Abstract
AIM: To summarize the relationship between p.Tyr113His and p.His139Arg polymorphisms in microsomal epoxide hydrolase (EPHX1) and risk for esophageal cancer (EC).
METHODS: The MEDLINE/PubMed and EMBASE databases were searched for studies of the association between EPHX1 polymorphisms and EC risk that were published from the database inception date to April 2013. A total of seven case-control studies, including seven on p.Tyr113His (cases, n = 1118; controls, n = 1823) and six on p.His139Arg (cases, n = 861; controls, n = 1571), were included in the meta-analysis. After data extraction by two investigators working independently, the meta-analyses were carried out with STATA 11.0 software. Pooled odds ratios and 95%CI were calculated using a fixed-effects model or a random-effects model, as appropriate.
RESULTS: The pooled EPHX1 p.Tyr113His polymorphism data showed no significant association with EC in any of the genetic models (OR = 1.00, 95%CI: 0.70-1.48 for Tyr/His vs Tyr/Tyr; OR = 1.10, 95%CI: 0.77-1.57 for His/His vs Tyr/Tyr; OR = 1.06, 95%CI: 0.75-1.49 for a dominant model; OR = 1.09, 95%CI: 0.89-1.34 for a recessive model). Similar results were obtained from the p.His139Arg polymorphism analysis (Arg/His vs His/His: OR = 1.02, 95%CI: 0.84-1.23; Arg/Arg vs His/His: OR = 0.96, 95%CI: 0.60-1.54; OR = 1.03, 95%CI: 0.78-1.37 for the dominant model; OR = 0.97, 95%CI: 0.61-1.56 for the recessive model). Subgroup analyses for ethnicity, subtype of EC, and source of controls (population-based or hospital-based) showed trends that were consistent with the pooled analysis (reported above), with no significant associations found.
CONCLUSION: This meta-analysis suggests that the p.Tyr113His and p.His139Arg polymorphisms in EPHX1 may not be associated with EC development.
Keywords: Esophageal cancer, Squamous cell carcinoma, Adenocarcinoma, EPHX1, Polymorphism, Meta-analysis
Core tip: A meta-analysis was performed to determine if the p.Tyr113His and p.His139Arg polymorphisms in microsomal epoxide hydrolase (EPHX1) are associated with an increased risk for esophageal cancer (EC). A total of seven studies of the association between EC risk and the EPHX1 polymorphisms (p.Tyr113His in seven and p.His139Arg in six) were included in the analysis. No significant association was found in any of the genetic models for the p.Tyr113His polymorphism in EPHX1 and EC. Similar results were obtained from the p.His139Arg polymorphism analysis. Subgroup analyses for ethnicity, subtype of EC, and source of controls also showed no significant association of EPHX1 polymorphisms with EC risk.
INTRODUCTION
Esophageal cancer (EC) is one of the most common fatal malignancies[1] and the sixth leading cause of cancer deaths worldwide[2]. Two histological subtypes of EC are characterized: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EADC). The pathogenesis of EC, however, remains poorly understood. Previous epidemiological studies have indicated that exposure to environmental carcinogens plays an important role in the development of EC[3,4].
Genetic susceptibility, in the form of phase I and phase II metabolizing enzyme polymorphisms, may also be associated with an increased risk for EC[5]. Microsomal epoxide hydrolase (EPHX1) plays a dual role in the response to environmental carcinogens, in that EPHX1 both activates and detoxifies toxins. In response to environmental carcinogens, EPHX1 not only produces trans-dihydrodiols that can be metabolized to mutagenic, poisonous and carcinogenic polycyclic hydrocarbon diol epoxides, but also generates products necessary for the detoxification reaction. It has been shown previously that EPHX1 can catalyze hydrolysis of alkene and arene oxides to water-soluble trans-dihydrodiols[6,7].
The EPHX1 gene, located on chromosome 1q42, is expressed in nearly all human tissues. EPHX1 activity varies widely among individuals, though the molecular basis of this variability is not fully understood. Genetic polymorphisms in exon 4 (A>G, p.His139Arg) and exon 3 (T>C, p.Tyr113His) of EPHX1, however, have been shown to alter the protein’s function. In exon 4, the 139Arg polymorphism enhances EPHX1 activity by 25%. Alternatively, the 113His allele in exon 3 has a negative effect on enzymatic activity, reducing it by at least 50%[8]. Polymorphisms that alter EPHX1 enzyme activity may then lead to inter-individual differences in sensitivity to chemical carcinogens.
