Abstract
MicroRNAs are small non coding RNAs that typically inhibit the translation and stability of messanger RNAs (mRNAs), controlling genes involved in cellular processes such as inflammation, cell cycle regulation, stress response, differentiation, apoptosis, and migration. Thus, miRNAs have been implicated in the regulation of virtually all signaling circuits within a cell and their dysregulation has been shown to play an essential role in the development and progression of cancer. Here, after a brief description of the miRNA genomics, biogenesis and function, we discuss the effects of miRNA deregulation in the cellular pathways that lead to the progressive conversion of normal cells into cancer cells and the potential to develop new molecular miRNA-targeted therapies.
Keywords: microRNAs, miRs, non coding RNAs, cancer
Introduction
The first microRNA (miRNA) has been discovered in 1993 when two independent studies identified the real nature of the C. elegans heterochronic gene lin-4 as small non-coding RNA (ncRNA) (1). At that time, this small ncRNA was considered a specific tool, employed by the worms, to control somehow the gene expression of the heterochronic coding genes. After seven years, Reinhart et al. (2000) showed that another C. elegans heterochronic gene, let-7, was also represented by a small ncRNA and together with lin-4 RNA were capable, through an RNA–RNA interaction with the 3′ untraslated region (UTR) of the target genes, to start the temporal cascade of regulatory heterochronic genes (2). This discovery triggered scientist curiosity to look for other small ncRNAs and, in 2001, three independent groups identified these small ncRNAs also in other different organisms and cellular systems providing evidence for the existence of a large class of small ncRNAs with potential regulatory roles that was named microRNAs (3–5). Since then, miRNAs have been discovered in virtually all plant and animal species, and the latest version of the miRNA database (miRBase), released in August 2012, hits 2,042 mature miRNAs in human, and approximately 1,281 in mouse (6). Animal miRNAs are phylogenetically conserved (~55% of C. elegans miRNAs have homologues in humans) and mammalian miRNA genes have also multiple isoforms (paralogues), which are probably generated by duplication (human let-7 gene accounts for 8 different isoforms distributed in 11 genomic loci) (Figure 1). The majority of miRNA genes are located in regions quite distant from other annotated genes suggesting that they represents single transcriptional units (Figure 1). Over 50% of miRNA genes are clustered and normally transcribed as multicistronic RNA transcripts. Generally, but not always, miRNA paralogues are clustered together and, since they have identical seed regions, they are thought to act redundantly. For instance, miR-17/92 cluster is produces a single polycistronic precursor that is maturated in six individual miRNAs (7), which are categorized into four miRNA families: miR-17 (miR-17 and 20), miR-18, miR-19 (miR-19a and 19b), and miR-92. A minority of miRNAs is instead considered intragenic, since they are located inside another gene and have the same transcriptional orientation (Figure 1). The host gene can be a coding or a non-coding gene: ~40% of the intragenic miRNAs are located in the intronic regions of the non coding transcripts, whereas ~10% are placed in the exonic regions. MiRNAs located in coding transcripts are generally located in introns. Most miRNA genes are transcribed by RNA polymerase II (Pol II) (8,9), and retain mRNA features such as 5′ cap structure and 3′ poly(A) tail. However, a minor group of miRNAs that are associated with Alu repeats has been reported to be transcribed by Pol III (10). MiRNAs are defined as a single stranded ncRNAs of 19–25 nucleotides in length, generated from endogenous hairpin transcripts (11). They negatively regulate gene expression by base-pairing to partially complementary sites on the target mRNAs, usually in the 3′UTR. Binding of a miRNA to the target mRNA typically leads to translational repression and exonucleolytic mRNA decay, although highly complementary targets can be cleaved endonucleolytically. Computational and experimental approaches indicate that a single miRNA may repress, on average, more than a hundred mRNAs. Furthermore, over 60% of human protein-coding genes are predicted to contain miRNA-binding sites within their 3′ UTRs (12). Thus, miRNAs constitute one of the most abundant classes of gene-regulatory molecules in animals and given the wide impact of miRNAs on gene expression, it is not surprising that they exert a uniquely important role in disease phenotypes, such as cancer. In this review, we synthesize our current understanding of the roles of miRNAs in human cancer and we analyze the new regulatory elements of the miRNA language and their emerging themes of action to interpret their contribution to the pathogenesis of cancer.
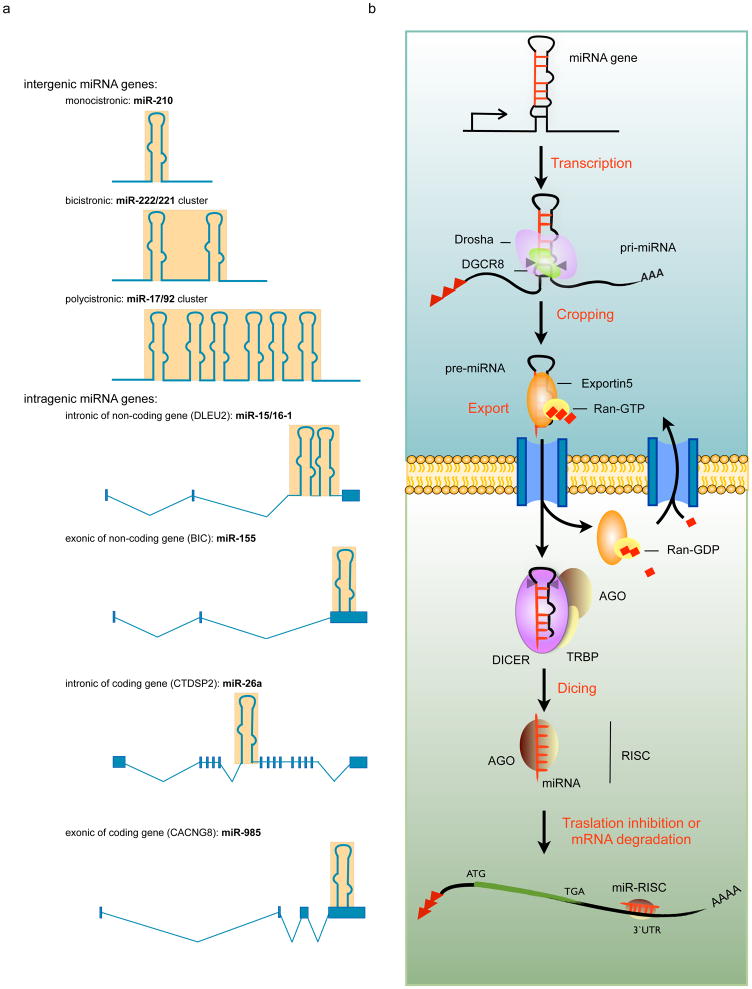
Figure 1. miRNAs genomics, biogenesis and function.
a) miRNAs can be divided in two main classes based on their genomic organization: intergenic, they are independent transcription unit; intragenic, located inside another gene and transcribed in the same orientation of the host gene. Intergenic miRNA gene can be single-, bi- or poly-cistronic. Intragenic miRNAs are generally located in the introns of the host genes but in a relative small percentage they are located in exons. The host genes can be coding or non-coding genes. The blue boxes represent the exon of the host genes and the hairpin-structures symbolize the miRNA sequence location. b) miRNA biogenesis is a multistep process that can be divided in transcription, nuclear cropping, export to cytoplasm and cytoplasmic dicing. MiRNA genes are generally transcribed in long capped and polyadenilated RNA transcripts (pri-miRNA) by RNA polymerase II (pol-II) and subjected to a first processing operated by the nuclear enzyme of the RNase III family, DROSHA. The resulting hairpin-loop RNAs (pre-mRNAs) are recognized by the Exportin 5 (XPO5) /RanGTP transporter and exported to the cytoplasm where a second enzyme of the RNase III family, Dicer, catalyses the second processing step (dicing) to produce miRNA duplexes. Dicer, TRBP, and Argonaute proteins (AGO) mediate the processing of pre-miRNA and the assembly of the RISC (RNA-induced silencing complex) in humans. One strand of the duplex remains on the Ago protein as the mature miRNA, whereas the other strand is degraded. Ago is thought to be associated with Dicer in the dicing step as well as in the RISC assembly step.
