Abstract
Accurate and in-depth mapping of antibody responses is of great value in vaccine and antibody research. Using hepatitis C virus (HCV) as a model, we developed an affordable and high-throughput microarray-based assay for mapping antibody specificities to continuous antibody epitopes of HCV at high resolution. Important parameters in the chemistry for conjugating peptides/antigens to the array surface, the array layout, fluorophore choice and the methods for data analysis were investigated. Microscopic glass slide pre-coated with N-Hydroxysuccinimide (NHS)-ester (Slide H) was the preferred surface for conjugation of aminooxy-tagged peptides. This combination provides a simple chemical means to orient the peptides to the conjugation surface via an orthogonal covalent linkage at the N- or C-terminus of each peptide. The addition of polyvinyl alcohol to printing buffer gave uniform spot morphology, improved sensitivity and specificity of binding signals. Libraries of overlapping peptides covering the HCV E1 and E2 glycoprotein polypeptides (15-mer, 10 amino acids overlap) of 6 major HCV genotypes and the entire polypeptide sequence of the prototypic strain H77 were synthesized and printed in quadruplets in the assays. The utility of the peptide arrays were confirmed using HCV monoclonal antibodies (mAbs) specific to known continuous epitopes and immune sera of rabbits immunized with HCV antigens. The methods developed here can be easily adapted to studying antibody responses to antigens relevant in vaccine and autoimmune research.
Keywords: HCV, E1E2 glycoprotein, peptide array
1. Introduction
Hepatitis C virus (HCV) infection is a major public health problem with an estimated 170 million infected people worldwide (Shepard et al., 2005). It is a leading cause of chronic liver disease, cirrhosis and hepatocellular carcinoma. Therapeutic antibodies and vaccines have been successfully developed to protect at-risk populations against many viral diseases, but, so far, have not been successful for HCV.
Early studies in chimpanzee using HCV envelope glycoproteins E1 and E2 demonstrated a correlation between antibody response and protection against virus challenge (Choo et al., 1994; Rosa et al., 1996). However, complete protection was only observed with homologous but not heterologous virus challenge. This highlights the need of eliciting cross-neutralizing antibodies in any candidate HCV vaccines to be broadly effective against this antigenically diverse virus. Recently, a phase I clinical trial of a HCV vaccine candidate composed of recombinant E1E2 formulated in MF59 adjuvant resulted in only weakly neutralizing antibody responses in humans (Ray et al., 2010; Meyer et al., 2011). Ray and coworkers performed antibody mapping using 4 biotinylated peptides in a standard ELISA format to study antibody responses to the E1 neutralizing epitope aa313-327 (Meunier et al., 2008), the E2 hypervariable region 1 (HVR1) aa384-411 important for antibody neutralization and escape (Weiner et al., 1992), the conserved E2 antigenic site aa412-419 (Kong et al., 2012a; Kong et al., 2012b; Potter et al., 2012) and the antigenic E2 aa434-446 region (Zhang et al., 2009). It has been suggested that antibodies to E2 aa434-446 can interfere with virus neutralization by antibodies to aa412-423 (Zhang et al., 2009), although neutralizing antibodies to E2 aa434-446 have recently been reported by multiple groups (Keck et al., 2012; Morin et al., 2012; Tarr et al., 2012; Deng et al., 2013). Clearly, better tools in the analysis of global antibody responses are needed to provide information for the improvement of HCV immunogens. Because of the polyclonal nature of antibody responses and the extreme genetic diversity of HCV field isolates with genomic sequences differing by up to 35% (Kuiken and Simmonds, 2009), existing analytic methods for monitoring antibody responses, e.g. ELISA, are difficult to address the viral diversity problem in assay design.
With the growing interest in anti-HCV antibodies for vaccine development, it would be highly desirable to develop sophisticated tools to improve the detail and throughput in mapping HCV antibody responses. Traditional methods are labor intensive, time consuming, requiring large amount of coating antigens in the μg/ml range and low in throughput. ELISA and other assays rely on the adsorption of macromolecules either by electrostatic forces or hydrophobic interactions. Several strategies have been developed to improve these assays, which include sandwich or indirect ELISA, competition ELISA, direct covalent attachment of peptides to modified plate surface (Niveleau et al., 1995), using a biotinylated peptide with a streptavidin modified plate (Selo et al., 1996), or using polystyrene-binding peptide tags (Kogot et al., 2012). However, the cost of these methods are prohibitive and highly labor-intensive if large number of features (>1,000) are to be studied.
Microarray technology has been widely used in the study of gene expression and regulatory profiles for over two decades (Schena et al., 1995) and has now been expanded to various formats including protein, peptide, and glycan arrays (MacBeath, 2002; Reimer et al., 2002; Reineke et al., 2002; Blixt et al., 2004; Sun et al., 2004; Andresen and Bier, 2009; Pejchal et al., 2011; Walker et al., 2011; Zhu et al., 2012). We herein describe a novel low cost peptide array assay for mapping antibody specificities at high resolution. We investigated the use of an oxyamine linkage for peptide–surface chemistry for conjugation of analyte to surface. We demonstrate the utility of this method using HCV envelope glycoproteins as an experimental model.