To date, a number of studies have investigated the association between EPHX1 polymorphisms and EC risk in different populations[5,9-16]. The results, however, have been conflicting. In order to establish a comprehensive estimation of the association between EPHX1 polymorphisms and EC risk, we conducted a meta-analysis of all available published studies.
MATERIALS AND METHODS
Data sources and search strategy
We searched the MEDLINE/PubMed and EMBASE databases for all articles published on the association between EPHX1 polymorphisms and EC risk from the database inception date to April 2013. The following search terms were used: microsomal epoxide hydrolase, EPHX1, esophageal cancer, and polymorphism. No restrictions were applied. The references section of reviews and retrieved articles were searched in an effort to identify any additional eligible studies. If identified articles overlapped or were duplicated, only the most recent, largest or most complete study was selected.
Inclusion and exclusion criteria
We reviewed titles and abstracts of all citations and retrieved studies. The inclusion criteria were as follows: (1) case-control studies conducted to evaluate the association between EPHX1 (p.Tyr113His and/or p.His139Arg) polymorphisms and EC risk; (2) sufficient genotype or allele data were presented to calculate the odds ratios (ORs) with 95% confidence intervals (CIs); and (3) the paper clearly described the sources of cases and controls. The following exclusion criteria were applied due to insufficient data for analysis: reviews, editorials, commentaries, and duplicated studies.
Data extraction
Two investigators (Li QT and Kang W) independently extracted relevant data from all eligible publications meeting the inclusion criteria. Disagreements were resolved by discussion. The characteristics of each included study were collected as follows: the first author’s name, year of publication, the country of participants, participants’ ethnicity, number of cases and controls, source of control group [e.g., population-based (PB) or hospital-based (HB)], and genotypes of cases and controls. For our analysis, a PB case-control study was defined by the use of controls obtained from the general population, while an HB case-control study had obtained controls from a hospitalized patient population. In addition, we contacted the authors to collect further information when necessary.
Statistical analysis
The strength of the association between EC risk and EPHX1 polymorphisms was estimated using ORs, with corresponding 95%CIs. For the EPHX1 p.Tyr113His polymorphism, we assessed the association using a co-dominant model (His/His vs Tyr/Tyr and Tyr/His vs Tyr/Tyr), a dominant model (His/His + Tyr/His vs Tyr/Tyr), and a recessive model (His/His vs Tyr/His + Tyr/Tyr). The same models were used in the EPHX1 p.His139Arg analysis.
Both the Cochran’s Q test for heterogeneity[17] and the I2 test to quantify the proportion of the total variation due to heterogeneity[18] were calculated. A P value less than 0.10 for the Q statistic indicated that heterogeneity was observed across studies and a random-effects model (the DerSimonian and Laird method) was used[19]; otherwise, a fixed-effects model (the Mantel-Haenszel method) was applied[20]. Subgroup analyses for ethnicity, subtype of EC, and source of controls (HB or PB) were conducted. Sensitivity analyses were conducted to estimate the stability of the results such that each study was omitted one at a time to reflect the influence of the individual data set on the pooled OR.
Potential publication bias was assessed by visual inspection of funnel plot asymmetry, the Begg’s rank correlation method[21] and the Egger’s weighted regression method[22]. All statistical analyses were conducted using the STATA software, version 11.0 (STATA Corp., College Station, TX, United States). All P values were two-sided.
RESULTS
Study selection for the meta-analysis
We preliminarily identified 13 studies based on the search terms applied. After the abstracts were screened and the full text of each article was assessed, a total of nine articles met the inclusion criteria. However, one article by Wang et al[12] was excluded due to insufficient data and another study conducted by Casson et al[16] was excluded because of the use of overlapping subjects. As a result, seven case-control studies[8-10,12-15] were included in this meta-analysis (Figure 1).
Figure 1.
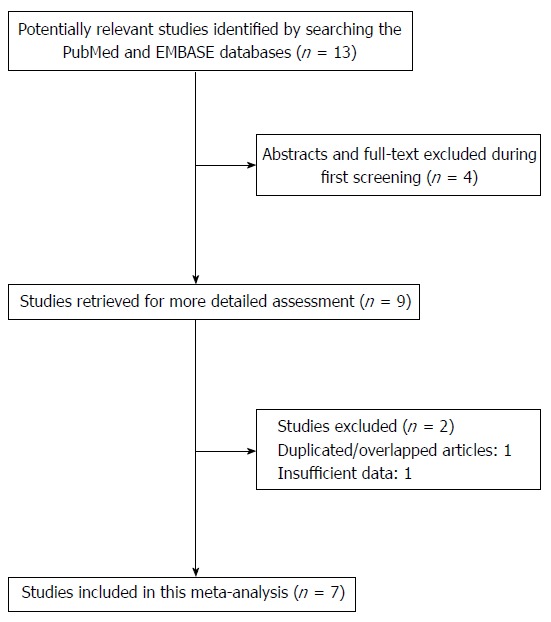
Flow diagram of the study selection strategy for this meta-analysis.
