these adaptations occur over time spans that would exceed the life history of a tumor in a patient, they would provide little information with respect to the tumors from which they were initially derived.
2. Materials
2.1. Tissues Digest Components
Media 199 (Invitrogen)
Blendzyme TM mix (Roche)
Blendzyme TH mix is added (Roche)
70 μm nylon mesh
Razor blade
ACK buffer (Gibco)
Heat Inactivated FBS (Omega Scientific)
HBSS (Cellgro)
Hemocytometer (Hausser Scientific, Horsham, PA)
2.2. Cell Staining Components
HBSS/2% FBS buffer
Blocking Reagent: Mouse IgG from mouse serum (Sigma)
Human Lineage Antibodies: CD45-Pacific Blue (Invitrogen), CD2-Pacific Blue (Biolegend), CD3-Pacific Blue (Biolegend), CD31-Pacific Blue (Biolegend), CD235ab-Pacific Blue (Glycophorin A) (Biolegend), CD326-Pacific Blue (EpCAM) (Biolegend).
Mouse Lineage Antibodies: H2kd-FITC(BD), mCD45-FITC (BD), Ter119-FITC (BD).
Melanoma Stem Cell Antibody CD271-Alexa 647 (BD) or conjugated antibodies specific for additional candidate melanoma stem cell surface markers.
2.3. Cell Transplantation Components
Regular Matrigel (BD Pharmingen)
31-gauge insulin syringes (BD Pharmingen)
Immunocompromised Mice: 4–6-weeks-old Rag2−/− γc−/− DKO (RG) or NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice.
Forceps
3. Methods
3.1. Preparation of Single Cell Suspension from Melanoma Tissue
Work under sterile conditions of tissue culture hood with all necessary precautions of biohazard material handling while processing human tissue.
Upon obtaining surgical sample place it on ice and use scalpel to trim any evident stromal, fat, and necrotic portions of the tumor.
Remaining tumor tissue is finely minced with a razor blade on a petri dish (this is critical step to make sure no large pieces >1–2 mm remain in the dish).
The resulting slurry is placed into the tube containing 10–30 ml of Media 199 with added liberase Blendzyme TM mix at the final concentration 60 μg/ml.
The tube is placed into the 37°C incubator chamber on the agitating or rocking platform for constant mixing. Every 15 min the tube is taken out and using 5 ml pipette the digest is mixed 10–15 times to prevent clump formation. Majority of melanoma samples will require 45–60 min incubation time; proceed to step 6 if no clumps or tissue pieces remain in the digest buffer at the end of this time period.
In some cases if significant proportion of tumor digest is still in clumps, liberase Blendzyme TH mix is added at concentration 60 μg/ml into the same tube and digest is placed back into 37°C for another 30 min (mixing every 15 min as described above).
Solution containing dissociated tissue is filtered through 70 μm nylon mesh into 50 ml tube and 30 ml of HBSS containing 2% Heat Inactivated FBS is added to neutralize enzyme activity.
Centrifuge at 258 × g for 5 min at 5°C. Discard supernatant and add 30 ml of HBSS containing 2% FBS for a second time to wash out remaining enzyme. Centrifuge as above. Discard supernatant and if no significant red blood cell contamination is present in cell pellet proceed to step 9.
If initial surgical tissue sample contained significant amount of red blood cells resuspend cell pellet in 2 ml of ACK buffer and incubate 1 min on ice; add 30 ml of HBSS containing 2% FBS and centrifuge at 258 × g for 5 min at 5°C.
Resuspend cell pellet in 500 μl of HBSS containing 2% FBS. Count 10 μl of cells mixed with 10 μl of trypan blue in hemocytometer under light microscope to determine the number of viable cells.
3.2. Tumor Cell Antibody Staining and Separation by Flow Cytometry
Centrifuge digested single cell mixture from previous step at 450 × g for 5 min at 5°C and resuspended cell pellet in 100 μl per 106 cells or less of cold HBSS containing 2% FBS.
Add blocking reagent mouse IgG 1 mg/ml and incubated on ice for 10 min.
Aliquot cells into separate tubes and prepare control (isotype) and functional (TSC marker) stains in the separate tubes.
All staining are performed in 100 μl volume of cold HBSS containing 2% FBS. For the control stain add the following lineage antibodies: CD45, CD31, CD2, CD3, Glycophorin A, EpCAM (all conjugated to pacific Blue) and diluted 1:50 (i.e., 2 μl for 100 μl of staining volume) for melanomas resected from human patients. Alternatively, for the tumors that had been passaged in mice the following mouse strain specific lineage antibodies should be used: anti-H2kd (diluted 1:100) anti-mCD45 and mTer119 (diluted 1:50) all conjugated to FITC. Add isotype of the same class and conjugated to the same fluorochromes as a MTSC marker antibody used in step 5.