miRNA biogenesis and function
miRNAs are canonically produced through a multistep process that, starting in the nucleus and ending into the cytoplasm, is composed by three main events: cropping, export and dicing (13–16). First, miRNA genes are transcribed in primary capped and polyadenylated precursors of miRNA (pri-miRNAs) by RNA polymerase II (8). These several kilobases long transcripts are characterized by stem–loop structures that harbor the mature miRNA in the 53 or 33 half of the stem. The mature miRNA is first “cropped” into the nucleus by the nuclear Drosha/DGCR8 heterodimer which cleaves at the stem of the pri-miRNAs and releases a hairpin-structured pre-miRNA of 60–100nt (17). DGCR8 is a dsRNA binding protein that recognizes the proximal ~10 bp of stem of the pri-miRNA hairpin, positioning the catalytic sites of the RNase III enzyme Drosha (18). The ~2 nt 3′ overhangs of pre-miRNA hairpins are recognized by Exportin-5 (XPO5) and its partner Ran-GTP, enabling their nuclear export (14). Pre-miRNAs are cleaved in the cytoplasm near the terminal loop by the RNAseIII Dicer, releasing the ~22 nt miRNA duplexes (19, 20). Human Dicer interacts with two related proteins TRBP (TAR RNA-binding protein) (21) and PACT (also known as PRKRA) (22). Although TRBP and PACT are dispensable for the processing activity of Dicer, they seem to contribute to activity of miRNAs, but the biochemical mechanism is still unknown. When miRNA duplexes are produced, they associate with Argonaute (Ago) proteins, forming the RNA-induced silencing complex (RISC) (23, 24). One strand of the ~22 nt RNA duplex remains in Ago as a mature miRNA (the guide strand or miRNA), whereas the other strand (the passenger strand or miRNA*) is degraded. miRNAs direct Ago proteins to target mRNAs by interacting with sites of imperfect complementarity. The seed sequences of the miRNA are considered essential for the selection of the target mRNAs and are essential to trigger with a controversial mechanism the accelerated degradation and reduced translation of the targeted mRNA (25, 26). Since miRNAs exert their function by regulating target mRNAs, identification of miRNA targtes is essential to understand their function. Effective prediction of miRNA targets remains challenging due to the interaction complexity and a limited knowledge of rules governing these processes. The seed sequence, the number of sites into the target, thermodynamic of the interaction, conservation and the context of the surrounding sequence in the mRNAs are some of the parameters that currently computational algorithms take in consideration for the prediction of a miRNA targets. The available algorithms can be classified into two categories established on the basis of the use or non-use of conservation comparison. The algorithms based on conservation criteria are for example the following: miRanda (27), PicTar (28), TargetScan (29), DIANA-microT (30); while PITA (31) and rna22 (32) belong to the algorithms using other parameters, such as free energy of binding or secondary structures of 3′UTRs that can promote or prevent miRNA binding. Despite a consistent number of miRNA–target are being identified, the fraction of validated messenger RNA targets is still small, given that miRNAs have been predicted to regulate 60% of human genes. Identification of new miRNA targets will improve our understanding of the rules of miRNA targeting allowing the development of better algorithm for the prediction of miRNA targets.
Decalogue of miRNAs in cancer
The initial indication that miRNAs play important roles in human disease came from high-throughput and functional studies in cancer cells. A seminal study by Croce and colleagues showed that miR-15a/16-1 cluster is frequently deleted in chronic lymphocytic leukemia (CLL), implicating these miRNAs as tumor suppressors (33). After this discovery, a myriad studies were published defining a role for miRNAs in the pathogenesis of cancer. In the following section, we synthesize our current understanding of the functional role of miRNAs in tumorigenesis. Rather than reporting a comprehensive analyses of all the main studies that have connected miRNAs to cancer, we will try to define some of the main emerging principles that have been identified and characterize the activity of miRNAs in cancer.
1. “miRNome”
The most striking theme in the study of miRNAs and cancer is the large alteration of miRNA expression in malignant cells compared to their normal counterpart. All the tumors analyzed have reported a specific miRNA signature, “miRNome”, that characterizes the malignant state and defines some of the clinico-pathological features of the tumors (grade, stage, sex, age, aggressiveness, vascular invasion, proliferation index, etc) (34). miRNA profiles of tumor tissues parallel the developmental origins of the malignant tissues and differences in miRNA expression reflect single developmental lineage and mechanisms of transformation. For examples, tumors of epithelial origin presented a different miRNA expression from haematopoietic malignancies (35); miRNA profiles can also subgroup acute lymphoblastic leukaemia (ALL) specimens into three major groups: one containing all t(9;22) BCR/ABL- and t(12;21) TEL/AML1-positive samples; a second group containing T-cell ALL samples; and a third group containing the MLL gene rearrangement (35).
2. Global downregulation
Despite the global dysregulation, most miRNAs have been found repressed in cancers relative to normal tissue counterparts, as indication of the general loss of differentiation of tumor cells (35). In agreement with these observations, global depletion of miRNAs by genetic deletion of the miRNA-processing machinery favorites cell transformation and tumorigenesis in vivo (36, 37). This highlights that miRNA alteration is not simply an effect of tumorigenesis but it has a causative role in cancer development. Despite the general reduction of miRNAs in cancers, several miRNAs are upregulated, some of which undoubtedly play oncogenic roles (38).
3. OncomiRs and Tumor-suppressor miRs
miRNAs are up- or down-regulated in malignant tissues compared to the normal counterpart, and they can be considered as oncogenes or tumor-suppressors, respectively (Table 1 and 2). Since miRNAs have a very restricted tissue-specific expression, it is important to observe that the apparent miRNA modulation in cancer tissues may represent the manifestation of a different cell population in the tumor compared to the normal tissue. Therefore, it is essential to experimentally identify the functional link between the tumor and the miRNA modulation. Extensive analyses have highlighted the causative role of many miRNAs in cancer by using either human cancer cells or genetically engineered animal models. For example, transgenic expression of miR-155 (39) or miR-21 (40) and deletion of miR-15a/16-1 are sufficient to initiate lymphomagenesis in mice. Conversely, systemic delivery of selected miRNAs including let-7, miR-26a, miR-34a, and miR-143/145 inhibits tumor progression in vivo (41–43).
Table 1.
Tumor-suppressor miRNAs.
| miR | Chr | Tumor | Target | Notes |
|---|---|---|---|---|
| miR-15/16 | 13q14.2 3q25.33 |
CLL | BCL2 | miR-15/16 target BCL2 inducing apoptosis in a leukemic cell line model. |
| Colon cancer | COX-2 | MiR-16 targets COX-2; elevated levels of HuR antagonize miR-16 function. | ||
| Follicolar lymphoma | CHEK1 | MiRNA profiles are associated with an increased proliferation and a “late” germinal center B-cell phenotype. | ||
| Fibroblast | CEBPβ, CDC25a, CCNE1 | Upon cell-cycle re-entry, rapid decay of miR-16 alleviates repression of target genes, allowing proper resumption of the cell cycle | ||
| Fibroblast | VEGF, VEGFR2, FGFR1 | MiR-16 plays important roles in regulating cell-intrinsic angiogenic activity of endothelial cells | ||
| Cancer associated fibroblast | FGF2, FGFR1 | Downregulation of miR-15 and miR-16 in cancer-associated fibroblasts (CAFs) promoted tumor growth and progression | ||
| - | CCNE1 | MiR-15 and miR-16 families as novel transcriptional targets of E2F. | ||
| Multiple myeloma | FGFR1, PI3KCa, MDM4, VEGFa | Deletion of miR-15/16 is observed in early stages of multiple myeloma. | ||
| Breast Cancer | WIP1 | MiR-16 regulates Wip1 phosphatase in the DNA damage response and mammary tumorigenesis. | ||
| Ovarian cancer | BMI-1 | miR-15a/16 target Bmi-1 is leading to low proliferation and clonal growth. | ||
| Lung cancer | CCND1, CCND2, CCNE1 | Overexpression of miR-15/16 induces arrest in G(1)-G(0). | ||
|
| ||||
| miR-34 | 1p36.22 11q23.1 |
Colon cancer | SIRT1 | MiR-34 targets SIRT1 leading to apoptosis only in p53 wildtype context. |
| Gastric cancer | BCL2, NOTCH, HMGA2 | MiR-34 targets Bcl-2, Notch, and HMGA2. | ||
| Fibroblast | MYC | During senescence, miR-34a targets MYC and controls a set of cell cycle regulators. | ||
| Lung cancer | AXL | MiR-34a and -199a/b target Axl receptor; both miRs are silenced by promoter methylation. | ||
| Ovarian cancer | MET | MiR-34 targets MET. | ||
| Embryonic fibroblast | NANOG, SOX2, MYCN | MiR-34a targets pluripotency genes and suppresses reprogramming. | ||
| Colon cancer | SNAIL | A new link between p53, miR-34, and Snail1 in the regulation of cancer cell EMT programs. | ||
|
| ||||
| Let-7 family | 9q22.32 Xp11.22 22q13.31 21q21.1 19q13.41 11q24.1 3q21.1 12q14.1 |
Lung cancer | KRAS | The let-7 family negatively regulates let-60/RAS in C. elegans and lung tumors. |
| - | HMGA2 | Chromosomal translocations disrupt repression of HMGA2 by let-7 miRNA. | ||
| Burkitt lymphoma | MYC | Dysregulation of let-7 participates in genesis and maintenance of Burkitt lymphoma and other MYC-dysregulated cancers. | ||
| - | IMP-1 | Let7-oncofetal proteins could be novel therapeutic targets and potential biomarkers for cancer treatment. | ||
| - | DICER | Existence of a regulatory loop to regulate the equilibrated state of Dicer and various miRNAs. | ||
| Fibroblast | CDC-34 | Let-7 represses Cdc34, stabilizes Wee1 kinase, and increases a fraction of cells in G(2)/M in primary fibroblasts | ||
| Breast cancer | Il6 | Inflammation activates a positive feedback loop that maintains the epigenetic transformed state. | ||
| Prostate cancer | E2F2, CCND2 | Let-7a targets E2F2 and CCND2 acting as a tumor suppressor in prostate cancer. | ||
| Liver cancer | BCL-XL | Let-7 targtes Bcl-xL and potentiates sorafinib-induced apoptosis. | ||
|
| ||||
| miR-200 family | 1p36.33 12p13.31 |
Breast cancer | ZEB1, ZEB2 | Downregulation of the miR-200 family may be an important step in tumor progression |
| Bladder cancer | ERRFI-1 | MiR-200 restores EGFR dependency. | ||
| Nasopharyngeal carcinoma | ZEB1, CTNNB1 | MiR-200a inhibits cell growth, migration and invasion. | ||
| Pancreatic cancer | BMI-1 | ZEB1 links EMT and stemness-maintenance by suppressing the miR-200 family and thereby promotes migration | ||
| Breast cancer | PLCγ1 | TmiR-200 negatively regulates EGF-driven invasion, viability, cell cycle progression. | ||
| FAP1 | MiR-200c sensitizes cells to apoptosis mediated by CD95. | |||
| Breast cancer | SUZ12 | miR-200b-Suz12-cadherin pathway sustains cancer stem cell growth and invasiveness. | ||
| Lung cancer | FLT1/VEGFR1 | MiR-200 suppresses metastasis by targeting Flt1. | ||
| - | JAG1, MALM2, MALM3 | These findings explain increased Notch signaling in some types of cancers, where mutations in Notch pathway genes are rare. | ||
| Breast and endometrial cancer | FN1, LEPR, NTRK2, ARHGAP19 | MiR-200c inhibits cell motility and anoikis resistance. | ||
| Ovarian cancer | p38α | MiR200a-dependent stress signature correlates with improved survival and response to treatment. | ||
Table 2.
