Abstract
Miniaturized microneedle devices are being developed for painlessly targeting vaccines to the immune cell populations in skin. As skin immunization studies are generally restricted to animal models however, where skin architecture and immunity is greatly different to human, surprisingly little is known about the local human response to intradermal (ID) vaccines. Here we use surgically excised human skin to explore for the first time the complex molecular and cellular host responses to a candidate influenza vaccine comprising nanoparticulate virus-like-particles (VLPs), administered via conventional hypodermic injection or reduced scale microneedles. Responses at the molecular level are determined by microarray analysis (47,296 discrete transcripts) and validated by quantitative PCR (96 genes). Cellular response is probed through monitoring migration of dendritic cells in viable skin tissue. Gene expression mapping, ontological analysis and qPCR reveal up-regulation of a host of genes responsible for key immunomodulatory processes and host viral response, including cell recruitment, activation, migration and T cell interaction following both ID and microneedle injection of VLPs; the response from the microneedles being more subtle. Significant morphological and migratory changes to skin dendritic cells are also apparent following microneedle VLP delivery. This is the first study displaying the global, multifaceted immunological events that occur at the site of vaccine deposition in human skin and will subsequently influence the degree and nature of innate and adaptive immune responses. An increased understanding of the detailed similarities and differences in response against antigen administered via different delivery modalities will inform the development of improved vaccines and vaccine delivery systems.
Keywords: Intradermal, Microneedle, Human skin, Gene expression, Influenza, Virus-like-particles
1. Introduction
Intradermal (ID) delivery of vaccines exploits the network of dendritic cells (DCs) that reside within skin and act as primary antigen presenting cells (APCs). DCs are critical in initiating adaptive immune response and the development of tolerance,[1] making them key targets for immunization and immunotherapy. Indeed, ID delivery of vaccines provides efficient and robust immunogenicity,[2-6] often at low antigen dose.[7] The ID route has been used to convey protective immunity against a range of diseases, including rabies,[6,8] hepatitis B,[9,10] tuberculosis,[11] smallpox[12] and influenza;[4,5,7,13,14] where the first commercially available ID-specific influenza vaccine has been approved for use against seasonal strains.[5] In some situations, for example when individuals are at high risk of disease complications, vaccines with enhanced immunogenicity may be essential but not currently achievable with intra-muscular immunization, whereby the vaccine encounters fewer DCs, macrophages and lymphocytes.[15]
Healthcare professionals currently deliver ID immunizations via the Mantoux technique, an administration procedure requiring specialist expertise. New ID delivery systems that enable simple and/or self-administration of the vaccine in an efficient, reproducible and pain-free manner with reduced infection risk and sharps disposal issues could provide significant advantages where access to healthcare is limited and needles are inappropriately managed. Microporation of the skin using microneedles represents one such minimally-invasive vaccine delivery procedure;[16-19] the microscopic length of the microneedles and nanoscale dimensions of the sharp tip enabling efficient and targeted vaccine delivery to the immune cells in the skin epidermis and dermis with minimum trauma.[20,21] Pre-clinical and clinical studies have demonstrated that microneedles can successfully deliver a wide range of vaccines, including influenza, hepatitis B, hepatitis C, rabies and measles[7,13,19,22-25] to and through skin to facilitate seroconversion and protective immune response.[25,26] Given the clear advantages of reduced scale microneedle delivery, compared with macroscale hypodermic injection, for both the patient (less pain, no bleeding, self-administration) and the clinical team (ease of use, storage, transport and disposal)[27] it is unsurprising that this new administration procedure is now at an advanced state of development.[5,25]
Despite this increasing emphasis towards ID immunization strategies,[28] very little detail is known about the human response to ID vaccines. Pre-clinical testing of ID vaccines is generally restricted to animal models, where skin structure[29] and immunology[30] is considerably different; this issue being amplified in microneedle delivery as the device is specially engineered to target the antigen cargo to specific layers of human skin. There has recently been a desire to address this balance through more basic research on human immunology.[31,32] In this study we begin to characterise and understand the degree and nature of human immune stimulation through analysis of differential gene expression in viable human skin following both ID injection and microneedle delivery of an experimental influenza (H1N1) virus-like-particle (VLP) vaccine. VLPs are artificial nanoparticle (approximately 100 nm in diameter) constructs that can be rapidly manufactured to retain the highly immunogenic components of the virus whilst excluding the potentially unsafe viral genome and replication machinery.[7] Our study aims to provide novel and essential understanding of the early events that occur at a molecular level within human skin following conventional and microneedle immunization with VLPs, providing new insight into how and why the body interacts with antigens and how vaccine type, formulation and delivery method can be modified to adapt immune respons
2. Results
2.1. Preliminary cluster analysis - inter-patient and inter-treatment variation
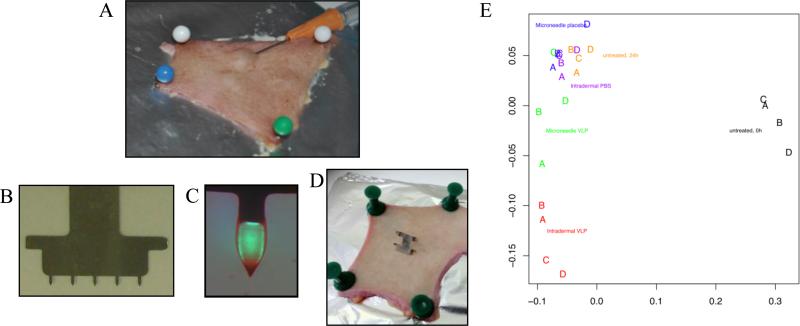
H1N1 VLP vaccines were delivered to viable human breast skin using either the Mantoux ID injection method (Figure 1A) or via VLP-coated microneedles (Figure 1B,C,D). Control samples were untreated skin, skin receiving ID injection of PBS and skin receiving microneedles coated with excipients only. Following a culture period of 24 h, and isolation of RNA from the skin, Illumina's Sentrix Beadchip microarrays was used to monitor changes in the gene expression profile of 47,296 discrete transcripts at the site of vaccine deposition. Classic multidimensional scaling shows that the expression profiles of the control samples were similar, and clearly distinct to the samples that received the vaccine (Figure 1E). All of the samples cultured for 24 h were clearly different from uncultured skin where RNA was isolated within a few hours of surgery. Figure 1E also suggests that one of the skin samples receiving the microneedle VLP vaccine (Donor C) did not respond above control, due to either biological/immunological inactivity or ineffective delivery of the VLPs with the microneedles. Nevertheless, preliminary results from the cluster analyses confirmed that a paired, two-class approach to the analysis of differential gene expression was suitable for further analyses.