For functional stain add the same lineage antibodies as in step 4 and add MTSC marker antibody CD271 (Alexa Fluor647-conjugated) at 1:50 dilution or conjugated antibodies specific for additional candidate melanoma stem cell surface markers.
Cover the tubes and incubate on ice in the dark for 30 min.
Wash the antibody staining cell suspension by adding 3 ml of HBSS containing 2% FBS incubate for 1 min on ice in the dark.
Centrifuge stained cells at 258 × g for 5 min at 5°C
Resuspend cell pellet in 0.5 ml HBSS containing 2% FBS and propidium iodide to allow exclusion of nonviable cells.
Perform flow cytometry analysis and cell sorting on BD FACSAria (Becton Dickinson) or similar instrument under 20 psi with a 100-micron nozzle using gating strategy as previously described (6). Cells are sorted into the tubes containing HBSS with 2% FBS.
3.3. Tumor Cell Transplantation into Immunocompromised Mice
Mix 10 μl of FACS-sorted tumor cells with 10 μl of trypan blue and load the mixture into in hemocytometer; count the number of viable cells under light microscope.
Prepare tubes on ice containing HBSS and Matrigel. Aliquot graded number (10-100-1000 etc.) of sorted cells from each population into separate tubes in the volume such that the final concentration of matrigel is equal to 30% and the final volume of cells–buffer–matrigel mixture should be equal to 50 μl.
Shave the flanks of RG or NSG mice.
Anesthetize mice with Isoflurane–O2 mixture.
Use 31-gauge insulin syringe to draw Cell–Matrigel mixture from the tube.
Use forceps to raise the shaved portion of mouse skin and inject intradermally the content of the syringe.
Mice are labeled accordingly to each injected cell population and checked periodically for palpable tumor formation.
4. Results
Depending on the tumor latency and patient disease characteristics palpable tumors should appear within 12 months after cell transplantation. Tumor formation frequency is determined as the ratio of tumor incidence relative to the number of injections. Tumor-initiating cell population is successfully identified when frequency of tumor formation is significantly increased (p value using the Fisher exact test is less than 0.05) after transplantation of FACS sorted cells homogeneous for candidate marker expression (5, 6, 8). Further, tumor-initiating cell frequencies and respective confidence intervals can be calculated using the L-Calc statistical software program for limiting dilution analysis (Stemcell Technologies). However, as discussed in details above, this number has to be interpreted with extreme care due to the surrogate nature of transplantation assays involving human cells and mouse hosts.
Additional experiments to determine whether identified MTSCs are capable of differentiation (i.e., give rise to heterogeneous cell populations reminiscent to those present in the initial surgical patient sample) and self-renewal in vivo (i.e., serially passaged each time giving rise to tumors) have to be performed to complete characterization of melanoma stem cells.
References
- 1.Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Belle PAV, Xu X, Elder DE, Herlyn M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65:9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 2.La Porta C. Cancer stem cells: lessons from melanoma. Stem Cell Rev. 2009;5:61–65. doi: 10.1007/s12015-008-9048-7. [DOI] [PubMed] [Google Scholar]
- 3.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, Fuhlbrigge RC, Kupper TS, Sayegh MH, Frank MH. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan KS, Espinosa I, Chao M, Wong D, Ailles L, Diehn M, Gill H, Presti J, Chang HY, Rijn MVD, Shortliffe L, Weissman IL. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc Natl Acad Sci U S A. 2009;106:14016–14021. doi: 10.1073/pnas.0906549106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boiko AD, Razorenova OV, van de Rijn M, Swetter SM, Johnson DL, Ly DP, Butler PD, Yang GP, Joshua B, Kaplan MJ, Longaker MT, Weissman IL. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 2010;466:133–137. doi: 10.1038/nature09161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexander CM, Puchalski J, Klos KS, Badders N, Ailles L, Kim CF, Dirks P, Smalley MJ. Separating stem cells by flow cytometry: reducing variability for solid tissues. Cell Stem Cell. 2009;5:579–583. doi: 10.1016/j.stem.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalerba P, Dylla SJ, Park I-K, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, Shelton AA, Parmiani G, Castelli C, Clarke MF. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishizawa K, Rasheed ZA, Karisch R, Wang Q, Kowalski J, Susky E, Pereira K, Karamboulas C, Moghal N, Rajeshkumar NV, Hidalgo M, Tsao M, Ailles L, Waddell T, Maitra A, Neel BG, Matsui W. Tumor-initiating cells are rare in many human tumors. Cell Stem Cell. 2010;7:279–282. doi: 10.1016/j.stem.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]