Characteristics of the included studies
The included studies were all reported in English. Five articles utilized Asian populations and two utilized Caucasian populations. Five of the studies obtained DNA for genotyping from blood samples, while two studies obtained DNA from EC tissue samples. To analyze the ESCC subtype, six studies were eligible with a total sample size of 804 cases and 1147 controls. To analyze the EADC subtype, two studies were pooled for analysis and included 314 cases and 676 controls. The main characteristics of all included studies are presented in Table 1.
Table 1.
Characteristics of the studies included in the meta-analysis
| Study | Year | Country | Ethnicity | Cases | Controls | Source of controls | Type of cancer | DNA source |
| Dura et al[9] | 2012 | The Netherlands | Caucasian | 349 | 581 | PB | ESCC and EADC | Blood and tissue samples |
| Ihsan et al[10] | 2010 | India | Asian | 142 | 185 | PB | ESCC | Blood samples |
| Jain et al[11] | 2008 | India | Asian | 107 | 320 | PB | ESCC | Blood samples |
| Casson et al[16] | 2006 | Canada | Caucasian | 56 | 95 | HB | EADC | Blood samples |
| Lin et al[13] | 2006 | China | Asian | 145 | 352 | PB | ESCC | Blood samples |
| Zhang et al[14] | 2003 | China | Asian | 257 | 252 | HB | ESCC | Blood samples |
| Wang et al[15] | 2003 | China | Asian | 62 | 38 | PB | ESCC | Tissue samples |
EPHX1: Microsomal epoxide hydrolase; ESCC: Esophageal squamous cell carcinoma; EADC: Esophageal adenocarcinoma; PB: Population-based controls; HB: Hospital-based controls.
EPHX1 p.Tyr113His polymorphism and EC risk
To analyze the EPHX1 p.Tyr113His polymorphism in relation to EC risk, seven studies with a total of 1118 cases and 1823 controls were pooled for analysis. The combined results showed that no significant association was observed in any of the genetic models applied (Figure 2; Table 2). The subgroup analysis for ethnicity, EC subtype, and source of controls also showed no evidence of association in any of the genetic models (Table 2).
Figure 2.
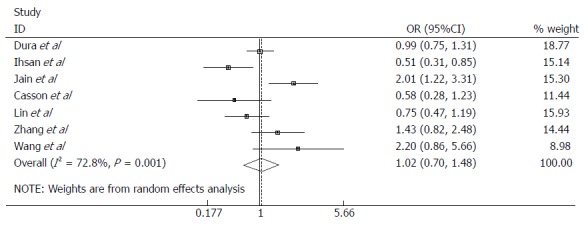
Odds ratios with 95%CI for the microsomal epoxide hydrolase p.Tyr113His polymorphism and risk of esophageal cancer (Tyr/His vs Tyr/Tyr).
Table 2.