2. Methods
2.1. Peptide preparation
A library of peptides, consisting of 15 amino acids in length with an offset of 5 amino acids, corresponding to the amino acid sequences of HCV E1 and E2 envelope glycoproteins representing 6 major HCV genotypes (genotype 1a, H77; 1b, UKN1b12.6; 2a, J6E3; 2b, UKN2B2.8; 3a, UKN3A1.28; 4, UKN4.11.1; 5, UKN5.15.7; and 6, UKN6.5.34), were custom synthesized in-house at 25-35 μg/well using a MultiPep RS automated peptide synthesizer. The entire H77 isolate polypeptide sequence was synthesized similarly as above for testing human samples. The peptides had a β-alanine C-terminus and a 2.5 polyethylene glycol (PEG) aminooxy N-terminus to ensure optimal epitope orthogonal attachment and presentation. The aminooxy group has greater reactivity kinetics to surface functional groups than lysine or other amine side chains at pH 8.
Synthesized peptides were suspended in 12.5 μl DMSO and 12.5 μl of ultra-pure water. Immediately prior to printing, suspended peptides were diluted 1:4 in a custom protein printing buffer [Saline Sodium Citrate (SSC) 300 mM sodium citrate, pH 8.0, containing 1 M sodium chloride supplemented with 0.1% Polyvinyl Alcohol (PVA) and 0.05% Tween 20], in a 384-well non-binding polystyrene assay plate. Two positive control peptides, hemagglutinin A (HA) (YPYDVPDYA) and FLAG tag (DYKDDDDK), were initially included in the print to guide proper grid placement and peptide ID alignment, and also served as controls for the assays. They were later replaced with a permanent fluorescent dye at 488 nm. The controls were used in the initial optimization of the array printing conditions.
2.2. Microarray printing
All peptide samples were printed in quadruplicatic at an approximate density of 1 ng/spot, on epoxy or N-hydroxysuccinimide ester (NHS-ester) derived glass slides (Slide E or H, Schott AG) using a Microgrid II (DigilabGlobal) microarray printing robot, equipped with solid steel (SMP4, TeleChem) microarray pins. Humidity was maintained at 50% during the print. Immediately prior to interrogating the arrays, slides were blocked for 1 h with ethanolamine buffer to quench any unreacted NHS-ester or epoxide amine residues on the slide. All slides were used within 2 months of printing and stored at -20°C (see Figure 1 for the attachment chemistry). The slides were stable for over 6 months in a -20°C fridge with no apparent loss of activity.
Figure 1.
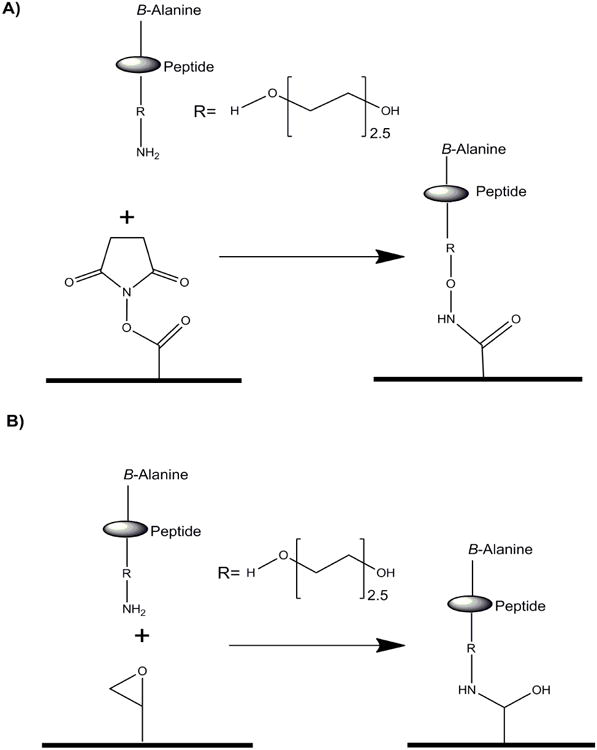
Peptide chemistry for Slide H (A) and E (B).
2.3. Immunolabeling with Alexa based detection system
Incubation area was circumscribed around the printed grids using a Peroxidase Anti-Peroxidase (PAP) hydrophobic marker pen (Research Products International Corp) and the subsequent steps done in a humidified chamber at room temperature on a rotator. Control monoclonal antibodies anti-HA and -FLAG were assayed at a concentration of 10 μg/ml whilst sera was diluted to 1:300 in PBS-TM and incubated for 1 h followed by three washes in PBS buffer. The arrays were then incubated for 1 h with goat antimouse IgG with Alexa-Fluor® tag (Invitrogen) or relevant secondary antibody. Arrays were washed three times in PBS-T, two times in PBS, and another two times in deionized water and centrifuged dry at 200 × g for 5 mins.