Oncogenic miRNAs.
| miR | Chr | Tumor | Target | Notes |
|---|---|---|---|---|
| miR-17-92 | 13q23.1 | Colon | TSP-1, CTGF | Upregulated in colonocytes coexpressing K-Ras, c-Myc and p53 impaired activity. |
| Prostate cancer Burkitt lymphoma Testis carcinoma |
E2F2, E2F3 | Autoregulatory feedback loop between E2F factors and miR17/92. | ||
| Myc induced lymphoma | BIM PTEN |
Transgenic mice with higher expression of miR-17/92 in lymphocytes. | ||
| Lung cancer | HIF1α | Intricate and finely tuned circuit involving c-myc, miR-17/92, and HIF1α. | ||
| Cervix tumor cell line | PTPRO | E2F1 and miR-17/92 controls PTPRO. | ||
| Myeloid cells | p63 | MiR-92 increases cell proliferation by repressing p63. | ||
| T-cell acute lymphoblastic leukaemia | BIM PTEN PRKAA1 PPP2R5e |
Functional genomics reveals a repression of regulators of PI3K survival signals by miR-19. | ||
| Endothelial cells | JAK1 | MiR-17/92 family provide a therapeutic perspective to enhance therapeutic angiogenesis | ||
| Breast cancer | HBP1 | MiR-17/92 inhibits HBP1 regulates invasion activating Wnt/β-catenin. | ||
| Ras induced senescent-fibroblasts | p21(WAF1) | Disruption of senescence by miR-17/92. | ||
| Glioblastoma | TGFβII SMAD4 |
MiR-17/92 suppress TGFβ stimulating angiogenesis and tumor cell growth. | ||
| Prostate | MnSOD GPX2 TRXR2 |
MiR-17/92 suppress tumorigenicity by inhibiting mitochondrial antioxidant enzymes. | ||
|
| ||||
| miR-222/221 | Xp11.3 | Glioblastoma, prostate and thyroid carcinoma | p27(Kip1) | High miR-222/221 maintain low p27(Kip1) and stimulate proliferation |
| Normal fibroblast | p57 (Kip2) | Up-regulation of miR-222/221 initiates S phase with growth factor signaling pathways that stimulate cell proliferation. | ||
| Non small cell lung cancer and hepatocellular carcinoma | PTEN, TIMP3 | MiR-222/221 target PTEN and TIMP3; induce TRAIL resistance and enhance cellular migration. MET oncogene activates miR-222/221 through the c-Jun transcription factor. | ||
| Breast cancer | FOXO3A | MiR-222/221 target FOXO3A to suppress p27(Kip1) also at a transcriptional level. | ||
| Endotelial cells | KIT | miR-222 targets c-Kit controlling the ability of endothelial cells to form new capillaries. | ||
| Breast cancer | ESR1 | Modulation of ERα is associated with anti-estrogen therapy. | ||
| Glioblastoma | PUMA | MiR-221/222 directly regulate apoptosis by targeting PUMA. | ||
| Breast cancer | TRSP1 | MiR-221/222 promote EMT contributing to the more aggressive clinical behavior of basal- like breast cancers. | ||
| Glioblastoma | PTPμ | MiR-221/222 target PTPμ and regulate glioblastoma tumorigenesis. | ||
| Breast cancer | DICER | miR-221/222 repress Dicer in ERa negative breast cancers. | ||
| Non small cell lung cancer | APAF1 | MiR-221/222 are activated by EGFR and MET; by targeting APAF1, miR-221/222 are responsible of gefitinib resistance | ||
|
| ||||
| miR-21 | 17q23.1 | Cholangiocarcinoma | PTEN | MiR-21 modulates gemcitabine-induced apoptosis by PTEN-dependent activation of PI3K. |
| Breast cancer | TPM1 | Suppression of mir-21 inhibits tumor growth | ||
| Breast cancer | PDCD4 | miR-21 suppresses PDCD4 to control apoptosis. | ||
| - | SPRY1 | MiR-21-null mice showed a significant reduction in papilloma formation compared with wild-type mice. | ||
| Glioblastoma | RECK, TIMP3 | Inhibition of miR-21 provides a novel therapeutic approach for “physiological” modulation of multiple proteins whose expression is deregulated in cancer. | ||
| Glioblastoma | p63, JMY, TOPORS, TP53BP2, DAXX, HNRPK, TGFβRII | MiR-21 targets multiple important components of p53, TGF-β and mitochondrial apoptosis tumor-suppressive pathways. | ||
| Prostate cancer | MARKS | MiR-21 promotes apoptosis resistance, motility, and invasion. | ||
| Prostate cancer | ANP32A, SMARCA4 | - | ||
|
| ||||
| miR-155 | 21q21.3 | Breast cancer | SOCS1 | miR-155 may serve as a bridge between inflammation and cancer. |
| AML | CEBPB, PU.1, CUTL1 PICALM | MiR-155 as a contributor to physiological GM expansion during inflammation and to certain pathological features associated with AML. | ||
| - | BACH1, ZIC3 | Induction of miR-155 by EBV contributes to EBV-mediated signaling. | ||
| Human cord blood CD34+ | ETS1, MEIS1 | MiR-155 is required for megakaryocytic proliferation and differentiation. | ||
| Lymphocytes | C-MAF | Bic/microRNA-155 plays a key role in the homeostasis and function of the immune system. | ||
| Diffuse large B-cell lymphoma | HGAL | Cell dissemination and aggressiveness is a phenotype of DLBCL typically expressing high levels of miR-155 and lacking HGAL expression. | ||
| Nasopharyngeal carcinoma | JMJD1A | LMP1 and LMP2A activates miR-155 to repress JMJD1A. | ||
| Breast cancer | WEE1 | MiR-155 targets WEE1 and enhances mutation rates by decreasing the efficiency of DNA safeguard mechanisms. | ||
| Pancreatic cancer | TP53INP1 | - | ||
| - | SMAD1, SMAD5, HIVEP2, CEBPB, RUNX2, MYO10 | Role for miR-155 in controlling BMP-mediated cellular processes | ||
| Breast cancer | FOXO3a | Molecular links between miR-155 and FOXO3a affect cell survival and response to chemotherapy. | ||
| Colon cancer | hMSH2, hMSH6, and hMLH1 | Inactivation of mismatch repair is induced by miR-155 | ||
| Diffuse large B-cell lymphoma | SMAD5 | - | ||
4. Hallmarks of cancer
miRNAs affect all of six hallmarks of malignant cells: 1) self-sufficiency in growth signals (let-7 family), 2) insensitivity to anti-growth signals (miR-17-92 cluster), 3) evasion from apoptosis (miR-34a), 4) limitless replicative potential (miR-372/373 cluster), 5) angiogenesis (miR-210), and 6) invasion and metastases (miR-10b).
5. Target Redundancy
A recurrent theme in miRNA biology is their redundancy. As previously described, miRNAs are genomically redundant. Redundancy is also a feature of the miRNA targeting. miRNAs control the expression of a transcript by binding with their seed region a miRNA responsive motif generally located in the 3′ UTR of the target mRNA. Each miRNA is predict to repress the expression of thousand mRNAs but, in turn, each mRNA can be targeted by several hundred different miRNAs.
6. Context specificity
miRNAs should not be simply described as oncogenes or tumor suppressor genes, unless the tissue or cell type involved in their action is specified. In fact, several miRNAs can be considered as oncogenes or tumor suppressor depending on cell types where they will be expressed. For example, miR-221 and miR-222 target an oncogene, KIT, and inhibit the growth of erythroblastic leukaemia (44), and therefore function as tumor suppressors in erythroblastic cells. But they also target at least four important tumor suppressors - phosphatase and tensin homologue (PTEN), p27, p57 and tissue inhibitor of metalloproteinases 3 (TIMP3) - and function as oncogenic miRNAs by suppressing these tumor suppressors in various human solid tumors (45).
7. 3′ UTR shortening
About half of mammalian genes use alternative cleavage and polyadenylation (APA) to generate multiple mRNA isoforms differing in their 3′ UTRs. In malignant tissues, 3′ UTRs are frequently shortened by alternative polyadenylation site choice (46,47). This reduction of the 3′ UTR length can determine a widespread loss of miRNA inhibitory activity, increasing the heterogeneity and plasticity of the tumor cell population. For instance, the loss of the extended isoform of Hip2 3′ UTR in activated activated T cells releases the negative regulation mediated by miR-21 and miR-155 on this transcript.