Distribution of epoxide hydrolase p.Tyr113His genotypes in controls and esophageal cancer patients
| Variable | Studies | Cases/controls | OR (95%CI) | P value | P for heterogeneity | I2 | Model |
| Tyr/His vs Tyr/Tyr | |||||||
| Total | 7 | 1118/1823 | 1.00 (0.70-1.48) | 0.928 | 0.001 | 72.8 | R |
| Caucasian | 2 | 405/676 | 0.92 (0.71-1.20) | 0.555 | 0.192 | 41.2 | F |
| Asian | 5 | 713/1147 | 1.14 (0.65-2.00) | 0.638 | 0.001 | 79.9 | R |
| ESCC | 6 | 804/1147 | 1.08 (0.69-1.69) | 0.746 | 0.001 | 75.6 | R |
| EADC | 2 | 314/676 | 0.95 (0.71-1.26) | 0.729 | 0.160 | 49.2 | F |
| PB | 5 | 805/1476 | 1.04 (0.66-1.66) | 0.857 | 0.001 | 78.3 | R |
| HB | 2 | 313/347 | 0.95 (0.39-2.28) | 0.902 | 0.057 | 72.4 | R |
| His/His vs Tyr/Tyr | |||||||
| Total | 7 | 1118/1823 | 1.10 (0.77-1.57) | 0.592 | 0.046 | 53.3 | R |
| Caucasian | 2 | 405/676 | 0.74 (0.48-1.13) | 0.165 | 0.172 | 46.5 | F |
| Asian | 5 | 713/1147 | 1.23 (0.94-1.60) | 0.137 | 0.120 | 45.3 | F |
| ESCC | 6 | 804/1147 | 1.18 (0.92-1.52) | 0.196 | 0.158 | 37.2 | F |
| EADC | 2 | 314/676 | 0.72 (0.45-1.15) | 0.163 | 0.183 | 43.7 | F |
| PB | 5 | 805/1476 | 1.18 (0.78-1.78) | 0.423 | 0.079 | 52.1 | R |
| HB | 2 | 313/347 | 0.78 (0.24-2.58) | 0.688 | 0.034 | 77.7 | R |
| His/His + Tyr/His vs Tyr/Tyr | |||||||
| Total | 7 | 1118/1823 | 1.06 (0.75-1.49) | 0.753 | 0.001 | 74.0 | R |
| Caucasian | 2 | 405/676 | 0.76 (0.42-1.38) | 0.367 | 0.088 | 65.6 | R |
| Asian | 5 | 713/1147 | 1.23 (0.77-1.98) | 0.382 | 0.001 | 77.8 | R |
| ESCC | 6 | 804/1147 | 1.15 (0.78-1.70) | 0.484 | 0.002 | 74.0 | R |
| EADC | 2 | 314/676 | 0.77 (0.41-1.46) | 0.421 | 0.074 | 68.6 | R |
| PB | 5 | 805/1476 | 1.13 (0.74-1.70) | 0.575 | 0.002 | 77.0 | R |
| HB | 2 | 313/347 | 0.87 (0.33-2.26) | 0.769 | 0.017 | 82.4 | R |
| His/His vs Tyr/His + Tyr/Tyr | |||||||
| Total | 7 | 1118/1823 | 1.09 (0.89-1.34) | 0.416 | 0.188 | 31.4 | F |
| Caucasian | 2 | 405/676 | 0.77 (0.51-1.16) | 0.213 | 0.281 | 13.9 | F |
| Asian | 5 | 713/1147 | 1.23 (0.97-1.55) | 0.093 | 0.388 | 3.3 | F |
| ESCC | 6 | 804/1147 | 1.20 (0.95-1.50) | 0.121 | 0.469 | 0.0 | F |
| EADC | 2 | 314/676 | 0.91 (0.29-2.84) | 0.878 | 0.041 | 76.1 | R |
| PB | 5 | 805/1476 | 1.14 (0.89-1.45) | 0.294 | 0.210 | 31.8 | F |
| HB | 2 | 313/347 | 0.97 (0.66-1.43) | 0.888 | 0.111 | 60.6 | F |
EPHX1: Microsomal epoxide hydrolase; OR: Odds ratio; R: Random-effects model; F: Fixed-effects model; ESCC: Esophageal squamous cell carcinoma; EADC: Esophageal adenocarcinoma; PB: Population-based controls; HB: Hospital-based controls.
EPHX1 p.His139Arg polymorphism and EC risk
Six studies with 861 cases and 1571 controls were eligible to analyze the association of the EPHX1 p.His139Arg polymorphism and EC risk. The EPHX1 p.His139Arg polymorphism analysis showed that the Arg allele had no significant association with EC susceptibility when compared to the His wild-type allele (Table 3). Similar results were obtained from the subgroup analysis for ethnicity, EC subtype, and source of controls; the results are summarized in Table 3.
Table 3.