2.4. Analysis of array data
The processed slides were scanned using a ProScanArray HT (Perkin Elmer) microarray scanner and images saved as TIF files. The median feature and background pixel intensities for each antigen spot were determined by Imagene® 6.1 microarray analysis software (BioDiscovery). The fluorescence signal was digitalized and exported as comma-delimited text files into Excel for further analysis.
2.5. Animal immunization
Two rabbits were immunized with a peptide whose sequence corresponds to the HCV1 mAb epitope HCV amino acids 412-423 (QLINTNGSWHIN) three times, three weeks apart using complete and incomplete Freund's adjuvant (Invitrogen). Bleeds were taken one week pre- and post-immunization and the sera pooled. The sera was interrogated in the array to see specific immune responses to the epitope and as well as the quality of immune responses in terms of cross-binding with other HCV genotypes in addition to the homologous H77 sequence (Kuiken et al., 2006) used in the immunization regiment.
2.6. Human sera analysis
The antibody responses in HCV-positive human sera and a normal donor serum were compared using the peptide arrays. The infected sera are a mixture of five HCV-positive human sera known to be HCV neutralizing. The samples were diluted 1:300 in PBS-TM buffer and tested on the peptide array consisting of E1 to E2 regions (Fig 6A) and the entire H77 polypeptide sequence from Core to NS5 (Fig 6B).
Figure 6.
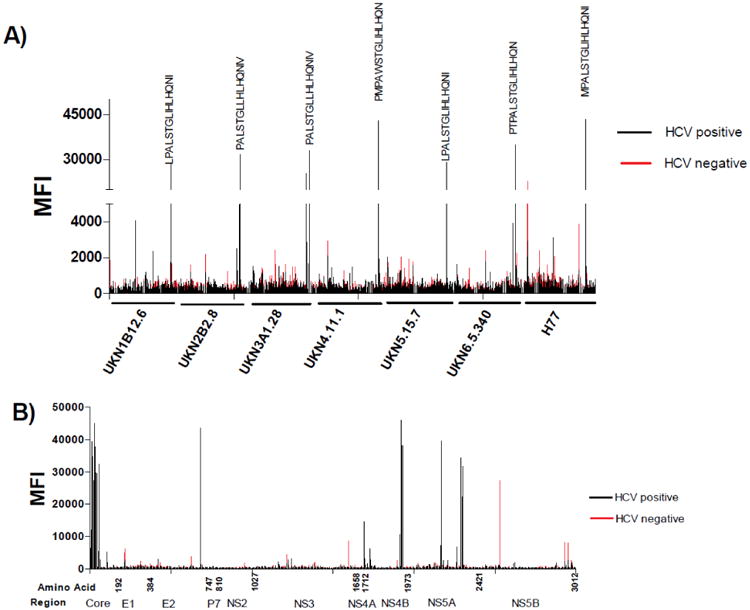
Peptide array analysis of anti-HCV antibody responses in sera of infected patients and normal blood donor. A) Responses of normal and infected sera to the E1 and E2 regions of 6 major HCV genotypes. B) A snap shot of antibody responses to the entire polypeptide sequence of the prototypic genotype 1a isolate H77. The arrays were incubated with human sera samples diluted at 1:300, followed by an Alexa-Fluor 633 conjugated anti-human IgG secondary antibody. Data was processed as described in Figure 4.
3. Results and Discussion
Peptide array performance is affected by various environmental conditions including degradation and denaturation of the peptides during synthesis, printing and storage, it is important to fully optimize and validate the experimental conditions for arrays to be useful as high throughput analytical assays. Additional experimental considerations are the print surfaces, peptide chemistry, print buffers, print conditions, array format, slide storage, assay format, detection system, image capture and data analysis. Orthogonal covalently attached peptides in a microarray format offers potential improvement to traditional immunoassay formats, e.g. ELISA.
A peptide array consisting of 15mer peptides, 10 amino acids overlap of the entire E1E2 region of the HCV glycoprotein covering the 6 major HCV genotypes (Simmonds et al., 2005; Kuiken and Simmonds, 2009) was developed. The peptides were synthesized from C- to N-terminus. For all peptides, there was a beta-alanine at both C- and N-terminus to protect peptides from exopeptidase degradation (Galati et al., 2003). The N-terminal β-alanine was linked to a 2.5 PEG linker enabling the peptides to be extended away from the conjugation surface thus maximizing accessibility and presentation of the antibody epitopes. Finally, an aminooxy group was added to the PEG linker. The unique properties of aminooxy group present an opportunity for chemo selective site-specific immobilization of peptides (Adamczyk et al., 2001). Conjugation of the peptides to the NHS-activated slide was performed at pH 8. The pKa of aminooxy groups is in the range of 5–6, while primary amines have a pKa >9. Thus at pH 8, aminooxy, but not amine functional groups in the epitopes (lysine or arginine), will be present at a nucleophilic protonated state and be used preferably in the conjugation reaction (Lees et al., 2006). Based on such solution chemistry, the kinetics of an aminooxy group will out-compete secondary amines on the peptide thus ensuring specific orientation of HCV peptides.