8. Loop activity
Participation in negative and positive feedback loops is another common mechanism of miRNA action in cancer cells. This is well illustrated by miR-146a, which is transactivated by the NF-kB pathway and negatively feeds back on this signaling cascade by targeting two upstream activators of the pathway, TRAF6 and IRAK1 (48). Another example of miRNA looping action is the transactivation mediated by NF-KB upon LIN28B, an RNA binding protein, which accelerates the turnover of let-7 precursors (49). Lin28B-dependent downregulation of let-7 causes upregulation of IL-6, a let-7 target, which further stimulates NF-kB (49).
9. RNAs sponge
It has been hypothesized that some genes with conserved miRNA-binding sites act as bona fide targets in one cell type or condition but as miRNA-sequestering agents (“pseudotargets”) in other cell types or conditions (50). Recently there have been reports of endogenous transcripts that appear to act as natural miRNA decoys, called “competitive endogenous RNAs” (ceRNAs) (51–55). Pandolfi laboratory demonstrated, by using an integrated computer analysis and an experimental validation process, that some of these ceRNAs are regulated by the same set of miRNAs that regulate PTEN and have similar expression profiles to PTEN. Knockdown of two ceRNAs, VAPA and CNOT6L, resulted in reduced levels of PTEN. Importantly, the link between PTEN, VAPA and CNOT6L was lost in cells that had defective miRNA processing, indicating that this regulatory network works by affecting the abundance of miRNAs that are able to bind a specific set of mRNAs.
10. Circulatory miRNAs
A new intriguing aspect of miRNA biology is their presence in numerous body fluids, including serum, plasma, saliva, urine and amniotic fluid (56). miRNAs in serum correlate with the presence of hematologic malignancies and solid tumors and have been reported to be of value for early detection of various types of cancer, preceding diagnosis by conventional methods (57). Active research is primarily focused in the understanding of the process of miRNA release and function. miRNAs are secreted from cells as lipoprotein complexes, small membranous vesicles known as exosomes. The majority of non vesicular miRNAs were reported to be present in plasma as Ago2 ribonucleo-protein complexes, suggesting functional miRNA-induced silencing complexes (58). How miRNAs are selected to be secreted is still unknown. The vesicular miRNAs fraction was recently reported to interact with the Toll-like receptors of immune cells to stimulate the production of prometastatic inflammatory cytokines and inducing the pro-tumor inflammatory processes (59). In this scenario, circulating miRNAs can act as signals for receptor activation, a function that is completely independent of their conventional role in post-transcriptional gene regulation.
Lesson from animals
The large number of experiments performed using genetically engineered mice has shown the dramatic influence of miRNAs in tumorigenesis. During these years, several strains of mice lacking or overexpressing cancer-associated miRNAs have been developed and characterized, including the germline transgenic or knockout mice of miR-155 (39), miR-17~92 and its paralogues (60–62), miR-21 (40), miR-221 (63), miR-15/-16-1 cluster (64), miR-122 (65, 66), and miR-34 paralogues (67). Additional information on the role of miRNAs in tumorigenesis have also been acquired by the analyses of phenotype generated by the conditional deletion of Dicer which mimics the general knockdown of miRNAs in tumors.
miR-155
In 2006, our laboratory described the first mouse model of miRNA overexpression, the E3-miR-155 mice, that provided the proof that a miRNA can contribute directly to the pathogenesis of a malignancy (39). miR-155 is overexpressed in aggressive CLL (68), pediatric Burkitt’s lymphoma (69), Hodgkin’s lymphoma (70), primary mediastinal non-Hodgkin’s lymphoma (70), AML (71), lung cancer (38) and breast cancer (72). Overexpression of miR-155 in preB-cells, obtained by using the immunoglobulin E3 enhancer, resulted in pre-leukemic expansion of the pre-B-cell population ultimately resulting in full-blown B cell tumors (39).
miR-17/92 cluster and parologues
Similar to miR-155, the miRNAs encoded by the miR-17–92 cluster were identified as highly overexpressed in cancer and located at a fragile site in the genome (38,73). Although miR-17~92 overexpression does not appear to be sufficient to initiate tumorigenesis per se, transgenic mice overexpressing this cluster in lymphocyte progenitor cells develop a lymphoproliferative disorder affecting both B and T cells that eventually results in autoimmunity (61). Retroviral overexpression of the cluster accelerated Myc-induced lymphoma formation (62) and in vivo dissection of the cluster activity demonstrated that miR-17-92 tumorigenic activity is dependent from the presence of miR-19 seed family (74, 75). Mice knockout of mir-17-92 confirmed the importance of the cluster for B cell development (61), whereas deletion of the paralogues, mir-106b~25 or mir-106a~363, had no obvious phenotypes (76). By contrast, overexpression of the mir-106b~25 cluster cooperated with overexpression of its host gene, mini-chromosome maintenance protein 7 (Mcm7) to induce prostatic intraepithelial neoplasia (77). This provides an example of overexpression of a single genetic locus contributing to two ‘oncogenic hits’: elevated MCM7 levels and an increased expression of oncogenic miR-106b~25 (77).
miR-21
Of particular interest is miR-21 which is overexpressed in most of the different solid tumors analysed (39). Genetic deletion of miR-21 reduced lung tumor burden following activation of a mutant KrasG12D allele and, in accordance, a miR-21 transgene resulted in increased tumor outgrowth (78). The final proof of miR-21 oncogenic activity came from the Slack laboratory who generated an inducible miR-21 overexpression which resulted in potent lymphoma development (40). This study furthermore established that tumors become addicted to miR-21 overexpression as transcriptional shut off of the transgene resulted in rapid tumor regression likely due to apoptosis.
miR-222/221 cluster
Another example of miRNA overexpression with oncogenic consequences is represented by the recent publication by Negrini laboratory (63). miR-222/221 cluster is one of the most commonly up-regulated miRNA in human cancer, including hepatocellular carcinoma where 70–80% of cases showed an increase in their levels (39). Liver-specific overexpression of miR-221 induced a strong predisposition to the development of liver tumors and, at 9 month of age, mice developed visible neoplastic lesions which range from liver adenomas to typical hepatocellular carcinomas characterized by an invasive trabecular growth and a high level of angiogenesis (63).
miR-15/16-1 cluster
miR-15 and miR-16 genomic locus (13q14) is heterozygously deleted in 68% of all patients with B-cell chronic lymphocytic leukemia (CLL) (79). The importance of genetic lesions reducing miR-15/16-1 cluster expression in CLL was furthermore confirmed by the identification of a point mutation located in the 3′ flanking region of miR-16 that reduces miR-16 expression in a naturally occurring CLL mouse model, the New Zealand black (NZB) mouse, that develops a hematological disorder similar to the human CLL (80). The final proof of the tumor suppressive effects of miR-15 and miR-16 came recently from the Dalla-Favera laboratory who proved the genetic deletion of miR-15/16-1 cluster in mice recapitulates after 18 month from birth the full spectrum of CLL-associated lymphoproliferations in humans, including CLL, CD5(+) monoclonal B-cell lymphocytosis, and CD5(−) non-Hodgkin lymphomas (64). Interestingly, the same authors also demonstrated that the size of 13q14 deletions, where the miR15/16-1 cluster is located, influences the phenotype of the lymphoproliferative disorders and potentially the severity of disease, suggesting a tumor-suppressor function for genetic elements besides miR-15a/16-1 and their host gene DLEU2 (81). In fact, an additional deletion of a 0.69 mb-large region telomeric to miR-15/16-1 increased the appearance of CLL and mice seemed to succumb to their disease faster than miR-15/16-1 deleted mice.
miR-122
miR-122 is a conserved liver-specific microRNA (miRNA) that constitutes 70% of the cloned hepatic miRNAs in the adult mouse (3). In vivo antisense-mediated inhibition of miR-122 is well tolerated in adult mice, reduces serum cholesterol and leads to the induction of genes that are normally repressed in adult liver (82), suggesting that this miRNA is important for the maintenance of the terminally differentiated hepatocyte gene expression program. Genetic deletion of miR-122 also lowers serum cholesterol and leads to accumulation of hepatic triglycerides in young mice that with age induces hepatic inflammation, progressive onset of fibrosis and, eventually, tumors resembling hepatocarcinoma (65, 66).
miR-34 paralogues
The miR-34 family consists of three highly related miRNAs, miR-34a, miR-34b and miR-34c, respectively. The miR-34 family has been shown to form part of the p53 tumor-suppressor network: their expression is directly induced by p53 in response to DNA damage or oncogenic stress (83). A big disappointment in the field was the report from two different laboratories that genetic deletion of all three miR-34 members are viable and fertile and do not display morphological defects and are not prone to spontaneous tumor formation (67). These findings are difficult to reconcile with reports placing miR-34 downstream p53 and illustrate the importance of genetic studies in understanding the significance of miRNA.