Distribution of the microsomal epoxide hydrolase p.His139Arg genotypes in controls and esophageal cancer patients
| Variable | Studies | Cases/controls | OR (95%CI) | P value | P for heterogeneity | I2 | Model |
| Arg/His vs His/His | |||||||
| Total | 6 | 861/1571 | 1.02 (0.84-1.23) | 0.869 | 0.176 | 34.8 | F |
| Caucasian | 2 | 405/676 | 0.97 (0.74-1.27) | 0.824 | 0.955 | 0.0 | F |
| Asian | 4 | 456/895 | 1.09 (0.69-1.73) | 0.709 | 0.060 | 59.4 | R |
| ESCC | 5 | 547/1319 | 0.99 (0.67-1.46) | 0.954 | 0.052 | 57.5 | R |
| EADC | 2 | 314/676 | 1.05 (0.79-1.40) | 0.718 | 0.755 | 0.0 | F |
| PB | 5 | 805/1476 | 1.02 (0.84-1.24) | 0.833 | 0.106 | 47.5 | F |
| HB | 1 | 56/95 | 0.95 (0.47-1.91) | NA | NA | NA | NA |
| Arg/Arg vs His/His | |||||||
| Total | 6 | 861/1571 | 0.96 (0.60-1.54) | 0.855 | 0.206 | 30.6 | F |
| Caucasian | 2 | 405/676 | 0.61 (0.31-1.21) | 0.158 | 0.308 | 3.9 | F |
| Asian | 4 | 456/895 | 1.59 (0.80-3.16) | 0.184 | 0.324 | 13.6 | F |
| ESCC | 5 | 547/1319 | 1.28 (0.72-2.30) | 0.402 | 0.346 | 10.6 | F |
| EADC | 2 | 314/676 | 0.60 (0.28-1.27) | 0.178 | 0.313 | 1.8 | F |
| PB | 5 | 805/1476 | 1.08 (0.66-1.76) | 0.765 | 0.251 | 25.5 | F |
| HB | 1 | 56/95 | 0.22 (0.03-1.90) | NA | NA | NA | NA |
| Arg/Arg + Arg/His vs His/His | |||||||
| Total | 6 | 861/1571 | 1.03 (0.78-1.37) | 0.810 | 0.095 | 46.6 | R |
| Caucasian | 2 | 405/676 | 0.92 (0.71-1.20) | 0.555 | 0.719 | 0.0 | F |
| Asian | 4 | 456/895 | 1.14 (0.71-1.81) | 0.593 | 0.042 | 63.5 | R |
| ESCC | 5 | 547/1319 | 1.02 (0.68-1.53) | 0.918 | 0.028 | 63.1 | R |
| EADC | 2 | 314/676 | 1.00 (0.75-1.31) | 0.976 | 0.550 | 0.0 | F |
| PB | 5 | 805/1476 | 1.07 (0.78-1.48) | 0.676 | 0.062 | 55.5 | R |
| HB | 1 | 56/95 | 0.82 (0.42-1.62) | NA | NA | NA | NA |
| Arg/Arg vs Arg/His + His/His | |||||||
| Total | 6 | 861/1571 | 0.97 (0.61-1.56) | 0.911 | 0.247 | 25.0 | F |
| Caucasian | 2 | 405/676 | 0.62 (0.31-1.21) | 0.163 | 0.307 | 4.0 | F |
| Asian | 4 | 456/895 | 1.66 (0.84-3.30) | 0.148 | 0.454 | 0.0 | F |
| ESCC | 5 | 547/1319 | 1.36 (0.76-2.45) | 0.297 | 0.480 | 0.0 | F |
| EADC | 2 | 314/676 | 0.58 (0.28-1.24) | 0.160 | 0.328 | 0.0 | F |
| PB | 5 | 805/1476 | 1.10 (0.67-1.80) | 0.704 | 0.312 | 16.1 | F |
| HB | 1 | 56/95 | 0.23 (0.03-1.91) | NA | NA | NA | NA |
EPHX1: Microsomal epoxide hydrolase; OR: Odds ratio; R: Random-effects model; F: Fixed-effects model; ESCC: Esophageal squamous cell carcinoma; EADC: Esophageal adenocarcinoma; PB: Population-based controls; HB: Hospital-based controls; NA: Not available.
Sensitivity analysis
We conducted a sensitivity analysis to assess the stability of this meta-analysis. When any one study was omitted, the results were not altered (data not shown). These data suggest that our results are stable and credible.
Publication bias
After performing the Begg’s test and the Egger’s test for publication bias, we observed no obvious bias in this meta-analysis (Begg’s test, P = 1.00; Egger’s test, P = 0.852) (Figure 3).
Figure 3.
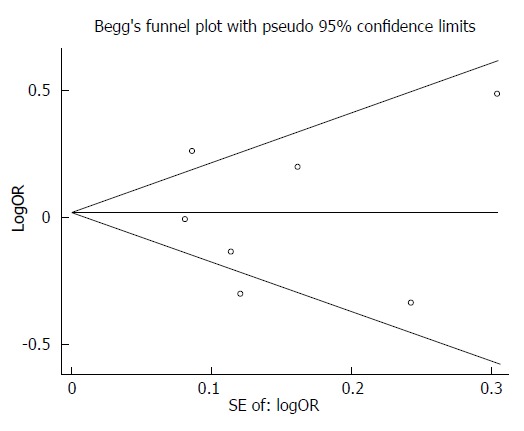
Funnel plot of the EPHX1 p.Tyr113His polymorphism to determine if publication bias was present (Tyr/His vs Tyr/Tyr).