Figure 1. Vaccine delivery and scaling of resulting gene expression profiles.
Delivery of H1N1 VLP vaccines to human skin explants via (A) ID or microneedle (D) injection. For microneedle injections 5 individual microneedles (B) were coated with VLPs (C; fluorescently labelled VLPs used for vizualisation of the coating in this image). (E) Classic multidimensional scaling on untransformed probe expression values using Euclidean distance.
2.2. Delivery of vaccines to human skin results in significant changes in gene expression
To determine the extent of changes in gene expression following vaccine delivery to skin several statistical comparisons were performed between various treatment and control groups. These comparisons were prepared as a list of arguments and the analysis performed in an iterative loop. Table 1 shows a summary of the differential gene expression detected by SAM analysis. The data confirms a significant degree of differential gene expression between untreated skin at 0 h compared to untreated but cultured skin at 24 h (205 probes at FDR = 0.1); this response being an inevitable consequence of skin excision and culture. In order to determine differential treatment responses against this significant background response all subsequent comparisons were made between treatment groups and their relevant controls following 24 h culture.
Table 1.
Numbers of differentially expressed genes detected by Significance Analysis of Microarrays (SAM) analysis> Delta values generating approximate false discovery rate (FDR) of 0.1, 0.25 and 0.33 were run and gene numbers tabulated for each FDR.
| Comparison | (0.1) | SAM (FDR) (0.25) | (0.33) |
|---|---|---|---|
| Skin 0h Vs 24h | 205 | - | - |
| MN VLP Vs skin 24h | 1 | 13 | 109 |
| ID VLP Vs Skin 24h | 188 | - | - |
| ID PBS Vs skin 24h | 2 | - | 7 |
| MN control Vs skin 24h | 0 | 0 | 8 |
| MN VLP Vs ID VLP | 8 | 203 | - |
| MN VLP Vs MN control | 7 | 207 | 321 |
| ID VLP Vs ID PBS | 393 | - | - |
| MN control Vs ID PBS | 0 | 0 | 2 |
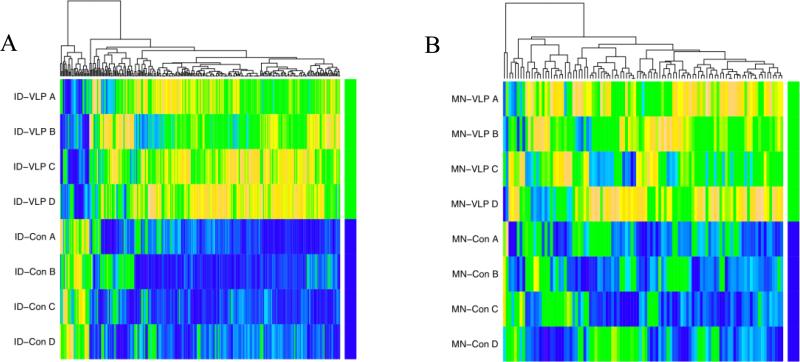
SAM analysis confirmed significant changes in the ID vaccinated group compared to ID PBS control and untreated skin at 24 h, with significant overlap between these two probes sets. Of the 393 (ID VLP versus ID PBS control) and 188 (ID VLP versus untreated skin at 24 h) probes identified as differentially expressed through SAM analysis (FDR 0.1) 127 probes were common. When an FDR of 0.33 was used 321 (microneedle VLP versus microneedle control) and 109 (microneedle VLP versus untreated skin at 24 h) probes were differentially expressed following microneedle vaccination, with a probe commonality of 50. Heat maps representing gene expression levels in probes differentially expressed only in the ID vaccination group (287 probes, Figure 2A) and only in the microneedle vaccination group (101 probes, Figure 2B) are shown in Figure 2. All of these probes have differential signals in the vaccination samples compared to control indicating that the VLP vaccine was having a demonstrable biological effect within excised skin.
Figure 2. Differential gene expression following ID and microneedle delivery of vaccine.
Expression heatmaps of gene probes differentially expressed in individual patients (A,B,C and D) following (A) H1N1 VLP ID injection and (B) microneedle H1N1 VLP delivery. In each case the terminal green bar represents the vaccine treated groups (termed ID-VLP and MN-VLP) and the blue bar represents the respective placebo control (termed ID-Con and MN-Con). Colour key for individual probes: blue = low, yellow = high expression.
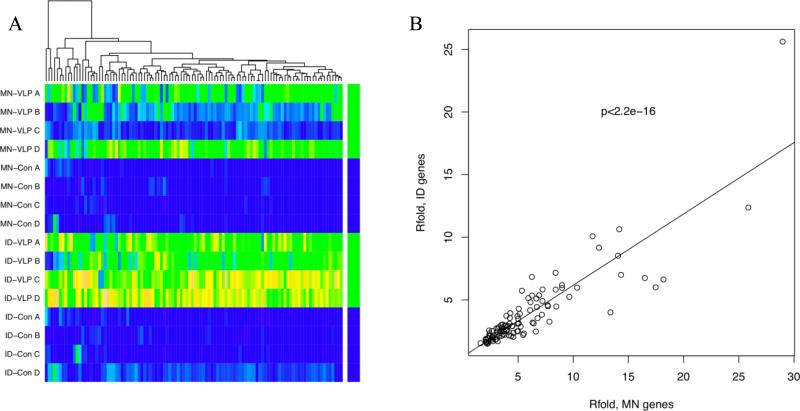
When differentially expressed probe sets from both ID and microneedle vaccinations were combined, there was significant overlap, suggesting that the probes detected by SAM analysis of the microneedle samples at 0.33 FDR are true positives. In this analysis 106 probes were found to be differentially expressed in both ID and microneedle vaccine groups; the combined heat map is shown in Figure 3A. All of these probes have significantly higher signals in the vaccine samples compared to respective control, although in two of the microneedle samples the increased signal was less pronounced. A plot of the fold-change of probes in the ID VLP compared with microneedle VLP samples demonstrates a strong correlation (p<2.2e-16, least squares regression; Figure 3B) demonstrating significant overlap in differentially expressed genes when vaccine is delivered via ID injection or microneedles.