Using this amine conjugation chemistry, the optimal surface for peptide immobilization was investigated. NHS-ester (Slide H) and Epoxide (Slide E) surfaces were evaluated and the NHS slide was found to produce better resolution with lower limit of detection and background compared to Slide E (Figure 2). This method allows a more stable and orthogonal attachment of the peptides compared to the standard method in peptide array synthesis onto nitrocellulose medium or coating peptides directly onto plastic-based ELISA microwells (Cretich et al., 2006). Attempts to use biotin conjugated peptides with streptavidin coated slides were unsuccessful due to low signals (data not shown).
Figure 2.
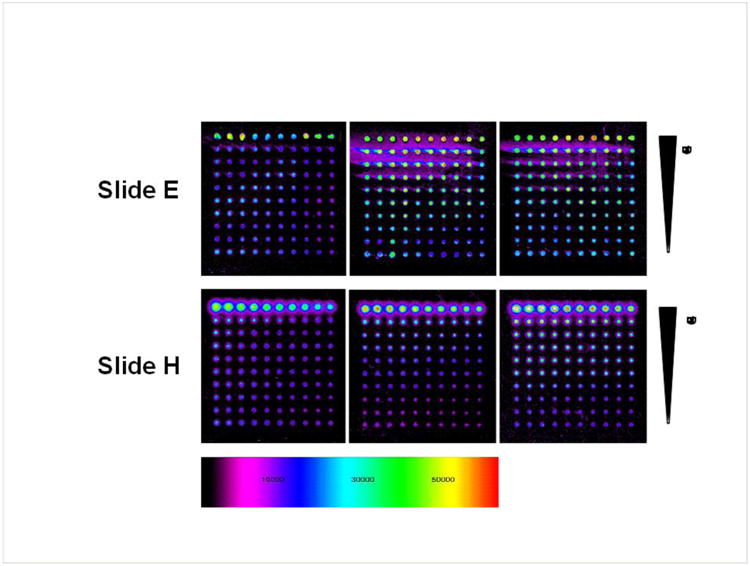
Slides E and H in triplicate were interrogated with a mouse anti-hemmaglutinin (HA) antibody titrated from 10 μg/ml by 2-fold dilution down to 0.0195 μg/ml. AlexaFluor488 conjugated anti-mouse-IgG (10 μg/ml) was used to develop the array. Slide H was the preferred surface for further assay development.
Printing buffer was also optimized for efficient spotting. Several solvent and detergent buffers were investigated to test their effect on spot morphology (Figure 3). Addition of PVA to the buffer (3× SSC + 0.1% PVA and 0.005% Tween-20) not only increases limit of detection, but also improves spot morphology and signal strength. In fact, addition of other commonly used additives in printing buffer, glycerol or PEG, seemed to reduce attachment/immobilization. The results are consistent with Wu et al. (Wu and Grainger, 2006) that among the various hydroxylated additives they investigated, PVA produced the most regular spot morphologies. The printing buffer also helps stabilize the droplet and ensures sufficient time for the attachment chemistry between peptides and surface to occur. Consequently, peptides were printed in PVA containing buffer in subsequent experiments as it gave the best spot morphology and uniformity.
Figure 3.
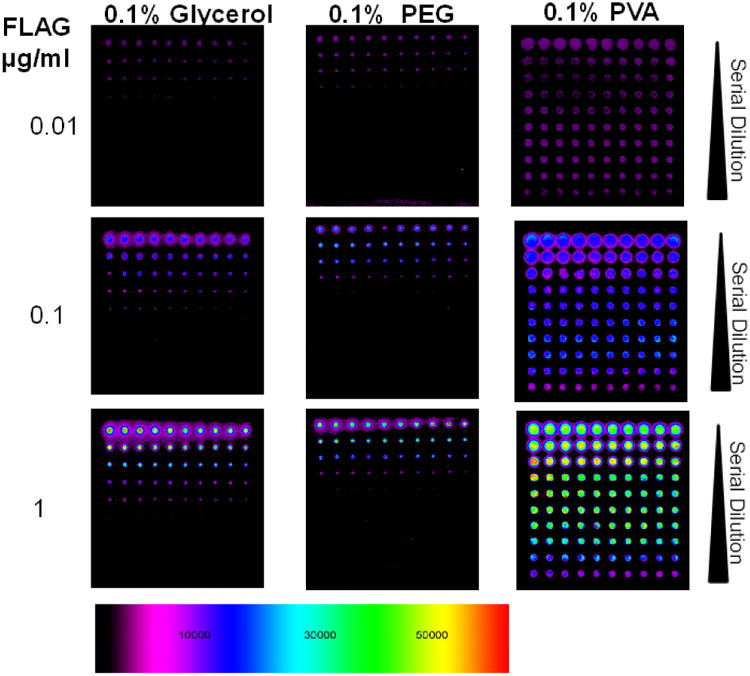
Print buffer Optimization. Three buffer systems; Glycerol, PEG and PVA were investigated and FLAG peptides were printed at different coating concentrations. The buffer was titrated down with the first row being 10 μg/ml of FLAG peptide in 25% H2O, 25% DMSO and 50% Buffer (Glycerol, PEG or PVA), followed by two-fold serial dilution of peptides in buffer.