Dicer
It is now clear, based on hundreds of expression profiling studies, that tumors ubiquitously exhibit dysregulated miRNA expression patterns to corresponding normal tissue, with a general decrease in the levels of the majority of miRNAs (35, 38). Two recent reports define one of the underlying genetic causes of impaired miRNA processing in human cancer (84, 85). Hemizygous deletion of the gene that encodes Dicer occurs in 27% of tumors of different tissue origins, whereas homozygous deletion has yet to be reported. Conditional Dicer mutation in different mouse models of cancer (e.g. K-Ras driven lung cancer (84), and Rb driven retinoblastoma (85)) downregulates miRNAs and accelerates the formation of early neoplastic lesions, which progress to aggressive and metastatic tumors only when Dicer is partially active. Therefore, Dicer is an example of a gene that functions as a tumor suppressor in the context of haploinsufficiency but not homozygous loss-of-function. However, since Dicer haploinsufficency accounts for only 27% of the human tumors, this condition may only partially explain the widespread loss of miRNAs in human tumors.
Causes of miRNA dysregulation in cancer
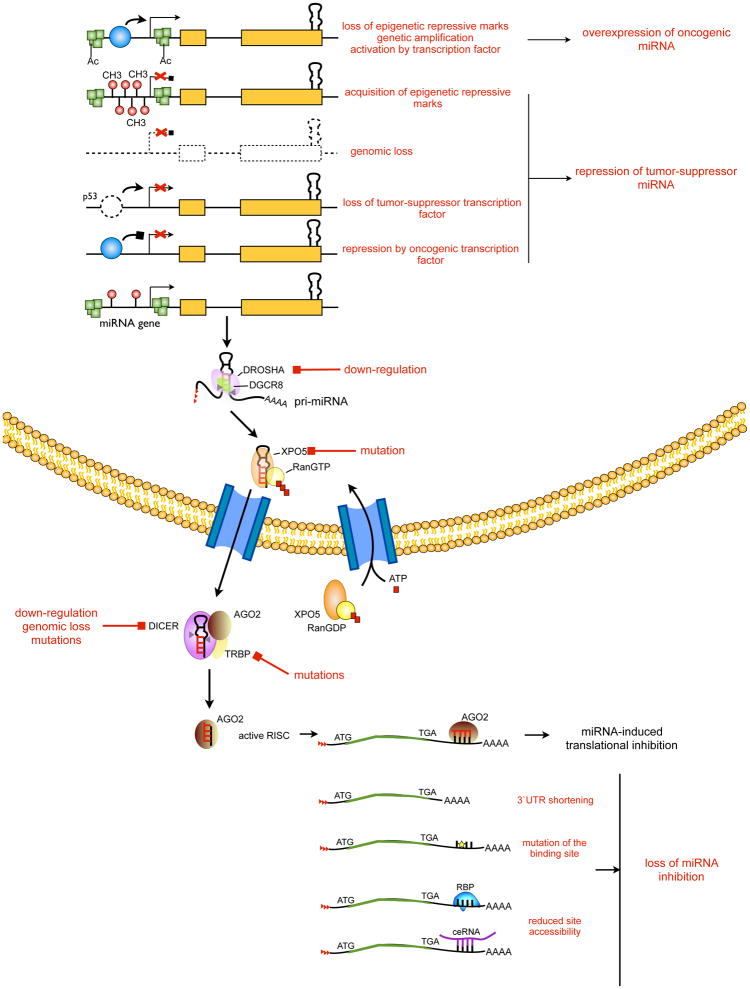
A precise control of miRNA levels is essential for maintaining normal cellular homeostasis. A daunting task in the miRNA field during the last 11 years has been the identification of mechanisms responsible for the dysregulation of miRNAs in cancer. In the following section, we describe some of the identified mechanisms (Figure 2).
Figure 2. miRNAs dysregulation in cancer.
The canonical miRNA biogenesis pathway and miR-RISC activity are schematically represented. In red, steps commonly deregulated in cancer are reported and visually associated to the miRNA dysregulation identified in cancer. RNA binding protein abbreviated as RBP. ceRNAs represented in blue. CH3, methylation; Ac, acetylation.
Genetic abnormalities and miRNAs
Chromosomal rearrangements, genomic amplifications, deletions or mutations, alter miRNA genes, much like they affect protein coding genes. In 2004, Calin and coworkers showed that more than half of miRNAs map to genomic regions that are frequently altered in cancer (73): 65 miRNAs map to loss of heterozygosity regions (LOH) (e.g. miR-15a/16-1), where the majority of tumor suppressor genes are located; 15 miRNAs are in amplified regions (e.g. miR-17-92 cluster, miR-155), which harbor most oncogenes; 61 miRNAs are in breakpoint regions and fragile sites (FRA) (e.g. let-7 family members), common sites for sister-chromatid exchange, translocation, deletion and tumor-associated viral integration. A clear example of genetic aberration in cancer is represented by the cluster miR-15a/16-1 located on chromosome 13q14.3, the most frequently deleted region in human CLL (33). Both miRNAs were identified as highly down-regulated in CLL patients that showed the deletion 13q14.3, demonstrating a tumor-suppressive role for the two miRNAs. The miR-15a/16-1 locus is also affected by mutations: 15% of analyzed CLL patients presented a germline mutation in the primary precursor of miR-15a/16-1 that reduces their accumulation (68); another point mutation, associated to low levels of both miRNAs, was identified in the 3′ flanking region of miR-16 in the New Zealand black mice that spontaneously develop CLL (80). Interestingly, miR-15/16-1 cluster is duplicated, and located on a different chromosome 3q25.33 (miR-15b/16-2); this redundancy could represent a safeguard mechanism against loss of the expression of the two miRNAs. Another genomic mechanism responsible for miRNA alteration in cancer is the altered levels or presence of mutations in the main enzymes of the miRNA biogenesis machinery, such as Dicer or Drosha. For instance, a decrease of 60% and 51% in the levels of Dicer and Drosha mRNAs have been identified in 39% of patients with ovarian cancer. (86). Moreover Dicer gene is frequently single-copy deleted in cancer, suggesting that it is a haploinsufficient tumor suppressor (84, 85). In contrast to this view, a very intriguing study reported the identification of a somatic missense mutation hotspot in Dicer genes in nonepithelial ovarian tumors (87). The recurrent and focal nature of these mutations and their restriction to nonepithelial ovarian tumors suggested a common oncogenic mechanism associated with a specific altered DICER1 function that is selected during tumor development in these cell types. In this way, a mutated DICER1 may modify its specificity for the recognition of miRNA precursors and generates a preferential processing that will create a new miRNome in cancer.
Epigenetics and miRNAs
Aberrant epigenetic changes are a well-known feature of cancer cells, such as DNA hypermethylation of tumor suppressor genes, extensive genomic DNA hypomethylation and alteration of histone modification patterns (88). In fact, epigenetic modifications represent another common mechanism related to the alteration of miRNA expression in cancer (89, 90). The majority of reports showed an increase in the methylation of tumor-suppressor miRNAs which in turn allows overexpression of the oncogenic targets. It is the case of miR-127 reported to be induced after DNA methylation and histone acetylation inhibitors treatment. Activation of miR-127, epigenetically repressed in 75% of primary prostate and bladder tumors, inhibits the expression of the proto-oncogene BCL6 (91), suggesting that miR-127 is a tumor suppressor. Similarly, the hypermethylation of miR-124a in colon cancer and allows the upregulation of the oncogene CDK6 kinase (92).
miRNAs can not only be modulated by epigenetics marks but, in turn, miRNAs can modify the epigenetic landmarks of the entire genome by targeting the main enzymes responsible for the epigenetic control. For instance, our laboratory identified miR-29 family as able to repress the de novo DNA methyltransferases DNMT3A and DNMT3B in lung (93) and AML cancer cells (94) inducing a general DNA demethylation and reactivation of tumor suppressor genes with further loss of tumorigenicity. In addition, we can consider the repression mediated by miR-101 of the histone methyltransferase EZH2 (95). Genetic deletion of one of both loci harboring miR-101 gene was somatically identified in 37.5% of clinically localized prostate cancer cells and 66.7% of metastatic disease cells. Loss of the miR-101 expression during cancer progression induces an increase in EZH2 expression, which contributes to the epigenetic silencing of target genes and regulates the survival and metastasis of cancer cells. SUZ12, another component of the polycomb repressor complex 2 (PRC2), of which EZH2 is the catalytic subunit, is targeted by the miR-200 family (96). This appears to be important in the formation of cancer stem cells where loss of miR-200 increases SUZ12 expression, which epigenetically mediates a Polycomb-mediated repression of the E-cadherin gene. In conclusion, epigenetic alterations of miRNA genes complemented by genetic inactivation due to mutation or deletion can partially account for the microRNA dysregulation in cancer.
Transcriptional control of miRNAs
The majority of miRNAs is transcribed by the RNA polymerase II and a myriad of PolII-associated factors are involved in this process, allowing an elaborate regulation of miRNA genes transcription in cells (8). One of the first example of cancer-involved transcriptional regulation of miRNA genes, it is the transcriptional activation of miR-17/92 cluster mediated by the MYC oncogene. O’Donnell and collaborators showed that MYC induces the expression of the polycistronic miR-17/92 cluster, which counterbalances the apoptotic activity of E2F1, allowing MYC mediated-proliferation (97). The miR-17-92 cluster and the paralogue miR-106a/363 are also upregulated after estradiol treatment in breast cancer cells (98). Another important transcriptional program that has been identified involves miR-34 family members and the tumor-suppressor protein p53 (83). Transcriptionally induced by p53, miR-34 recapitulates the biological effects of p53 activation including apoptosis or cellular senescence, induction of G1 arrest and loss migration. Mutational and loss of p53 function is one of the most frequent genetic abnormalities in cancer: in ovarian cancer accounts for 60–80% of both sporadic and familial cases (99). In accordance with the loss of p53, miR-34 paralogues (miR-34a, b, c) are repressed: miR-34a expression is decreased in 100% and miR-34b*/c in 72% of ephitelial ovarian cancer with p53 mutation (100). However, miR-34a is also downregulated in 93% of tumors with wild-type p53, indicating the presence of other mechanisms implicated in the suppression of miR-34a gene. In fact, methylation and reduced copy number at the mir-34a gene occur in 27% and 39% of ovarian cancer tissues, respectively.