Figure 3. Commonality of changes in gene expression following ID and microneedle delivery of vaccine.
A) Expression heatmap of differentially expressed probes common for both ID and microneedle H1N1 VLP samples; MN-VLP = Microneedle VLP delivery, MN-Con = Microneedle placebo control, ID-VLP = Intradermal injection of VLPs, ID-Con = Intradermal injection of PBS. Colour key for individual probes: blue=low, yellow=high expression. B) Least squares regression of R fold gene expression changes common to both ID and microneedle vaccination.
2.3. Gene ontology analysis reveals nature of innate immune response
To investigate the nature of human skin response to vaccine at the site of delivery, ontological analysis was initially performed on the 106 probes that were differentially expressed following both ID and microneedle injection of influenza VLP vaccine. This analysis provides strong evidence that skin delivery of the VLP vaccines stimulates the expression of a plethora of genes implicated in immune response. The 106 probes provided a total of 55 genes having ontology terms, which were tested in a gene universe of 5,973 genes expressed in these samples. A total of 118 ontology terms were found to be over-represented for these genes (p<0.01); the top 20 are shown in Table 2. The ontology analysis was further applied to the up-regulated probes resulting from either ID or microneedle VLP treatment. The full list of terms with p<0.01 are provided as supplementary data (Supp 1). Supp 2 shows the similar features of the hierarchal relationship of the top 10 ontological terms returned for either the ID (Supp 2A) or microneedle (Supp 2B) VLP treatments in the form of directed acyclic graphs.
Table 2.
Ontological analysis Top 20 ontology terms, with respective P values, overrepresented for 106 probes up-regulated following both ID and microneedle H1N1 VLP delivery.
| Rank | Term | P value |
|---|---|---|
| 1 | Response to type I interferon | 3.995e-29 |
| 2 | Type I interferon-mediated signalling pathway | 3.995e-29 |
| 3 | Response to virus | 1.169e-18 |
| 4 | Response to biotic stimulus | 1.513e-17 |
| 5 | Cellular response to organic substance | 1.742e-17 |
| 6 | Multi-organism process | 6.855e-13 |
| 7 | Response to interferon-gamma | 1.764e-12 |
| 8 | Interferon-gamma-mediated signalling pathway | 8.290e-11 |
| 9 | Response to chemical stimulus | 1.790e-08 |
| 10 | Interspecies interaction between organisms | 1.924e-08 |
| 11 | Signal transduction | 2.274e-07 |
| 12 | ISG15-protien conjugation | 2.937e-06 |
| 13 | Cellular response to interferon-alpha | 2.937e-06 |
| 14 | Regulation of immune system process | 7.063e-06 |
| 15 | Defence response to virus | 2.093e-05 |
| 16 | Innate immune response | 3.652e-05 |
| 17 | Immune response | 4.516e-05 |
| 18 | Cytosol to ER transport | 8.326e-05 |
| 19 | Cellular extravasation | 1.096e-04 |
| 20 | Viral reproduction | 1.405e-04 |
2.4.Quantitative PCR generally supports microarray data
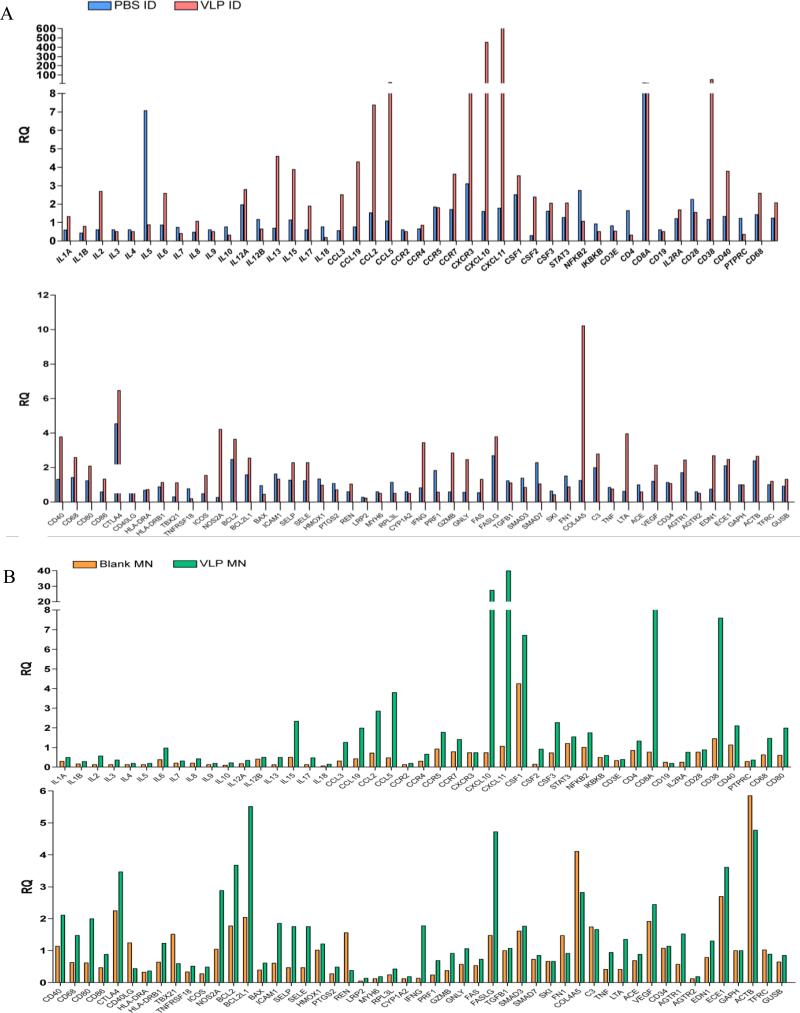
To validate and further rationalise the microarray data we subjected the RNA from donor samples to a gene signature panel containing genes implicated in immune and inflammatory response. Comparison of the treatment modalities suggests many similarities in the gene expression profiles, although as with the microarray study, changes are generally more pronounced following ID injection. PCR data showed that a number of cytokines and chemokines were up-regulated against control in both the ID VLP (Figure 4A) and microneedle VLP treatment groups (Figure 4B). Key up-regulated cytokines, chemokines, chemokine receptors and co-stimulatory molecules in both the ID and microneedle VLP treatment groups include CCL2, CCL3, CCL5, CXCL10, CXCL11, CCR7, IL2, IL6, IL13, IL15, CD38, CD40, CD68, CD80, CD86 and IFNγ; these, along with other markers, being indicative of a multi-faceted innate immune response to the VLP vaccine.