DISCUSSION
Summary of results
In recent years, there has been increased interest in investigating the potential association of EPHX1 polymorphisms and susceptibility to EC. Because the results of these studies have been inconsistent, it is necessary to perform a meta-analysis of the studies performed to date. In the meta-analysis presented herein, seven studies on EPHX1 polymorphisms were analyzed to provide the most comprehensive assessment to date of the association between EPHX1 polymorphisms and EC risk. We observed no significant association of the p.Tyr113His and p.His139Arg polymorphisms in EPHX1 with EC risk, even when a subgroup analysis for ethnicity, EC subtype, or source of controls was performed.
Explanations for absence of EC association with the EPHX1 polymorphisms
The ESCC and EADC EC subtypes are reflected histologically by the progression from metaplasia to dysplasia to carcinoma. The recent identification of molecular markers lends further insight into the molecular pathogenesis of the different EC subtypes and there is no distinction between ESCC and EADC.
We found that the EPHX1 p.Tyr113His and p.His139Arg polymorphisms were not associated with EC risk. Previous studies have also found no association between EC and polymorphisms in the cytochrome oxidase genes of CYP1A1, CYP1B1, CYP2A6 and CYP2E1, the glutathione S transferase genes of GSTM1 and GSTP1 or EPHX[4]. Concurrently, other studies have demonstrated that polymorphisms in EPHX1 are risk factors for hepatocellular carcinoma, colorectal cancer, lung cancer, and cervical cancer. Zhong et al[23], however, demonstrated that the p.His139Arg microsomal epoxide hydrolase genotype may not be associated with hepatocellular carcinoma, while Liu et al[24] provided data that the EPHX1 p.Tyr113His polymorphism had no association with colorectal cancer development.
We performed a subgroup analysis for ethnicity[5,25,26] and observed no difference in the association between the polymorphisms and EC risk in Caucasians or Asians. The lack of an observed association may be due to the limited number of studies included in this meta-analysis. The allele and genotype frequencies of polymorphisms and their effects on EC risk varied in different ethnicities. Larger and well-designed multi-center studies using Caucasian and Asian populations are needed to re-evaluate such an association. Moreover, different sources of controls may be a confounding factor that influenced the conclusion of our study.
Some studies in this meta-analysis used PB controls as the reference group, while others used HB controls as the reference group. In order to eliminate the potential bias from this confounding factor, subgroup analysis by source of controls was conducted. The pooled results indicated that no significant association between EPHX1 (p.His139Arg and p.Arg139His) polymorphisms and EC risk was observed in PB or HB studies. HB studies are prone to selection biases because some of the controls may actually be ill (so that they are more similar to the cases). HB controls are not representative of the general population, especially when the investigated genotypes were patient controls. A proper PB control subject may be the superior choice to reduce potential biases in genetic association studies.
Potential confounders
In the current study, a Begg’s funnel plot and the Egger’s test were performed to assess potential publication bias. The funnel plots were symmetric in shape and the statistical results did not reveal any publication bias. Moreover, the results were consistent when the sensitivity analysis was performed, which implied that the results were reliable.
Limitations
Several limitations should be acknowledged for this analysis. Heterogeneity is a potential problem when interpreting the results of a meta-analysis and the sources of heterogeneity are usually explored in most meta-analyses. EC is a multi-factorial disease and potential gene-gene and gene-environment interactions should be considered. Different ethnicities have diverse genetic backgrounds and varied environmental exposures. In the present study, significant heterogeneity was detected in overall comparisons for the EPHX1 polymorphisms. Although we performed a careful database search for published studies, used strict criteria for study inclusion, and performed precise data extraction and original data analysis, significant heterogeneity still existed in some of our comparisons. However, in the subgroup analysis for ethnicity, the significant heterogeneity persisted in some genetic models in both the European and Asian populations. Small sample size may have contributed to limited statistical power to estimate the possible EC risk with EPHX1 polymorphisms. A consortium based on thousands of individuals would be more ideal for this type of association study.
This meta-analysis suggests that the EPHX1 p.Tyr113His and p.His139Arg polymorphisms may be not associated with EC development. Further studies with a larger sample size are needed to further assess the presence of an association.