A print of 941 unique peptides in quadruple was designed based on the total length of HCV E1E2 glycoprotein primary amino acid sequence of the 6 major HCV genotypes and J6E3 plus H77 isolates. If the same number of features were done using a standard ELISA 96 well plate format, for same number of interactions totaling 3764 measurements, a total of 62 ELISA plates would be required at a cost of five times more than a NHS slide. The peptides were printed at 1 ng/spot in the peptide array, resulting in >100-fold reductions of necessary antigens, as an average of 250 ng of peptides are required for each microwell in ELISA. The experimental time for running peptide array usually takes 6 hours whilst running an ELISA with 40 plates require much more labor and it can takes as much as 3 days of a full-time technical staff. In addition, the peptide array has automated data capturing and processing making it less tedious than ELISA. Once printed the slides are stable for at least 6 months if stored at -20°C, without any significant loss in activity. Limitations of using peptide microarrays include the restriction to mimicking continuous epitopes and utility diminished in instances of complex three-dimensional antibody interactions.
To validate the above strategy, the specificity and sensitivity of the peptide array were investigated using HCV specific monoclonal antibodies and sera from rabbits immunized with KLH-conjugated with the HCV E2 peptide aa412-423. Various control monoclonals were interrogated to investigate the specificity and performance of the peptide arrays. Positive controls included mAb HCV1 (Broering et al., 2009), specific to the highly conserved E2 linear epitope aa 412-423 (Kong et al., 2012b), mAb A4 (Dubuisson et al., 1994), specific to the E1 region aa 197 to 207 (SSGLYHVTNDC) (extensively used as an anti E1 glycoprotein antibody control in HCV studies)(Triyatni et al., 2002) and the negative control anti-HIV mAb b6. Figure 4 shows that the assay is highly specific to mAb HCV1 which binds to a highly conserved E2 region, hence recognizing the corresponding epitopes in all viral genotypes tested. We were also able to show that mAb A4 binds to genotypes 1a (H77), 4 (UKN4.11.1), 5 (UKN5.15.7) and 6 (UKN6.5.34). This epitope sequence is not conserved across the genotypes. The integrity of the control monoclonal antibodies and peptides was also confirmed by ELISA. These data validate that the peptide array can be used to characterize antibody responses to diverse HCV genotypes with high specificity.
Figure 4.
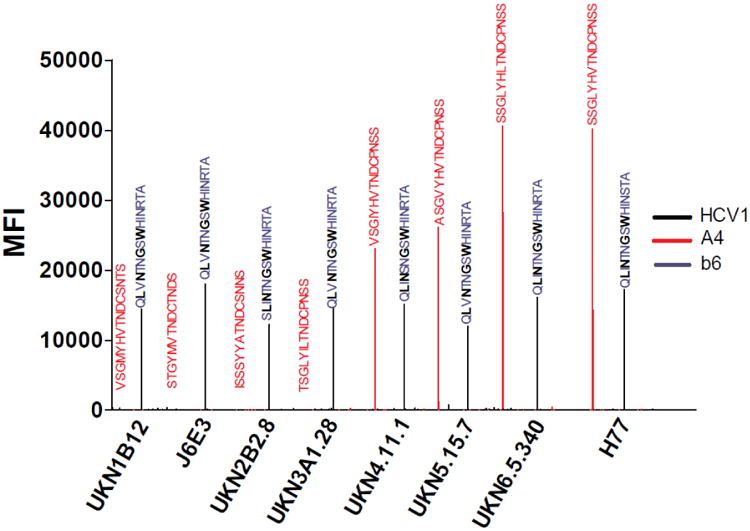
A plot of Mean Relative Fluorescence Intensity (MFI) vs Genotype for mAb b6 (negative control, no detectable signal observed), A4 (anti-E1) and HCV1 (anti-E2). For mAb HCV1 epitope critical residues L413, N415, G418 and W420 are shown in black bold (Kong et al., 2012b) An HCV peptide print consisted of the E1E2 glycoprotein overlapping peptide sequences for the 6 major HCV genotypes and J6E3 plus H77 isolates. The signals were obtained after development with Alexa-Fluor tag conjugated anti-IgG antibodies. The processed slides were scanned using a ProScanArray HT microarray scanner and images saved as TIF files. Fluorescence signal was digitalized and exported as comma-delimited text files. The median feature and background pixel intensities for each antigen spot were determined by Imagene 6.1 microarray analysis software.
The HCV peptide array assay can be used for probing immune responses to linear epitopes in immunization and for general antibody profiling experiments. The arrays were further validated using antisera from rabbits immunized 3 times with a 12-mer cyclic peptide conjugated to Keyhole limpet hemocyanin (KLH) carrier protein in Freund's complete/incomplete adjuvant system and demonstrated its specificity to detect antibodies to E2 aa412-423 from different HCV genotypes (Figure 5). The antisera did not react with other HCV linear epitopes non-specifically nor did the pre-immune sera.
Figure 5.