Figure 4. qPCR analysis following delivery of vaccine to human skin.
TaqMan® Human Inflammation Array quantification of differential gene expression (Relative Quantity (RQ) value) in human skin treated with A) influenza H1N1 VLPs delivered by ID injection, and B) influenza H1N1 VLPs delivered by surface-coated microneedles.
2.5. Immunohistochemistry in human epidermal sheets and tissue sections reveals DC migration
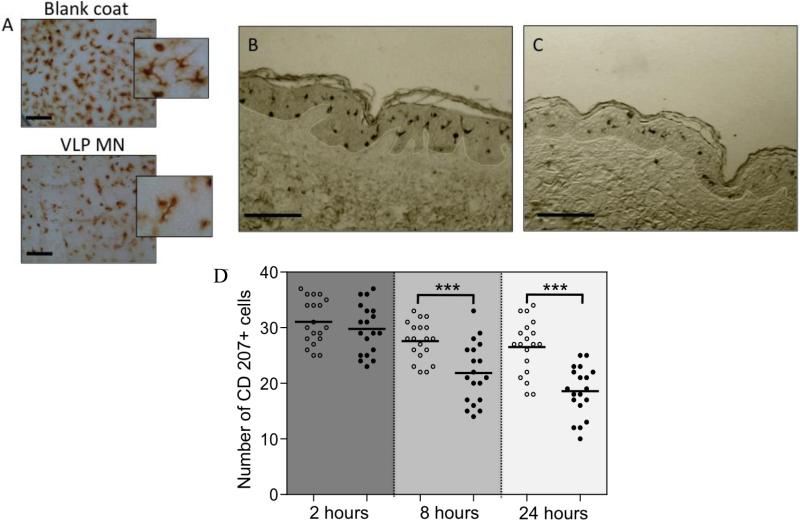
Visualisation of immune-responsive cells in the skin permitted further exploration of local human response to vaccine, in this case exclusively following microneedle delivery. Immuno-staining of control epidermal sheets (Figure 5A) highlighted the uniform network of dendritic Langerhans cells (LCs) distributed through the epidermis. Both the frequency and morphology of LCs in epidermal sheets changed 24 h after microneedle delivery of VLPs. The reduced number of LCs in VLP treated samples were much more likely to display either a rounded or elongated morphology[33] as opposed to non-treated skin samples where the majority of cells retained a typical multi-dendrite morphology at 24 h (Figure 5A inserts). Such morphological changes have been described previously, albeit in response to a different vaccine and delivery method,[33] and attributed to the stimulated cells preparing for migration in response to antigen. Tissue sections further highlight differences in the distribution of LCs within the epidermis (Figure 5B&C). Following microneedle-VLP treatment LCs were more dispersed within the epidermis, with greater numbers exhibiting protrusions in the direction of the basement membrane (BM). LCs within these samples were often observed in close proximity to the BM (Figure 5B), presumably during migration out of the epidermis, into the dermis and beyond. In contrast, LCs in control sections were generally distributed in central regions of the epidermis (Figure 5C). Changes in cell distribution were further supported when the total number of LCs within epidermal sheets were counted. This semi-quantitative analysis shows that epidermal sheets from microneedle VLP treated skin contained significantly fewer LCs per unit area compared with control (Figure 5A&D).
Figure 5. Morphological and migratory changes to dendritic cells following vaccine delivery.
Immunohistochemical study of LCs in human skin following microneedle delivery of H1N1 VLPs. A) Epidermal sheets isolated from skin 24 h after placebo (Blank coat) and VLP (VLP MN) delivery (Bar = 100μm). B, C) Immunohistochemically stained longitudinal sections of human skin treated with (B) VLP and (C) placebo (Bar = 200μm). D) Immuno-stained epidermal sheets were visualised by light microscopy 2, 8 and 24 h after microneedle delivery of H1N1 VLPs. The number of CD207+ cells observed in 200μm2 fields of view are shown for VLP treated samples (black circles) and placebo treated samples (white circles). A total of 3-5 replicate counts were taken from a total of 4 independent skin donors. ***denotes significant (P<0.01) difference between VLP and control samples at each timepoint.
3. Discussion
The data presented provide the first insight into the effects on global gene expression in human skin following skin vaccination, providing powerful evidence to make more informed predictions regarding the degree and nature of local immune response to a delivered antigen. Although work in animal models remains essential in understanding immunology and testing vaccinology, it is questioned whether the results can be extrapolated to the clinical situation.[34] We suggest that human skin testing can complement and validate studies in animal models; for example, in the case of microneedles, confirming that the protective immune responses seen in mice[7,26] is likely to arise from immune stimulation in the skin. Indeed, the use of human skin explants is now becoming more widespread, providing new data on the role of immunoresponsive skin cells with regard to vaccination.[35,36]
We must however acknowledge the limitations in using skin organ culture systems. Despite the fact that subjects were of similar age and health status in our study it is not surprising that there is significant inter-individual variation. As the medical histories of these patients is not known, due to ethical restraints, we are not aware whether patients have been immunized or had prior exposure to influenza strains. Regarding our patient profile we should also consider the influence of age and disease on immune function, possibly translating to diminished response to an ID vaccine. However, these limitations are reflective of the clinical situation and will possibly provide realistic interpretation of relevant immune responses to compliment and support studies performed in unexposed, naïve animals with an inbred, homogenous immune function.
We further acknowledge that the skin has been separated from the patient and therefore (i) we cannot explore the effects of underlying circulation or lymphatic drainage, and (ii) gene expression will alter after excision. The gross changes we observed in samples at 24 h compared to freshly excised skin (termed 0 h) can be attributed to the predictable responses that will occur during the 24 h culture period as the tissue reacts to surgical excision, extended culture and deterioration; including response to hypoxia, response to wounding, locomotion and cell migration; a baseline response we have previously observed histochemically in cultured skin.[33,37] To account for these responses all subsequent gene expression comparisons were against control tissue cultured for 24 h.