Post-transcriptional control of miRNAs
As described above, miRNA biogenesis is complex multistep process that integrates several enzymes and regulatory proteins to allow a correct maturation of the primary miRNA precursors. Many components of the miRNA biogenesis machinery could either be misexpressed in tumors or mutated, contributing to a dysregulation in the miRNA expression. The cropping step operated by Drosha is an important point of regulation of miRNA levels. Doxorubicin-induction of p53 increases the expression of several mature miRNAs that have growth-suppressive functions and are downregulated in cancer (such as miR-15, miR-16 and miR-145) (101). In fact, upon treatment, p53 associates with Drosha promoting a Drosha-mediated processing of certain tumor-suppressor miRNAs in response to DNA damage. Thus, the loss of p53 in cancer reduces the levels of miRNAs that are either transcriptionally regulated by p53 or regulated by p53-induced Drosha processing. As previously discussed, pre-miRNA processing occurs in the cytoplasm and they need to transit from the nucleus into the cytoplasm, a process that requires the nuclear export receptor XPO5. In cancers with microsatellite instability, which account for 15% of total colon cancer, mutation of XPO5 generates a truncated version of the protein that is unable to associate with its pre-miRNA cargo and exiting the nucleus (102). Reduction in the cytoplasmic pre-miRNA pools reduced the rate of the dicing step, resulting in decreased mature miRNA levels. Interestingly, although the heterozygous XPO5 mutation decreased accumulation of a large fraction (<20%) of detectable miRNAs, many others remained unaffected. It thus appears possible that XPO5 does not bind to pre-miRNAs indiscriminately but has certain substrate preferences, perhaps mediated by sequence or structure. Downregulated miRNAs might then be those that bind poorly to XPO5 and thus cannot compete well for export by limiting amounts of XPO5. Tumors with microsatellite instability are also characterized by inactivating mutations of the gene encoding TAR RNA-binding protein 2 (TARBP2 or TRBP), an essential functional partner of DICER1 (103). Loss of TRBP destabilizes DICER1 following impairment of miRNA processing and enhancement of cellular transformation. An important post-transcriptional mechanism of miRNA regulation is represented by the control of let-7 family members maturation mediated by Lin28. Despite the almost ubiquitous expression of let-7 primary precursor in normal and cancer cells, the mature miRNAs are abundantly produced exclusively in normal cells (104). Several laboratories have identified the RNA binding protein Lin28 able to interact in the cytoplasm with pre-let-7 in order to induce terminal uridylation of miRNA precursor through the recruitment of the terminal uridyl transferase (TUTase) Zcchc11 (105). The U tail (~14 nt) that is added to the 32 end of pre-let-7 blocks Dicer processing and facilitates the decay of pre-let-7. In fact, levels of Lin-28, an RNA binding inversely correlates with mature let-7 expression during development (106). Given the proto-oncogenic activity of Lin28, this pathway may restrict the expression of the tumor suppressor let-7 and contribute to its repression in cancer (107).
miRNA function in cancer
Cancer development is the result of a multistep process that involves multiple alterations in oncogenes and tumor suppressor genes over a period of time, often over several years. A great deal of data already establishes an important role for miRNAs among the many regulatory factors involved in the pathogenesis of cancer. Many miRNAs have been shown to function as oncogenes or tumor suppressor miRs in the majority of cancers profiled to date (Table 1 and 2).
Tumor suppressor
miR-15/16 cluster
miR-15 and -16 were the first to establish the link between miRNAs and cancer (33). Their expression has been shown to be lost and inversely correlated to BCL2 expression in CLL (108). Loss of miR-15a and miR-16-1 has also been observed in prostate cancer and multiple myeloma (109, 110). Musumeci et al. demonstrated that miR-15 and miR-16 are downregulated in stromal fibroblasts surrounding the prostate tumors in the majority of patients analyzed (111). Reconstitution of miR-15 and miR-16 impaired considerably the tumor-supportive capability of stromal cells in vitro and in vivo by targeting Fgf-2 and its receptor Fgfr1, which act on both stromal and tumor cells to enhance cancer cell survival, proliferation and migration. Pouliot et al. performed a high parallel high-throughput screens for miRNAs that could restore sensitivity to cisplatin-resistant epidermoid cells (112). They demonstrated that miR-15/16 sensitized cisplatin-resistant cells by controlling the expression of Wee1 and CHK1, two kinases commonly overexpressed in cisplatin-resistant cells. Together, these data suggest that miR-15a/16-1 genes are natural antisense interactors of BCL2 and other oncogenes and that they can be used to suppress tumor growth in therapeutic application for a variety of tumors.
Let-7 paralogues
The miRNAs that are encoded by the let-7 family, which include 12 human homologues, were considered as tumor suppressors because they map to fragile sites associated with lung, breast, urothelial and cervical cancers (73). More direct evidence, presented by Takamizawa et al., showed that transcripts of certain let-7 homologues were significantly downregulated in human lung cancer and that this correlated with a poor prognosis (113). In 2005, Johnson and collaborators reported that the loss of let-7 family members resulted in the constitutive overexpression of RAS, an oncogene that contributes to the pathogenesis of several types of human tumors (114). Intriguingly, the promoter activity of let-7a2 has been shown to be downregulated in A549 cells by CEBP-a, a transcription factor with oncogenic properties in lung cancer (115). Hu et al. reported that the expression levels of three let-7 family members, let-7a, let-7b, and let-7g, were significantly decreased in the breast cancer patients with lymph node metastasis compared to those without lymph node metastasis (116).
miR-34 paralogues
miR-34 family comprises three processed miRNAs that are encoded by two different genes: miR-34a is encoded by its own transcript, whereas miR-34b and miR-34c share a common primary transcript. The promoter region of miR-34a,-34b and -34c contains CpG islands and aberrant CpG methylation reduces miR-34 family expression in multiple types of cancer (100). In 2007, reports from several laboratories showed that members of the miR-34 family are direct p53 targets, and their upregulation induced apoptosis and cell-cycle arrest (83). BCL2 and MYCN were identified as miR-34a targets and likely mediators of the tumor suppressor phenotypic effect in neuroblastoma (117). Wang et al. reported that the expression of miR-34b was lower in NSCLC tissues compared to that in pericarcinous tissues of lung cancer (118). Additionally, they showed that lower miR-34b expression was correlated with higher lymph node metastasis. In vitro gain-of-function experiments indicated that miR-34b suppressed cell proliferation by inducing cell apoptosis. They proved also that miR-34b by downregulating Met increases the phosphorilation of p53 (phospho S392), and inversely p53 upregulates miR-34b in a feedback loop, which provides new insights into the roles of miR-34 family members in the regulation of signaling pathways of NSCLC.
miR-200 family
Members of the miR-200 miRNA family (miR-200a, miR-200b, miR-200c, miR-141, and miR-429) are downregulated in human cancer cells and tumors due to aberrant epigenetic gene silencing and play a critical role in the suppression of epithelial-to-mesenchymal transition (EMT), tumor cell adhesion, migration, invasion and metastasis, by targeting and repressing the expression of key mRNAs that are involved in EMT (119). Gregory and colleagues, reported that enforced constitutive expression of the miR-200 miRNAs in mesenchymal cells promoted mesenchymal epithelial transition (MET) (120). Conversely, inhibition of miR-200 induces a mesenchymal-like spindle cell morphology, accompanied by an increase in ZEB1 expression and cell migration. Furthermore, loss of miR-200 correlates with a lack of E-cadherin expression in invasive breast cancer cell lines and in breast tumor specimens, supporting an in vivo role for the miR-200 family in EMT repression (121). Overexpression of the individual miR-200 members or separate clusters represses EMT by directly targeting and downregulating ZEB1 and ZEB2 via miR-200-binding sites located within their 3′ UTRs, resulting in enhanced E-cadherin expression and inhibition of murine mammary tumor cell migration and cancer cell motility (121, 122). Interestingly, ZEB1 and SIP1 have been found to bind directly to an E-box proximal minimal promoter element and repress primary transcript and mature miR-200 miRNA expression in mesenchymal human breast cancer cells, demonstrating a potential double-negative feedback loop between ZEB1/SIP1 and the miR-200 family during EMT and tumorigenesis (123). In a recent study, DNA methylation was shown to play an important role in regulating normal cell and tissue-specific expression of the miR-200c-141 cluster (124). Iliopulos et al. investigating the role of the three Akt isoforms, found that the balance of Akt1/2 in transforming growth factor-beta (TGFbeta)-treated MCF10A cells controls the abundance of miR-200 (125). Akt1 knockdown in MCF10A cells promoted TGFb-induced epithelial-mesenchymal transition (EMT) and a stem cell–like phenotype. Mammary adenocarcinomas in MMTV-ErbB2/Akt1−/− mice showed increased invasiveness because of the low abundance of the miR-200 family and E-cadherin and high abundance of Zeb1. Importantly, the ratio of Akt1 to Akt2 and the abundance of miR-200 and of the messenger RNA encoding E-cadherin in a set of primary and metastatic human breast cancers were consistent with the hypothesis that in many cases, breast cancer metastasis may be under the control of the Akt–miR-200–E-cadherin axis. Together, these findings suggest a potential tumor suppressor role for miR-200 that is downregulated in human cancers leading to EMT and facilitating tumor cell invasion and metastasis, and loss of miR-200 family members is associated with an aggressive cancer cell phenotype. Cancer stem-like cells (CSCs) are a highly tumorigenic cell type present as a minority population in developmentally diverse tumors and cell lines (126). Using a genetic screen in an inducible model of CSC formation in a breast cell line, Polytarchou et al. identified several miRNAs that are down-regulated in CSCs and inhibit CSC growth (127). Among them members of the miR-200, let-7, miR-15/16, and miR-103/107 family. Strikingly, individual overexpression of these miRNAs inhibits the formation of mammospheres by at least 50%. These miRNAs affect common target genes that encode the Bmi1 and Suz12 components of the polycomb repressor complexes as well as the DNA-binding transcription factors Zeb1, Zeb2, and Klf4.