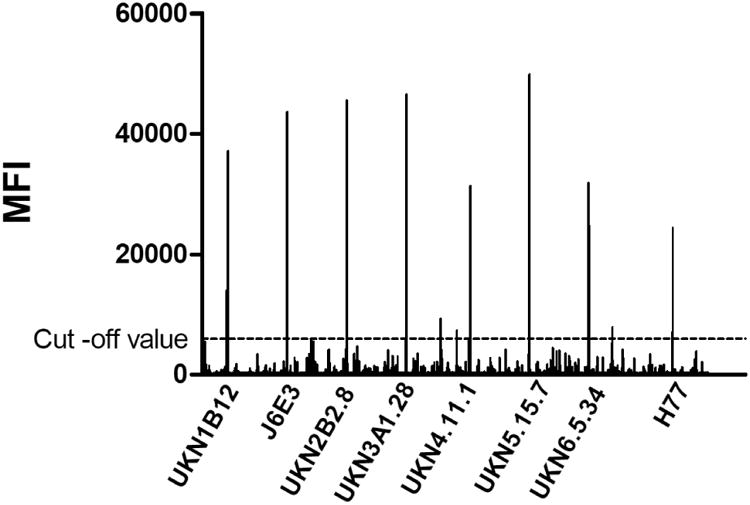
Peptide array analysis of anti-HCV antibody responses in rabbits immunized with KLH-conjugated peptide corresponding to HCV aa412-422 sequence. The array displayed clear sera specificity for the immunizing peptide (H77) and the corresponding peptides in the other HCV genotypes. The signal cut-off was defined as 10% of the maximum signal in the assay. The peaks are identical to the HCV1 epitopes shown in Figure 4. The array was incubated with rabbit sera diluted at 1:300, followed by an Alexa-Fluor 633 conjugated anti-rabbit IgG secondary antibody. Data was processed as described in Figure 4. Pre-immune sera did not produce any significant signals above the background cut-off value 5000 MFI (Data not shown).
To demonstrate potential usage of the peptide arrays, HCV-positive patient sera were assessed (Figure 6). Five HCV-neutralizing sera from chronic patients were mixed in equal volume and evaluated using the E1E2 peptide array above and another array containing the entire HCV polypeptide (Core to NS5) of the genotype 1a prototypic strain H77. In this preliminary assessment, the HCV-immune sera specifically detected peptides around the E2 region 680-695 from all 8 isolates of the 6 major genotypes (Figure 6A). This region is part of the membrane proximal external region that is targeted by one of the broadly neutralizing mAb AR4A (Giang et al., 2012). In comparison to antibody responses to E1E2, antibody responses to the entire polypeptide (Figure 6B) showed high reactivity towards the Core, NS4A/B and NS5A regions. The uninfected serum generally had low reactivity towards the HCV peptides but non-specific background reactivity could be observed in the E1, E2, NS3 and NS5B regions, particularly to the N-teriminus of E1 and NS5B of the H77 isolate. Interestingly, NS5B is typically excluded from HCV diagnostic assays as a detecting antigen because of its significant homology to other RNA viruses (Maertens et al., 1999). Further experiments are underway to determine peptides in the arrays that are prone to generate non-specific signals using a large panel of sera from normal blood donors. The results will identify the assay limitations for studying polyclonal antibodies, and help optimize the specificity and sensitivity of the assays.
Together these data support a robust, effective and rapid method to dissect immune responses to linear HCV epitopes in great details. The arrays can also be adapted for alanine scanning mutagenesis analysis for mapping epitope to single amino acid level. The method presented has broad applicability and adaptability as similar strategy can be used for other HCV structural and non-structural proteins and also in HIV, FLU and other infectious diseases studies.
Acknowledgments
This research was supported by funds from the National Institutes of Health (AI79031 to M.L. and AI71084 to D.R.B). We acknowledge Georg Lauer of Harvard Medical School and Massachusetts General Hospital for the HCV neutralizing human sera. This paper is subject to the NIH Public Access Policy. This is TSRI manuscript number 24054.