Neither the ID nor microneedle placebo control samples demonstrated significant levels of differential gene expression against untreated skin. This was somewhat surprising as it was anticipated that the invasive nature of an intradermal injection, resulting in a raised bleb at the injection site, would cause tissue damage and upregulation of stress and damage pathways. It is possible that such events occurred within a shorter time period following the injection and that the gene expression levels returned to baseline 24 h after injection. Indeed, we generally acknowledge that, due to a restricted supply of human donor tissue, this study is limited to observing events 24 h after injection and certain processes will occur at shorter time points. Given the dimensional reduction in invasiveness of the microneedle injection it was less surprising that gene expression changes were imperceptible against control. Microarray analysis did however reveal significantly altered gene expression profiles in skin exposed to the VLP vaccine. Whilst both ID injection and microneedle delivery of VLPs resulted in differential gene expression, the degree of response was subtler when microneedles were used to deliver the vaccine, differential expression being observed at an increased FDR. Regarding the nature of response, the top two ontological terms returned in both the ID and microneedle treated samples were ‘response to type I interferon (GO:0034340)’ and ‘type I interferon-mediated signalling pathway (GO:0060337)’. Type I interferons (IFNs) are key pleiotropic immunomodulatory factors responsible for innate immunity,[38] in particular host response to viral infection.[39] As type I IFNs are produced following initial contact with the antigen they are likely to be significant factors affecting the form and subsequent cascade leading to innate and adaptive immunity[38,39] and will play crucial roles in functional and phenotypic maturation of DCs and the development of CD4+ TH1 immunity.[40] In addition to type 1 IFNs, the ontological terms returned in this analysis revealed further evidence of viral response (e.g. ‘response to viruses’ GO:0009615; ‘defence response to virus’ GO:0051607) and the subsequent local innate immune response generated thereafter (e.g. ‘innate immune response’ GO:0045087). Cells within human skin appear therefore to react to the VLP vaccine as if it were an invading virus, rather than an artificial construct. VLPs are known to activate DCs,[41,42] leading to induction of T helper type 1 response and interferon.[43] Influenza hemagglutinin (HA) protein anchored on VLPs is also known to have an adjuvant effect on these responses[44] and it is also possible that a low level of nucleic acid components from baculovirus or insect cells may contribute to the interferon response. The ontological interrelationships and underlying biological processes revealed through microarray analysis suggest that the nature of these responses is similar, although not identical, whether the VLP vaccine is delivered via ID injection or via microneedles.
DCs respond to type I IFNs by up-regulating expression of MHC class I, T-cell costimulatory molecules, such as CD40, CD80 and CD86[45] and also chemokine receptors,[46] such as CCR7[47] which are required for migration into the lymphatics. Up-regulation of these genes within the treatments groups was confirmed by qPCR. While we cannot determine the cell types responsible for these changes in gene expression, morphological and spatial observation of LCs in epidermal sheets and tissue sections clearly shows that this particular DC population becomes mobilized in response to antigen delivery. The consistent up-regulation of co-stimulatory molecules CD40, CD80 and CD86, seen in the pPCR data set, implies that APCs within the skin, when exposed to the delivered antigen, have the ability to efficiently activate T and B cells. CTLA4 was also up-regulated following vaccine delivery. CTLA4 is an inhibitory signalling protein found on T cells, where it competes with CD28 for CD80/86 on the APC surface. This data could be suggestive of the resident T cells in the skin becoming primed for a regulatory role within the skin. Additionally, samples treated with VLP by ID injection showed a marked increase in levels of IL-13 and IL-17, which are associated with a Th17 response.[48] The role of this specific subset of T cells is debatable with seemingly dual roles of protection and immunopathology.[49] Taken together this data provides a molecular snapshot of the skin responding to antigen, producing signals that are both inflammatory and regulatory in nature and exemplifying the dynamic nature of the skin at the site of vaccine deposition.
Immunization is commonly described as a one-way process, i.e. the immune cells traffic the antigen from the delivery site to the lymphatics where it is presented to naïve T-cells to proffer protective immunity. This study, focussing on the local skin response at the vaccine delivery site, suggests multilateral processes, e.g. the antigen response acting as a beacon to attract cells from the circulation (‘cellular extravasation’ GO:0045123). An increase in expression of CCL19 was also observed by qPCR. CCL19 is over secreted by the thymus and lymph nodes to recruit activated lymphocytes and DCs.[50] Even more pronounced changes in gene expression observed following VLP delivery include up-regulation in CCL2 and CCL5, chemoattractants that signal innate immune cells including monocytes, eosinophils, basophils and DCs and CXCL10 and CXCL11, chemoattractants secreted by cells in response to interferon that signal leukocyte trafficking relating to chemotactic migration. The findings presented here also show similarities to the local skin response previously observed in mice to microneedles and an inactivated influenza vaccine,[51] where a local increase in cytokines important for recruitment of neutrophils, monocytes and DCs was demonstrated at the site of immunization.
Data from this study indicates that the nature of local skin response toward the vaccine is similar whether the vaccine is delivered via a solution (ID injection) or as a dried, solid formulation (microneedle injection), although the degree of response was generally more pronounced in the ID injection group. This is interesting given that ID injection of the vaccine results in delivery deep into the dermis, whereas microneedle delivery exposes the vaccine to the environment along the entire needle length, i.e to cells of both the epidermis and upper dermis; this is also possibly why PCR data showed a significant up-regulation of COL4A5, a gene encoding a subunit of type IV collagen, the major structural component of basement membranes, following ID, but not MN delivery of VLPs.
The more subtle responses generally observed in the microneedle group is likely a function of i) reduced delivered dose through incomplete needle coating and/or incomplete release in situ; ii) deposition in the vicinity of different APC subsets; iii) reduced physical adjuvancy through less insult; and iv) compromised VLP immunogenicity through coating, storage and subsequent rehydration within biological milieu. The organ culture system may provide a suitable environment in which to determine how each of these factors affect local response and assess further optimised formulations and devices.