Oncogenes
miR-155
miR-155, encoded by the ncRNA BIC (B-cell integration cluster), has been shown to function as oncomiR in the majority of solid and hematopoietic malignancies profiled to date. In humans, BIC RNA levels are low in normal lymphoid tissues but increase in various B-cell malignancies, including Hodgkin’s Lymphoma and only some subtypes of Non Hodgkin’s Lymphoma (128, 129). Specifically, high expression of BIC/miR-155 was observed in Reed-Sternberg cells of Hodgkin’s Lymphoma (130), and in the lymphocytic and histiocytic cells of the variant nodular lymphocyte-predominant Hodgkin’s Lymphoma (128). Quite recently, O’Connell and others, found increased expression of miR-155 in bone marrow blasts of patients with acute myeloid leukemia (AML) (131). Strong induction of miR-155 in mouse bone marrow after injection of bacterial lipopolysaccharide (LPS) correlated with granulocyte/monocyte (GM) proliferation. Furthermore, miR-155 repressed a subset of genes with reported roles in myeloid hyperplasia and/or hematopoiesis such as Bach1, Sla, Cutl1, Csf1r, Jarid2, Cebpβ, PU.1, Arntl, Hif1a, and Picalm. MiR-155 gene was found to be over expressed in several solid tumors, such as breast cancer (38, 77), colon cancer (38), cervical cancer (132), pancreatic ductal adenocarcinoma (PDAC) (133), thyroid carcinoma (134), and lung cancer (135), where it is considered to be a marker of poor prognosis.
miR-17/92 cluster and paralogues
miR-17-92 cluster is located within ~1 kb of an intron of the C13orf25 locus on human chromosome 13q31, a region frequently amplified in several types of lymphoma and solid tumors (136, 137). Overexpression of the cluster is observed in a variety of human cancers, including small cell lung cancer, colon cancer, neuroblastomas, medulloblastoma, gastric cancer and osteosarcoma (60). The miR-106b-25 polycistron is composed of the highly conserved miR-106b, miR-93, and miR-25 that accumulate in different types of cancer, including gastric, prostate, and pancreatic neuroendocrine tumors, as well as neuroblastoma and multiple myeloma (60). Petrocca and collaborators demonstrated that miR-106b-25 cluster, upregulated in a subset of human gastric tumors, is activated by E2F1 in parallel with its host gene, Mcm7 (138). In turn, miR-106b and miR-93 regulate E2F1 expression, establishing a miRNA-directed negative feedback loop. Furthermore, upregulation of these miRNAs induces the development of TGFbeta resistance in gastric cancer by suppressing p21 and Bim.
miR-222/221 cluster
miR-222/221 cluster is among the most dysregulated miRNAs implicated in cancer. Expression of miR-222/221 is highly upregulated in a variety of solid tumors, including, hepatocarcinoma (139), estrogen receptor negative breast cancer cells (140), thyroid cancer (141), and melanoma cells (142). Stinson et al. reported that miR-221 and miR-222 decreased expression of epithelial- and increased expression of mesenchymal-specific genes (143). They showed that the transcription factor FOSL1 directly stimulated the transcription of miR-221/222, which in turn by targeting TRPS1 (trichorhinophalangeal syndrome type 1), a Zeb2 (Zinc finger E-box-binding homeobox 2) inhibitor, reduce E-cadherin aboundance, contributing to the aggressive clinical behavior of basal-like breast cancer. Elevated miR-221&222 expression has been linked not only to proliferation (144) and migration (143) but also to apoptosis (145). We recently reported that miR-221/222, controlled by both MET and EGF receptors, play a pivotal role in TKIs (tyrosine kinase inhibitors) resistance in NSCLCs (145).
miRNAs and therapy
The development of new treatments has contributed substantially to increased 5-year survival and the reduction in overall mortality rates. However, although the classification of cancers has become increasingly diversified, the variety and specificity of treatment options has lagged behind. Because each miRNA regulates the expression of hundreds of different genes, miRNAs can function as master coordinators, efficiently regulating and coordinating multiple cellular pathways and processes. With the progress in profiling, cancer treatments can now be customized for each individual. The complexity of microRNA biology offers a novel mechanism of action for therapeutic intervention but also poses unique challenges for the development of therapeutic modulators as drugs. The therapeutic application of miRNAs involves two strategies: the first one aims to inhibit oncogenic miRNAs by using miRNA antagonists, such as anti-miRs, locked-nucleic acids (LNA) or antagomiRs (146). MiRNA antagonists are single-stranded RNA molecules of about 21–23 nucleotides in length which act through complementary base pairing with miRNAs. To achieve effective pharmacological inhibition of disease-associated miRNAs, miRNA antagonists contain chemical modifications to enhance binding affinity, confer nuclease resistance and facilitate cellular uptake (146). To increase nuclease resistance, most antimiRs harbour phosphorothioate backbone linkages which promote plasma protein binding, thereby reducing clearance of the antimiRs by glomerular filtration and urinary excretion, which facilitates tissue delivery of antimiRs in vivo (147, 148). AntimiRs containing cholesterol, conjugated via a 2′ Omethyl (2′ OMe) linkage, named “antigomiRs”, are fully complementary to the mature miRNA sequence and contain several phosphorothioate moieties to increase stability. The miRNA antagonist chemistries currently in development use unconjugated phosphorothioate antisense molecules with various additional high-affinity 2′ sugar modifications such as 2′ Omethoxyethyl (2′ MOE) or locked nucleic acid (LNA) which possess the highest affinity. Esau et al. showed, for the first time, the efficacy of 2′-MOE-modified antimiR122 in mice. miR-122 inhibition in normal mice reduces plasma cholesterol levels and cholesterol synthesis rates. MiR-122 inhibition in a diet-induced obesity mouse model resulted in decreased plasma cholesterol levels and a significant improvement in liver steatosis, accompanied by reductions in several lipogenic genes (149). LNA-modified antimiR122 has been shown to be efficacious in non-human primates (150). The liver-expressed microRNA-122 is essential for hepatitis C virus (HCV) RNA accumulation in liver cells. Landford and colleagues found that treatment of chronically infected chimpanzees with a locked nucleic acid (LNA)-modified oligonucleotide complementary to miR-122 led to long-lasting suppression of HCV viremia, with no evidence of side effects in the treated animals.
The second strategy, miRNA replacement, involves the re-introduction of a tumor suppressor miRNA mimic to restore a loss-of-function. (151). microRNA mimetics represent an additional level of complexity compared to anti-miRs. First, there are the RISC saturation effects described by Grimm and coworkers (152). On the other hand, there is the potential to induce unwanted effects when novel microRNAs are introduced into a cell. Even though these assumptions are understandable, in vivo evidence for toxicity induced by miRNA mimics is still lacking. Mouse studies that evaluated the therapeutic delivery of tumor suppressor miRNAs failed to find adverse events associated with the miRNA and suggest that delivery of miRNA to normal tissues was well tolerated (41, 154). First, administration of therapeutic miRNA mimics is only an insignificant incremental increase of what is already present in normal cells. Second, normal cells are not addicted to oncogenic pathways and manage to recover from the therapeutic dose used.
Conclusion
Since the discovery of miR-15a and miR-16-1 deletions in CLL, there has been an explosion of publications on miRNAs showing miRNA dysregulation in all tumors, including the most common, such as lung, breast, prostate and gastrointestinal cancers. Many of these have emphasized the role of miRNAs in cancer biology. miRNAs play a role in almost all aspects of cancer biology, including proliferation, apoptosis, invasion/metastasis, and angiogenesis. In addition to transcriptional regulation, post-transcriptional repression mediated by miRNAs seems contributing to the differentiated states by fine-tuning a large network of genes. Despite the large progress in field, many challenges are still ahead. One of the most important aspects is the identification of definitive miRNA signatures for all tumors by large and comprehensive profiling studies: these results will allow the identification of clear diagnostic and prognostic biomarkers that can help physicians in the patients evaluation and therapy. Another important point will be the identification of all targets of the miRNAs involved in cancer, establishing their contribution to malignant transformation. If these miRNA targets are crucial for the expression of the malignant phenotype and the cancer cells depend on their dysregulation for proliferation and survival, we can expect that the use of miRNAs or anti-miRNAs will result in tumor regression. Development of miRNA-based therapeutics is in its infancy. It is too soon to predict whether the promise of harnessing miRNAs for cancer treatment will be realized. The use of this new class of therapeutics represents a new way to tackle some of the more challenging diseases in medicine, including chronic infection, cancer, and neurodegenerative disease but further research is needed to determine the best formulation and precise delivery to the cells, avoiding unwanted miRNA effects that could result from targeting important genes in other healthy tissues.