Footnotes
Competing interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamczyk M, Gebler JC, Reddy RE, Yu Z. A chemoselective method for site-specific immobilization of peptides via aminooxy group. Bioconjug Chem. 2001;12:139–42. doi: 10.1021/bc0001239. [DOI] [PubMed] [Google Scholar]
- Andresen H, Bier FF. Peptide microarrays for serum antibody diagnostics. Methods Mol Biol. 2009;509:123–34. doi: 10.1007/978-1-59745-372-1_8. [DOI] [PubMed] [Google Scholar]
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A. 2004;101:17033–8. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broering TJ, Garrity KA, Boatright NK, Sloan SE, Sandor F, Thomas WD, Jr, Szabo G, Finberg RW, Ambrosino DM, Babcock GJ. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J Virol. 2009;83:12473–82. doi: 10.1128/JVI.01138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo QL, Kuo G, Ralston R, Weiner A, Chien D, Van Nest G, Han J, Berger K, Thudium K, Kuo C, et al. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc Natl Acad Sci U S A. 1994;91:1294–8. doi: 10.1073/pnas.91.4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretich M, Damin F, Pirri G, Chiari M. Protein and peptide arrays: recent trends and new directions. Biomol Eng. 2006;23:77–88. doi: 10.1016/j.bioeng.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Deng L, Zhong L, Struble E, Duan H, Ma L, Harman C, Yan H, Virata-Theimer ML, Zhao Z, Feinstone S, Alter H, Zhang P. Structural evidence for a bifurcated mode of action in the antibody-mediated neutralization of hepatitis C virus. Proc Natl Acad Sci U S A. 2013;110:7418–22. doi: 10.1073/pnas.1305306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuisson J, Hsu HH, Cheung RC, Greenberg HB, Russell DG, Rice CM. Formation and Intracellular-Localization of Hepatitis-C Virus Envelope Glycoprotein Complexes Expressed by Recombinant Vaccinia and Sindbis Viruses. J Virol. 1994;68:6147–60. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galati R, Verdina A, Falasca G, Chersi A. Increased resistance of peptides to serum proteases by modification of their amino groups. Z Naturforsch C. 2003;58:558–61. doi: 10.1515/znc-2003-7-819. [DOI] [PubMed] [Google Scholar]
- Giang E, Dorner M, Prentoe J, Dreux M, Evans MJ, Bukh J, Rice CM, Ploss A, Burton DR, Law M. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci U S A (2012) 2012;109:6205–10. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck ZY, Xia J, Wang Y, Wang W, Krey T, Prentoe J, Carlsen T, Li AY, Patel AH, Lemon SM, Bukh J, Rey FA, Foung SK. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. 2012;8:e1002653. doi: 10.1371/journal.ppat.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogot JM, Sarkes DA, Val-Addo I, Pellegrino PM, Stratis-Cullum DN. Increased affinity and solubility of peptides used for direct peptide ELISA on polystyrene surfaces through fusion with a polystyrene-binding peptide tag. BioTechniques. 2012;52:95–102. doi: 10.2144/000113810. [DOI] [PubMed] [Google Scholar]
- Kong L, Giang E, Nieusma T, Robbins JB, Deller MC, Stanfield RL, Wilson IA, Law M. Structure of hepatitis C virus envelope glycoprotein E2 antigenic site 412 to 423 in complex with antibody AP33. J Virol. 2012a;86:13085–8. doi: 10.1128/JVI.01939-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Giang E, Robbins JB, Stanfield RL, Burton DR, Wilson IA, Law M. Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc Natl Acad Sci U S A. 2012b;109:9499–504. doi: 10.1073/pnas.1202924109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken C, Combet C, Bukh J, Shin IT, Deleage G, Mizokami M, Richardson R, Sablon E, Yusim K, Pawlotsky JM, Simmonds P Los Alamos, H.I.V.D.G. A comprehensive system for consistent numbering of HCV sequences, proteins and epitopes. Hepatology. 2006;44:1355–61. doi: 10.1002/hep.21377. [DOI] [PubMed] [Google Scholar]
- Kuiken C, Simmonds P. Nomenclature and numbering of the hepatitis C virus. Methods Mol Biol. 2009;510:33–53. doi: 10.1007/978-1-59745-394-3_4. [DOI] [PubMed] [Google Scholar]
- Lees A, Sen G, LopezAcosta A. Versatile and efficient synthesis of protein-polysaccharide conjugate vaccines using aminooxy reagents and oxime chemistry. Vaccine. 2006;24:716–29. doi: 10.1016/j.vaccine.2005.08.096. [DOI] [PubMed] [Google Scholar]
- MacBeath G. Protein microarrays and proteomics. Nat Genet. 2002;32:526–32. doi: 10.1038/ng1037. Suppl. [DOI] [PubMed] [Google Scholar]
- Maertens G, Dekeyser F, Van Geel A, Sablon E, Bosman F, Zrein M, Pollet D. Confirmation of HCV Antibodies by the Line Immunoassay INNO-LIA HCV Ab III. Methods Mol Med. 1999;19:11–25. doi: 10.1385/0-89603-521-2:11. [DOI] [PubMed] [Google Scholar]