4. Conclusions
Many factors influence the innate and adaptive responses generated following skin immunization. Indeed, the cascade of interactions that occur to elicit an adaptive immune response may be dictated by the innate immune response that develops in the microenvironment formed at the initial point of contact between the antigen and the body.[38] In this study we explore for the first time the subtle and complex events that occur at the site of vaccine deposition in human skin following delivery of H1N1 influenza VLP vaccines administered either by ID injection or via minimally-invasive microneedle devices. Whilst our previous studies demonstrated that human DCs, such as LCs, are stimulated following ID injection and microneedle delivery of influenza VLPs,[33,37] this new study explores the subtle, multi-faceted molecular changes that occur, which could have a significant influence on the degree and nature of immune response. This approach, using ex vivo human skin, could be used to show which skin immunization approaches more closely mimic the response of a conventional ID injection and investigate safety and efficacy profiles of novel vaccine candidates within the correct biological context.
5. Experimental Section
Ethics Statement
Human skin was obtained under full ethical committee approval (South East Wales Research Ethics Committees Panel C: 08/WSE03/55) from anonymous donors undertaking surgical procedures. All patients provided written consent to participate in the study.
Preparation of swine origin 2009 H1 HA VLPs
Swine origin 2009 H1 HA VLPs were prepared as described previously.[52] Briefly, Sf9 insect cells were co-infected with recombinant baculovirus (rBV) expressing HA and matrix M1 protein respectively, both of which were derived from the 2009 H1N1 pandemic strain, A/California/09 virus. Culture supernatants containing released influenza VLPs were clarified using low speed centrifugation (6000 rpm, 20 min) to remove cell debris, and then purified by sucrose gradient ultracentrifugation (SW32 rotor, 28000 rpm, 60 min). The expression of HA and M1 on purified VPs was confirmed by western blot using mouse polyclonal antibodies raised by live virus infection with the 2009 H1N1 pandemic virus. The amount of HA in influenza VLPs was estimated to contain approximately 0.1μg HA (A/California/2009) per 1 μg of total protein of VLPs (~10%).
Human skin collection and processing
Excised human breast skin from surgical procedures was obtained from four separate female donors aged 62 (Donor A), 61 (Donor B), 54 (Donor C) and 57 (Donor D). Subcutaneous fat was removed by blunt dissection and the tissue was pinned, dermis side down, onto a dissection board for treatment.
Intradermal delivery of VLPs to human skin
Two methods of delivery were used to introduce VLPs into the skin: (i) ID injection: A 10μl volume of VLP suspension (1mg/ml in PBS) was injected into the dermal compartment using a 26G hypodermic needle. Successful delivery was confirmed by the formation of a distinct bleb at the injection site (Fig 1A). Control samples comprised ID injection of 10μl of PBS. (ii) Microneedle delivery: Two-dimensional microneedle arrays consisting of five individual solid microneedles of 750μm length were fabricated by cutting needle structures from stainless steel sheets (McMaster-Carr, Atlanta, GA) using an infrared laser (Resonetics Maestro, Nashua, NH) and finished by electropolishing. VLPs were combined with 1% (w/v) carboxymethylcellulose sodium salt (CMC, Sigma–Aldrich Chemical Company, Poole, UK), 0.5% (w/v) Lutrol F-68 NF (BASF, Ludwigshafen, Germany) and 15% (w/v) trehalose (Sigma–Aldrich Chemical Company, Poole, UK). Each microneedle array was coated with up to 10μg of VLP using a well-established dip-coating process, detailed previously.[37] Placebo coated microneedles were also prepared whereby PBS replaced the VLPs. Coated microneedles were applied to skin with a force of 0.2-0.5 N and left in situ for 10 mins before removal. Each donor received four repeat injections of each treatment and respective controls.
Human skin culture
Treated regions of skin were excised with a 6 mm punch and cultured at air-liquid interface in a modified Trowell-type organ culture system at 37°C and 5% CO2 for 24 hours.[53] After culture samples were immersed in RNAlater® (Life Technologies, Paisley, UK) and stored at −80°C.
RNA extraction and quantification
Total RNA was extracted using the commercially available RNeasy® kit (Qiagen, Crawley, UK) according to the manufacturer's instruction. RNA was pooled from the four repeat samples with purity and quality confirmed electrophoretically and spectrophotometrically at 260 nm.
Preparation of samples for microarray
High-throughput RNA amplification was achieved with an Illumina® TotalPrep™-96 RNA amplification kit (Illumina, Inc., CA, USA). This was subject to a whole-genome gene expression direct hybridization assay (Illumina, Inc., CA, USA) as directed by the manufacturer's protocol.
Microarray data analysis
A) Data Processing
Microarray data was analysed using methods from the specified Bio-conductor packages to perform the following tasks; beadarray to import and process the raw data from the chip images, implementing the BASH algorithm for detecting and managing spatial artefacts and quantile signal normalisation using negative and positive control probes. Summary data was exported as log transformed mean values of probe signals.
B) Cluster Analysis
Unsupervised hierarchical cluster analysis was performed to ascertain the quality of biological replicates and determine how the relationships between samples and treatments impact upon gene expression. Euclidean distance was calculated using Pearson's correlation on untransformed probe expression values and clustered by Ward's minimum variance method.
C) Differential Gene Expression Analysis
The log-transformed summary probe expression data was analysed using an implementation of the Significance Analysis of Microarrays (SAM) method in the package siggenes. A paired two-class analysis was used to identify differentially expressed genes. For heat maps, genes were ordered by a distance function using Euclidean distance of Spearman correlation.
D) Gene Ontology (GO) Analysis
Gene Ontology analysis was performed using an implementation of the Hypergeometric test in the package GOstats, using a significance cut-off of p=0.01. The term comparisons of the top 10 ontological terms for ID and microneedle injection of VLPs were displayed using QuickGO (http://www.ebi.ac.uk/QuickGO/).
Quantitative PCR (qPCR) with TaqMan® Low Density Array (TLDA)
The isolated RNA was also exposed to qPCR reactions performed with Applied Biosystems® TaqMan® Human Inflammation Array according to manufacturers’ recommendations. cDNA was generated with Ambion Reverse Transcription Kit (Life Technologies, Paisley, UK) according to manufacturer's protocol. 2 μg of RNA were used per sample in the RT-PCR runs. 400 ng (4 μl) cDNA was used in each reaction containing TaqMan Universal PCR Master Mix (Applied Biosystems, Paisley, UK). The mixture was equally divided over four sample-loading ports of each TLDA, centrifuged at 1300 RPM and sealed. qPCR amplification was performed with an Applied Biosystems Prism 7900HT sequence detection system with the following thermal cycler conditions: 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 30s at 97°C and 1 min at 60°C. Differential gene expression against control determined using the ΔΔCt method with DataAssist™ software (Applied Biosystems, Paisley, UK).