COMMENTS
Background
Esophageal cancer (EC) is the sixth leading cause of cancer death, but the underlying pathogenic mechanism is not yet fully elucidated. Several studies have investigated the potential association between polymorphisms in the microsomal epoxide hydrolase (EPHX1) gene and susceptibility to EC development. Polymorphisms in EPHX1 alter its enzymatic activity and may thus lead to inter-individual differences in sensitivity to chemical carcinogens. However, the association of these polymorphisms with genetic susceptibility is currently unclear.
Research frontiers
Over the past two decades, many studies have been performed in diverse populations to determine if associations exist between EPHX1 polymorphisms and risk for EC. The results, however, have been conflicting and no consistent conclusion has been reached.
Innovations and breakthroughs
The findings in this meta-analysis are of great value. The EPHX1 p.Tyr113His and p.His139Arg polymorphisms may be not associated with EC development. Subgroup analyses for ethnicity, subtype of EC, and source of controls [hospital-based (HB) or population-based (PB)] were conducted; yet, no significant association was observed for any of these subgroups. No evidence of publication bias was found.
Applications
EPHX1 p.Tyr113His and p.His139Arg polymorphisms, ethnicity, subtype of EC, and source of controls (HB/PB) may be not associated with EC development. An exploration of this association may not be relevant to EC development.
Terminology
EPHX1 is a critical biotransformation enzyme that converts epoxides to trans-dihydrodiols during aromatic compound degradation. The epoxides can be conjugated and excreted from the body. EPHX1 functions in both the activation and detoxification of epoxides.
Peer review
This manuscript presents a well-performed meta-analysis that assesses the association of two EPHX1 polymorphisms with esophageal cancer risk. The authors show clearly that neither of the EPHX1 polymorphisms, p.Tyr113His or p.His139Arg, is associated with an increased risk for EC.
Footnotes
P- Reviewers: Islami F, Kury S, Nishida T, Vieth M S- Editor: Zhai HH L- Editor: Wang TQ E- Editor: Wu HL
References
- 1.Sant M, Aareleid T, Berrino F, Bielska Lasota M, Carli PM, Faivre J, Grosclaude P, Hédelin G, Matsuda T, Møller H, et al. EUROCARE-3: survival of cancer patients diagnosed 1990-94--results and commentary. Ann Oncol. 2003;14 Suppl 5:v61–118. doi: 10.1093/annonc/mdg754. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2002;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Lu SH, Chui SX, Yang WX, Hu XN, Guo LP, Li FM. Relevance of N-nitrosamines to oesophageal cancer in China. IARC Sci Publ. 1991;(105):11–17. [PubMed] [Google Scholar]
- 4.Stoner GD, Gupta A. Etiology and chemoprevention of esophageal squamous cell carcinoma. Carcinogenesis. 2001;22:1737–1746. doi: 10.1093/carcin/22.11.1737. [DOI] [PubMed] [Google Scholar]
- 5.Casson AG, Zheng Z, Porter GA, Guernsey DL. Genetic polymorphisms of microsomal epoxide hydroxylase and glutathione S-transferases M1, T1 and P1, interactions with smoking, and risk for esophageal (Barrett) adenocarcinoma. Cancer Detect Prev. 2006;30:423–431. doi: 10.1016/j.cdp.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Harrison DJ, Hubbard AL, MacMillan J, Wyllie AH, Smith CA. Microsomal epoxide hydrolase gene polymorphism and susceptibility to colon cancer. Br J Cancer. 1999;79:168–171. doi: 10.1038/sj.bjc.6690028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lancaster JM, Brownlee HA, Bell DA, Futreal PA, Marks JR, Berchuck A, Wiseman RW, Taylor JA. Microsomal epoxide hydrolase polymorphism as a risk factor for ovarian cancer. Mol Carcinog. 1996;17:160–162. doi: 10.1002/(SICI)1098-2744(199611)17:3<160::AID-MC8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 8.Hassett C, Aicher L, Sidhu JS, Omiecinski CJ. Human microsomal epoxide hydrolase: genetic polymorphism and functional expression in vitro of amino acid variants. Hum Mol Genet. 1994;3:421–428. doi: 10.1093/hmg/3.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dura P, Bregitha CV, Te Morsche RH, Roelofs HM, Kristinsson JO, Wobbes T, Witteman BJ, Tan AC, Drenth JP, Peters WH. EPHX1 polymorphisms do not modify esophageal carcinoma susceptibility in Dutch Caucasians. Oncol Rep. 2012;27:1710–1716. doi: 10.3892/or.2012.1734. [DOI] [PubMed] [Google Scholar]