Acknowledgments
Our work on microRNAs was supported by grant from the National Institutes of Health (CA 152758) and Kimmel Award foundation.
DEFINITION LIST
- heterochronic genes
regulatory genes that are temporally controlled to specify the timing of developmental events in C. elegans
- seed regions
sequence of 6–8nt in length at the 5′ end determinant for target identification
- 5′ cap structure
found on the 5′ end consists of a guanine nucleotide connected to the mRNA via an unusual 5′ to 5′ triphosphate linkage
- polyA
it consists of multiple adenosine monophosphates and it is important for the nuclear export, translation, and stability of mRNA
- Tumor-suppressor
a gene that protects against cancer (e.g. p53)
- Oncogene
a gene that has the potential to cause cancer (e.g. MYC)
- Haploinsufficiency
only a single functional copy of a gene leads to cancer
Footnotes
Disclosure statement
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–06. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–58. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–64. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 6.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327–35. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y, Kim M, Han J, Yeom KH, Lee S, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–01. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 14.Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14:156–59. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han J, Lee Y, Yeom KH, Kim YK, Jin H, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeom KH, Lee Y, Han J, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–29. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–38. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 20.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–59. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–44. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y, Hur I, Park SY, Kim YK, Suh MR, et al. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–32. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 24.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–53. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.John B, Enright AJ, Aravin A, Tuschl T, Sander C, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 29.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 30.Maragkakis M, Vergoulis T, Alexiou P, Reczko M, Plomaritou K, et al. DIANA-microT Web server upgrade supports Fly and Worm miRNA target prediction and bibliographic miRNA to disease association. Nucleic Acids Res. 2011;39:W145–48. doi: 10.1093/nar/gkr294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 32.Loher P, Rigoutsos I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics. 2012;28:3322–23. doi: 10.1093/bioinformatics/bts615. [DOI] [PubMed] [Google Scholar]
- 33.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–29. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 35.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–38. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 36.Lambertz I, Nittner D, Mestdagh P, Denecker G, Vandesompele J, et al. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2010;17:633–41. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar MS, Pester RE, Chen CY, Lane K, Chin C, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–04. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–29. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 41.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10:1470–80. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–87. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. 2005;102:18081–86. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med. 2012;12:27–33. doi: 10.2174/156652412798376170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–47. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–84. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–86. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–06. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seitz H. Redefining microRNA targets. Curr Biol. 2009;19:870–73. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- 51.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–38. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–95. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–81. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–57. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, et al. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boeri M, Verri C, Conte D, Roz L, Modena P, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–18. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–08. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109:E2110–16. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–91. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–14. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Callegari E, Elamin BK, Giannone F, Milazzo M, Altavilla G, et al. Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic model. Hepatology. 2012;56:1025–33. doi: 10.1002/hep.25747. [DOI] [PubMed] [Google Scholar]
- 64.Klein U, Lia M, Crespo M, Siegel R, Shen Q, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 65.Hsu SH, Wang B, Kota J, Yu J, Costinean S, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–83. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–97. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, et al. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8:e1002797. doi: 10.1371/journal.pgen.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–01. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 69.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167–69. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 70.Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–49. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 71.Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–89. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 73.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–04. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mu P, Han YC, Betel D, Yao E, Squatrito M, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–11. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–49. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–93. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–29. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–86. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lia M, Carette A, Tang H, Shen Q, Mo T, et al. Functional dissection of the chromosome 13q14 tumor-suppressor locus using transgenic mouse lines. Blood. 2012;119:2981–90. doi: 10.1182/blood-2011-09-381814. [DOI] [PubMed] [Google Scholar]
- 82.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–99. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 83.He L, He X, Lim LP, de Stanchina E, Xuan Z, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–34. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumar MS, Pester RE, Chen CY, Lane K, Chin C, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–04. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lambertz I, Nittner D, Mestdagh P, Denecker G, Vandesompele J, et al. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2009;17:633–41. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–50. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, et al. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med. 2012;366:234–42. doi: 10.1056/NEJMoa1102903. [DOI] [PubMed] [Google Scholar]
- 88.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 89.Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 2012;31:1609–22. doi: 10.1038/onc.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fabbri M, Calore F, Paone A, Galli R, Calin GA. Epigenetic regulation of miRNAs in cancer. Adv Exp Med Biol. 2013;754:137–48. doi: 10.1007/978-1-4419-9967-2_6. [DOI] [PubMed] [Google Scholar]
- 91.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 92.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–29. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 93.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–18. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–99. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, et al. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761–72. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 98.Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci USA. 2009;106:15732–37. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9:415–28. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–38. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 101.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, et al. Modulation of microRNA processing by p53. Nature. 2009;460:529–33. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 102.Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–15. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 103.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–70. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 104.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–07. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–25. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol. 2005;25:9198–208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012;22:474–82. doi: 10.1016/j.tcb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2006;102:13944–49. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–70. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Roccaro AM, Sacco A, Thompson B, Leleu X, Azab AK, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;26:6669–80. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Musumeci M, Coppola V, Addario A, Patrizii M, Maugeri-Saccà M, et al. Control of tumor and microenvironment cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene. 2011;30:4231–42. doi: 10.1038/onc.2011.140. [DOI] [PubMed] [Google Scholar]
- 112.Pouliot LM, Chen YC, Bai J, Guha R, Martin SE, et al. Cisplatin sensitivity mediated by WEE1 and CHK1 is mediated by miR-155 and the miR-15 family. Cancer Res. 2012;72:5945–55. doi: 10.1158/0008-5472.CAN-12-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–56. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 114.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 115.Guan H, Zhang P, Liu C, Zhang J, Huang Z, et al. Characterization and functional analysis of the human microRNA let-7a2 promoter in lung cancer A549 cell lines. Mol Biol Rep. 2011;38:5327–34. doi: 10.1007/s11033-011-0683-8. [DOI] [PubMed] [Google Scholar]
- 116.Hu X, Guo J, Zheng L, Li C, Zheng TM, et al. The heterochronic microRNA let-7 inhibits cell motility by regulating the genes in the actin cytoskeleton pathway in breast cancer. Mol Cancer Res. 2013 doi: 10.1158/1541-7786.MCR-12-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17(15):1298–307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 118.Wang LG, Ni Y, Su BH, Mu XR, Shen HC, et al. MicroRNA-34b functions as a tumor suppressor and acts as a nodal point in the feedback loop with Met. Int J Oncol. 2013;42:957–62. doi: 10.3892/ijo.2013.1767. [DOI] [PubMed] [Google Scholar]
- 119.Mongroo PS, Rustgi AK. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol Ther. 2010;10:219–22. doi: 10.4161/cbt.10.6312548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–01. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 121.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–4. 22. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Park SM, Gaur AB, Lengyel E, Peter ME. The miR- 200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–54. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 124.Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5:e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Iliopoulos D, Polytarchou C, Hatziapostolou M, Kottakis F, Maroulakou IG, et al. MicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cells. Sci Signal. 2009;2:ra62. doi: 10.1126/scisignal.2000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–28. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 127.Polytarchou C, Iliopoulos D, Struhl K. An integrated transcriptional regulatory circuit that reinforces the breast cancer stem cell state. Proc Natl Acad Sci USA. 2012;109:14470–75. doi: 10.1073/pnas.1212811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kluiver S, Poppema D, de Jong T, Blokzijl G, Harms G, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–49. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 129.Eis PS, Tam W, Sun L, Chadburn A, Li Z, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:2632–27. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.van den Berg A, Kroesen BJ, Kooistra K, de Jong D, Briggs J, et al. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer. 2004;39:167–69. doi: 10.1002/gcc.10186. [DOI] [PubMed] [Google Scholar]
- 131.O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–94. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang X, Tang S, Le SY, Lu R, Rader JS, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–54. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNAexpression profiling of thyroid tumors: biological significance and diagnosticutility. J Clin Endocrinol Metab. 2008;93:1600–08. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, et al. UniquemicroRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 136.Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–95. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 137.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 138.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–86. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 139.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–61. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 140.Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, et al. MicroRNA cluster 221–222 and estrogen receptor ainteractions in breast cancer. J Natl Cancer Inst. 2010;102:706–21. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13(2):497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 142.Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, et al. The promyelocytic leukemia zinc finger–microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–54. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 143.Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, et al. miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1) promotes epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4:ra41. doi: 10.1126/scisignal.2002258. [DOI] [PubMed] [Google Scholar]
- 144.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–08. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Garofalo M, Romano G, Di Leva G, Nuovo G, Jeon YJ, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2011;18:74–82. doi: 10.1038/nm.2577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 146.Krützfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, et al. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–92. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–89. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 148.Levin AA. A review of the issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim Biophys Acta. 1999;1489:69–84. doi: 10.1016/s0167-4781(99)00140-2. [DOI] [PubMed] [Google Scholar]
- 149.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, et al. miR122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 150.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–01. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–30. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 153.Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–30. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]