- Meunier JC, Russell RS, Goossens V, Priem S, Walter H, Depla E, Union A, Faulk KN, Bukh J, Emerson SU, Purcell RH. Isolation and characterization of broadly neutralizing human monoclonal antibodies to the E1 glycoprotein of hepatitis C virus. J Virol. 2008;82:966–73. doi: 10.1128/JVI.01872-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Banerjee A, Frey SE, Belshe RB, Ray R. A weak neutralizing antibody response to hepatitis C virus envelope glycoprotein enhances virus infection. PLoS One. 2011;6:e23699. doi: 10.1371/journal.pone.0023699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin TJ, Broering TJ, Leav BA, Blair BM, Rowley KJ, Boucher EN, Wang Y, Cheslock PS, Knauber M, Olsen DB, Ludmerer SW, Szabo G, Finberg RW, Purcell RH, Lanford RE, Ambrosino DM, Molrine DC, Babcock GJ. Human monoclonal antibody HCV1 effectively prevents and treats HCV infection in chimpanzees. PLoS Pathog. 2012;8:e1002895. doi: 10.1371/journal.ppat.1002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niveleau A, Bruno C, Drouet E, Brebant R, Sergeant A, Troalen F. Grafting Peptides onto Polystyrene Microplates for Elisa. J Immunol Methods. 1995;182:227–34. doi: 10.1016/0022-1759(95)00053-d. [DOI] [PubMed] [Google Scholar]
- Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter JA, Owsianka AM, Jeffery N, Matthews DJ, Keck ZY, Lau P, Foung SK, Taylor GL, Patel AH. Toward a hepatitis C virus vaccine: the structural basis of hepatitis C virus neutralization by AP33, a broadly neutralizing antibody. J Virol. 2012;86:12923–32. doi: 10.1128/JVI.02052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray R, Meyer K, Banerjee A, Basu A, Coates S, Abrignani S, Houghton M, Frey SE, Belshe RB. Characterization of antibodies induced by vaccination with hepatitis C virus envelope glycoproteins. J Infect Dis. 2010;202:862–6. doi: 10.1086/655902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer U, Reineke U, Schneider-Mergener J. Peptide arrays: from macro to micro. Curr Opin Biotechnol. 2002;13:315–20. doi: 10.1016/s0958-1669(02)00339-7. [DOI] [PubMed] [Google Scholar]
- Reineke U, Ivascu C, Schlief M, Landgraf C, Gericke S, Zahn G, Herzel H, Volkmer-Engert R, Schneider-Mergener J. Identification of distinct antibody epitopes and mimotopes from a peptide array of 5520 randomly generated sequences. J Immunol Methods. 2002;267:37–51. doi: 10.1016/s0022-1759(02)00139-4. [DOI] [PubMed] [Google Scholar]
- Rosa D, Campagnoli S, Moretto C, Guenzi E, Cousens L, Chin M, Dong C, Weiner AJ, Lau JY, Choo QL, Chien D, Pileri P, Houghton M, Abrignani S. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc Natl Acad Sci U S A. 1996;93:1759–63. doi: 10.1073/pnas.93.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–70. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Selo I, Negroni L, Creminon C, Grassi J, Wal JM. Preferential labeling of alpha-amino N-terminal groups in peptides by biotin, application to the detection of specific anti-peptide antibodies by enzyme immunoassays. J Immunol Methods. 1996;199:127–38. doi: 10.1016/s0022-1759(96)00173-1. [DOI] [PubMed] [Google Scholar]
- Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–67. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, Halfon P, Inchauspe G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin IT, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–73. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- Sun L, Rush J, Ghosh I, Maunus JR, Xu MQ. Producing peptide arrays for epitope mapping by intein-mediated protein ligation. BioTechniques. 2004;37:430–6. 438, 440 passim. doi: 10.2144/04373RR01. [DOI] [PubMed] [Google Scholar]
- Tarr AW, Urbanowicz RA, Jayaraj D, Brown RJ, McKeating JA, Irving WL, Ball JK. Naturally occurring antibodies that recognize linear epitopes in the amino terminus of the hepatitis C virus E2 protein confer noninterfering, additive neutralization. J Virol. 2012;86:2739–49. doi: 10.1128/JVI.06492-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triyatni M, Vergalla J, Davis AR, Hadlock KG, Foung SK, Liang TJ. Structural features of envelope proteins on hepatitis C virus-like particles as determined by anti-envelope monoclonal antibodies and CD81 binding. Virology. 2002;298:124–32. doi: 10.1006/viro.2002.1463. [DOI] [PubMed] [Google Scholar]
- Walker LM, Sok D, Nishimura Y, Donau O, Sadjadpour R, Gautam R, Shingai M, Pejchal R, Ramos A, Simek MD, Geng Y, Wilson IA, Poignard P, Martin MA, Burton DR. Rapid development of glycan-specific, broad, and potent anti-HIV-1 gp120 neutralizing antibodies in an R5 SIV/HIV chimeric virus infected macaque. Proc Natl Acad Sci U S A. 2011;108:20125–9. doi: 10.1073/pnas.1117531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AJ, Geysen HM, Christopherson C, Hall JE, Mason TJ, Saracco G, Bonino F, Crawford K, Marion CD, Crawford KA, et al. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci U S A. 1992;89:3468–72. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Grainger DW. Comparison of hydroxylated print additives on antibody microarray performance. Journal of proteome research. 2006;5:2956–65. doi: 10.1021/pr060217d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhong LL, Struble EB, Watanabe H, Kachko A, Mihalik K, Virata-Theimer ML, Alter HJ, Feinstone S, Major M. Depletion of interfering antibodies in chronic hepatitis C patients and vaccinated chimpanzees reveals broad cross-genotype neutralizing activity. Proc Natl Acad Sci U S A. 2009;106:7537–41. doi: 10.1073/pnas.0902749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, McBride R, Nycholat CM, Yu W, Paulson JC, Wilson IA. Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic Acid receptors. J Virol. 2012;86:13371–83. doi: 10.1128/JVI.01426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