Immunohistochemistry in epidermal sheets and tissue sections
Human epidermal sheets were prepared[36,37] at 2 h, 8 h and 24 h following administration of VLP-coated microneedles to additional skin donors (aged 62, 66, 70, 72). Approximately 10μg of VLP was coated per microneedle array. Briefly, skin samples were immersed in 3.8% ammonium thiocyanate in PBS for 30 min at room temperature. The epidermis was peeled away from the dermis with forceps, rinsed in PBS and fixed in acetone at −20°C for 20 min. Epidermal LCs were immunohistochemically stained with a mouse monoclonal antibody CD207 primary antibody (ABcam, Cambridge, UK) and EnVision+ system-HRP (DAB) kit (Dako UK Ltd., Cambridgeshire, UK). Immuno-stained sheets were visualised via light microscopy with CD207+ cells counted in random 200μm2 fields of view. A total of 3-5 replicate counts were taken with a total of 4 biological replicates. A t-test determined significant (P<0.01) difference at each timepoint.
Additional skin samples were rinsed in PBS and fixed in formalin for 24-36 hours. Samples were dehydrated in an ethanol gradient, and cleared in chloroform before embedding in paraffin wax. Immunohistochemistry was preformed on histological sections of 4μm, following rehydration and antigen retrieval (Tris-EDTA buffer at 95°C for 40 mins). The previously described anti-CD207 mouse monoclonal antibody and secondary detection system was used to visualise LC distribution.[36]
Supplementary Material
Acknowledgements
The authors acknowledge the contribution of Professor Mark Prausnitz and Dr Vladimir Zarnitsyn, Georgia Institute of Technology for manufacture and supply of microneedles, Dr Fu-Shi Quang and Dr William Weldon, Emory University for preparing VLPs and reviewing the gene expression data respectively, and Dr Claudia Consoli, Cardiff University for assistance with qPCR studies. This work was funded by the National Institutes of Health Grant EB006369.
Footnotes
Supporting Information is available under http://www.small-journal.com or from the author.
References
- 1.Steinman RM. Pathol. Biol. 2003;51:59. doi: 10.1016/s0369-8114(03)00096-8. [DOI] [PubMed] [Google Scholar]
- 2.Kis EE, Winter G, Myschik J. Vaccine. 2012;30:523. doi: 10.1016/j.vaccine.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 3.Lambert PH, Laurent PE. Vaccine. 2008;26:3197. doi: 10.1016/j.vaccine.2008.03.095. [DOI] [PubMed] [Google Scholar]
- 4.Paccalin M, Weinberger B, Nicolas JF, Van Damme P, Mégard Y. Eur. Geriatr. Med. 2010;1:82. [Google Scholar]
- 5.Icardi G, Orsi A, Ceravolo A, Ansaldi F. Hum. Vaccin. Immunother. 2012;8:67. doi: 10.4161/hv.8.1.18419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sudarshan MK, Gangaboraiah B, Ravish HS, Narayana DH. Hum. Vaccin. 2010;6:562. doi: 10.4161/hv.6.7.11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quan FS, Kim YC, Compans RW, Prausnitz MR, Kang SM. J. Control. Rel. 2010;147:326. doi: 10.1016/j.jconrel.2010.07.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gherardin A, Lau S. Med. J. Australia. 2007;187:58. doi: 10.5694/j.1326-5377.2007.tb01128.x. [DOI] [PubMed] [Google Scholar]
- 9.Miller KD, Gibbs RD, Mulligan MM, Nutman TB, Francis DP. The Lancet. 1983;322:8365. doi: 10.1016/s0140-6736(83)90800-0. [DOI] [PubMed] [Google Scholar]
- 10.Bryan JP, Sjogren MH, Perine PL, Legters LJ. Clin Infect Dis. 1992;14:697. doi: 10.1093/clinids/14.3.697. [DOI] [PubMed] [Google Scholar]
- 11.Hiraishi Y, Nandakumar S, Choi SO, Lee JW, Kim YC, Posey JE, Sable SB, Prausnitz MR. Vaccine. 2011;29:2626. doi: 10.1016/j.vaccine.2011.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moss B. Immunol. Rev. 2011;239:8. doi: 10.1111/j.1600-065X.2010.00975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernando GJ, Chen X, Prow TW, Crichton ML, Fairmaid EJ, Roberts MS, Frazer IH, Brown LE, Kendall MA. PLos One. 2010;5:e10266. doi: 10.1371/journal.pone.0010266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Damme P, Arnou R, Kafeja F, Fiquet A, Richard P, Thomas S, Meghlaoui G, Samson SI, Ledesma E. Infect. Dis. 2010;10:134. doi: 10.1186/1471-2334-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raz E, Carson DA, Parker SE, Parr TB, Abai AM, Aichinger G, Gromkowski SH, Singh M, Lew D, Yankauckas MA, Baird SM, Rhodes Proc GH. Nat. Acad. Sci. USA. 1994;91:9519. doi: 10.1073/pnas.91.20.9519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill HS, Prausnitz J MR. Diabetes, Sci. Tec. 2007;1:725. doi: 10.1177/193229680700100517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martanto W, Moore JS, Kashlan O, Kamath R, Wang PM, O'Neal JM, Prausnitz MR. Pharm. Res. 2006;23:104. doi: 10.1007/s11095-005-8498-8. [DOI] [PubMed] [Google Scholar]
- 18.Birchall JC. Expert Rev. Med. Devices. 2006;3:1. doi: 10.1586/17434440.3.1.1. [DOI] [PubMed] [Google Scholar]
- 19.Kim YC, Park JH, Prausnitz MR. Adv. Drug Deliv. Rev. 2012;64:1547. doi: 10.1016/j.addr.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haq MI, Smith E, John DN, Kalavala M, Edwards C, Anstey A, Morrissey A, Birchall JC. Biomed Microdevices. 2009;11:35. doi: 10.1007/s10544-008-9208-1. [DOI] [PubMed] [Google Scholar]