- 10.Ihsan R, Chattopadhyay I, Phukan R, Mishra AK, Purkayastha J, Sharma J, Zomawia E, Verma Y, Mahanta J, Saxena S, et al. Role of epoxide hydrolase 1 gene polymorphisms in esophageal cancer in a high-risk area in India. J Gastroenterol Hepatol. 2010;25:1456–1462. doi: 10.1111/j.1440-1746.2010.06354.x. [DOI] [PubMed] [Google Scholar]
- 11.Jain M, Tilak AR, Upadhyay R, Kumar A, Mittal B. Microsomal epoxide hydrolase (EPHX1), slow (exon 3, 113His) and fast (exon 4, 139Arg) alleles confer susceptibility to squamous cell esophageal cancer. Toxicol Appl Pharmacol. 2008;230:247–251. doi: 10.1016/j.taap.2008.02.023. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, Tang L, Sun G, Tang Y, Xie Y, Wang S, Hu X, Gao W, Cox SB, Wang JS. Etiological study of esophageal squamous cell carcinoma in an endemic region: a population-based case control study in Huaian, China. BMC Cancer. 2006;6:287. doi: 10.1186/1471-2407-6-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin YC, Wu DC, Lee JM, Hsu HK, Kao EL, Yang CH, Wu MT. The association between microsomal epoxide hydrolase genotypes and esophageal squamous-cell-carcinoma in Taiwan: interaction between areca chewing and smoking. Cancer Lett. 2006;237:281–288. doi: 10.1016/j.canlet.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Zhang JH, Jin X, Li Y, Wang R, Guo W, Wang N, Wen DG, Chen ZF, Kuang G, Wei LZ, et al. Epoxide hydrolase Tyr113His polymorphism is not associated with susceptibility to esophageal squamous cell carcinoma in population of North China. World J Gastroenterol. 2003;9:2654–2657. doi: 10.3748/wjg.v9.i12.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang LD, Zheng S, Liu B, Zhou JX, Li YJ, Li JX. CYP1A1, GSTs and mEH polymorphisms and susceptibility to esophageal carcinoma: study of population from a high- incidence area in north China. World J Gastroenterol. 2003;9:1394–1397. doi: 10.3748/wjg.v9.i7.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casson AG, Zheng Z, Chiasson D, MacDonald K, Riddell DC, Guernsey JR, Guernsey DL, McLaughlin J. Associations between genetic polymorphisms of Phase I and II metabolizing enzymes, p53 and susceptibility to esophageal adenocarcinoma. Cancer Detect Prev. 2003;27:139–146. doi: 10.1016/s0361-090x(03)00033-3. [DOI] [PubMed] [Google Scholar]
- 17.Vangel MG, Rukhin AL. Maximum likelihood analysis for heteroscedastic one-way random effects ANOVA in interlaboratory studies. Biometrics. 1999;55:129–136. doi: 10.1111/j.0006-341x.1999.00129.x. [DOI] [PubMed] [Google Scholar]
- 18.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 20.MANTEL N, HAENSZEL W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 21.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 22.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong JH, Xiang BD, Ma L, You XM, Li LQ, Xie GS. Meta-analysis of microsomal epoxide hydrolase gene polymorphism and risk of hepatocellular carcinoma. PLoS One. 2013;8:e57064. doi: 10.1371/journal.pone.0057064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu F, Yuan D, Wei Y, Wang W, Yan L, Wen T, Xu M, Yang J, Li B. Systematic review and meta-analysis of the relationship between EPHX1 polymorphisms and colorectal cancer risk. PLoS One. 2012;7:e43821. doi: 10.1371/journal.pone.0043821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haufroid V, Merz B, Hofmann A, Tschopp A, Lison D, Hotz P. Exposure to ethylene oxide in hospitals: biological monitoring and influence of glutathione S-transferase and epoxide hydrolase polymorphisms. Cancer Epidemiol Biomarkers Prev. 2007;16:796–802. doi: 10.1158/1055-9965.EPI-06-0915. [DOI] [PubMed] [Google Scholar]
- 26.Hengstler JG, Arand M, Herrero ME, Oesch F. Polymorphisms of N-acetyltransferases, glutathione S-transferases, microsomal epoxide hydrolase and sulfotransferases: influence on cancer susceptibility. Recent Results Cancer Res. 1998;154:47–85. doi: 10.1007/978-3-642-46870-4_4. [DOI] [PubMed] [Google Scholar]