- 21.Coulman SA, Birchall JC, Alex A, Pearton M, Hofer B, O'Mahony C, Drexler W, Považay B. Pharm. Res. 2011;28:66. doi: 10.1007/s11095-010-0167-x. [DOI] [PubMed] [Google Scholar]
- 22.Hirschberg H, van Kuijk S, Loch J, Jiskoot W, Bouwstra J, Kersten G, Amorij JP. Eur. J. Pharm. Sci. 2012;46:1. doi: 10.1016/j.ejps.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 23.Prausnitz MR, Mikszta JA, Cormier M, Andrianov AK. Curr. Top. Microbiol. Immunol. 2009;333:369. doi: 10.1007/978-3-540-92165-3_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laurent PE, Bourhy H, Fantino M, Alchas P, Mikszta JA. Vaccine. 2010;28:5850. doi: 10.1016/j.vaccine.2010.06.062. [DOI] [PubMed] [Google Scholar]
- 25.Kang SM, Song JM, Kim Expert Rev YC. Vaccines. 2012;11:547. doi: 10.1586/erv.12.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koutsonanos DG, Vassilieva EV, Stavropoulou A, Zarnitsyn VG, Esser ES, Taherbhai MT, Prausnitz MR, Compans RW, Skountzou I. Sci. Rep. 2012;2:357. doi: 10.1038/srep00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birchall JC, Clemo R, Anstey A, John Pharm DN. Res. 2011;28:95. doi: 10.1007/s11095-010-0101-2. [DOI] [PubMed] [Google Scholar]
- 28.Hickling JK, Jones KR, Friede M, Zehrung D, Chen D, Kristensen D. Bulletin of the World Health Organization. 2011;89:221. doi: 10.2471/BLT.10.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmook FP, Meingassner JG, Billich A. Int. J. Pharm. 2001;215:51. doi: 10.1016/s0378-5173(00)00665-7. [DOI] [PubMed] [Google Scholar]
- 30.Mestas J, W Hughes J CC. Immunol. 2004;172:2731. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 31.Davis MM. Sci. Transl. Med. 2012;4:117fs2. doi: 10.1126/scitranslmed.3003385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis Immunity MM. 2008;29:835. doi: 10.1016/j.immuni.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearton M, Kang SM, Song JM, Anstey AV, Ivory M, Compans RW, Birchall PLoS One JC. 2010;5:e12410. doi: 10.1371/journal.pone.0012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis HL. Hum. Vaccin. 2008;4:246. doi: 10.4161/hv.4.3.5318. [DOI] [PubMed] [Google Scholar]
- 35.Schneider LP, Schoonderwoerd AJ, Moutaftsi M, Howard RF, Reed SG, de Jong EC, Teunissen MB. Vaccine. 2012;30:4216. doi: 10.1016/j.vaccine.2012.04.051. [DOI] [PubMed] [Google Scholar]
- 36.Liard C, Munier S, Joulin-Giet A, Bonduelle O, Hadam S, Duffy D, Vogt A, Verrier B, Combadière B. J. Invest. Dermatol. 2012;132:615. doi: 10.1038/jid.2011.346. [DOI] [PubMed] [Google Scholar]
- 37.Pearton M, Kang SM, Song JM, Kim YC, Quan FS, Anstey A, Ivory M, Prausnitz MR, Compans RW, Birchall JC. Vaccine. 2010;28:6014. doi: 10.1016/j.vaccine.2010.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paladino P, Cummings DT, Noyce RS, Mossman KL, Immunol J. 2006;177:8008. doi: 10.4049/jimmunol.177.11.8008. [DOI] [PubMed] [Google Scholar]
- 39.Stark GR. Annu. Rev. Biochem. 1998;67:227. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 40.Longhi MP, Trumpfheller C, Idoyaga J, Caskey M, Matos I, Kluger C, Salazar AM, Colonna M, Steinman RM. J. Exp. Med. 2009;206:1589. doi: 10.1084/jem.20090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buonaguro L, Tornesello ML, Tagliamonte M, Gallo RC, Wang LX, Kamin-Lewis R, Abdelwahab S, Lewis GK, Buonaguro FM, Virol J. 2006;80:9134. doi: 10.1128/JVI.00050-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sailaja G, Skountzou I, Quan FS, Compans RW, Kang SM. Virology. 2007;362:331. doi: 10.1016/j.virol.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang SM, Yoo DG, Kim MC, Song JM, Park MK, E. O, Quan FS, Akira S, Compans RW. J. Virol. 2011;85:11391. doi: 10.1128/JVI.00080-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao Q, Zhang R, Guo L, Li M, Chen C, Immunol J. 2004;173:1951. doi: 10.4049/jimmunol.173.3.1951. [DOI] [PubMed] [Google Scholar]
- 45.Hoebe K, Janssen E, Beutler B. Nature Immuol. 2004;5:971. doi: 10.1038/ni1004-971. [DOI] [PubMed] [Google Scholar]
- 46.Rot A, Andrain HV. Ann. Rev. Immunol. 2004;22:891. doi: 10.1146/annurev.immunol.22.012703.104543. [DOI] [PubMed] [Google Scholar]
- 47.Penna G, Vulcano M, Sozzani S, Adorini L. Hum. Immunol. 2002;63:1164. doi: 10.1016/s0198-8859(02)00755-3. [DOI] [PubMed] [Google Scholar]
- 48.Korn T, Bellelli E, Oukka M, Kuchroo VK. Ann. Rev. Immunol. 2009;27:485. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 49.Bettelli E, Korn T, Kuchroo VK. Curr. Opin. Immunol. 2007;19:652. doi: 10.1016/j.coi.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katou F, Ohtani H, Nakayama T, Nagura H, Yoshie O, Motegi K, Pathol J. 2003;199:98. doi: 10.1002/path.1255. [DOI] [PubMed] [Google Scholar]
- 51.del Pilar Martin M, Weldon WC, Zarnitsyn VG, Koutsonanos DG, Akbari H, Skountzou I, Jacob J, Prausnitz MR, Compans RW. MBio. 2012;3:e00012–12. doi: 10.1128/mBio.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quan FS, Yoo DG, Song JM, Clements JD, Compans RW, Kang SM, Virol J. 2009;83:4489. doi: 10.1128/JVI.02035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ng KW, Pearton M, Coulman S, Anstey A, Gateley C, Morrissey A, Allender C, Birchall J. Vaccine. 2009;27:5948. doi: 10.1016/j.vaccine.2009.07.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.