Abstract
In mammals, genetic recombination during meiosis is limited to a set of 1- to 2-kb regions termed hotspots. Their locations are predominantly determined by the zinc finger protein PRDM9, which binds to DNA in hotspots and subsequently uses its SET domain to locally trimethylate histone H3 at lysine 4 (H3K4me3). This sets the stage for double-strand break (DSB) formation and reciprocal exchange of DNA between chromatids, forming Holliday junctions. Here we report genome-wide analyses of PRDM9-dependent histone modifications using two inbred mouse strains differing only in their PRDM9 zinc finger domain. We show that PRDM9 binding actively reorganizes nucleosomes into a symmetrical pattern, creating an extended nucleosome-depleted region. These regions are centered by a consensus PRDM9 binding motif, whose location and identity was confirmed in vitro. We also show that DSBs are centered over the PRDM9 binding motif within the nucleosome-depleted region. Combining these results with data from genetic crosses, we find that crossing-over is restricted to the region marked by H3K4me3. We suggest that PRDM9-modified nucleosomes create a permissible environment that first directs the location of DSBs and then defines the boundaries of Holliday junction branch migration.
A hallmark of sexual reproduction is the exchange of genetic material between chromatids through the process of meiotic recombination. This exchange is essential for proper chromatid segregation during meiosis, and improper segregation can lead to germ cell death or the production of abnormal gametes that result in spontaneous abortion and developmental disorders (Paigen and Petkov 2010). Rather than randomly located along chromosomes, mammalian recombination sites are restricted to 1- to 2-kb regions, termed hotspots, whose locations are determined by the meiosis-specific protein PRDM9 (Baudat et al. 2010; Berg et al. 2010; Myers et al. 2010; Parvanov et al. 2010; Berg et al. 2011). PRDM9 is a DNA-binding, zinc finger protein that also trimethylates histone H3K4 (Hayashi et al. 2005), a chromatin modification enriched at mammalian hotspots (Buard et al. 2009; Grey et al. 2011; Billings et al. 2013).
An irreversible commitment to meiotic recombination occurs when DNA double-strand breaks (DSBs) are introduced at hotspots by the conserved enzyme SPO11 (Keeney 2008; Cole et al. 2010a). These DSBs are preferentially repaired using a homologous chromatid as the template and either involve the reciprocal exchange of DNA, leading to crossing-over, or to repair without exchange, resulting in gene conversion (Kohl and Sekelsky 2013). After introduction of DSBs, the DNA is processed to produce single-stranded ends that are bound by the meiotic recombinase DMC1 to facilitate strand invasion of the homologous chromatid. Repair of DSBs involving reciprocal exchange of DNA proceeds by formation of double Holliday junctions which undergo enzymatically assisted branch migration, allowing the exact positions of crossing-over to be moved from the original site of the DSB (Neale and Keeney 2006; Handel and Schimenti 2010). Physical confirmation that sites of PRDM9 binding determine where recombination can occur came with the first genome-wide maps of DSBs, by mapping DMC1-associated DNA, showing that nearly all DSBs overlap with sites of PRDM9-dependent H3K4me3 (Smagulova et al. 2011; Brick et al. 2012). In the absence of Prdm9, DSBs are redirected to other H3K4me3-modified loci (Brick et al. 2012). These ectopic DSBs are inefficiently repaired, leading to meiotic arrest (Hayashi et al. 2005), thus highlighting another important role of PRDM9.
Prdm9 is highly polymorphic in many mammalian species including primate and mouse populations (Thomas et al. 2009; Berg et al. 2010; Buard et al. 2014; Kono et al. 2014). These polymorphisms are almost entirely constrained to variation in the number of C-terminal zinc finger repeats and, within each zinc finger, nonsynonymous substitutions found at the three amino acids that contact DNA. Sequencing large populations of both wild-caught and laboratory strains of Mus, Kono et al. (2014) found 57 different alleles in a sample of 116 mice, while Buard et al. (2014) found 113 alleles in their sample of 257 mice. These allele-specific combinations of zinc fingers dictate the nucleotide sequences at which PRDM9 can bind DNA and locally trimethylate hotspot nucleosomes, ultimately marking the position of meiotic recombination (Grey et al. 2011; Billings et al. 2013).
To facilitate genome-wide identification of trimethylated hotspots, we used co-isogenic mouse strains carrying two different Prdm9 alleles, both in the C57BL/6J (B6) background (Fig. 1A). One strain was B6 itself, containing Prdm9Dom2. The other strain was constructed by replacing exon 12 of Prdm9Dom2, which encodes the entire zinc finger domain, with the corresponding exon from the Prdm9Cst allele found in the CAST/EiJ (CAST) strain. The only genetic difference between these strains is the identity of the PRDM9 zinc finger array.
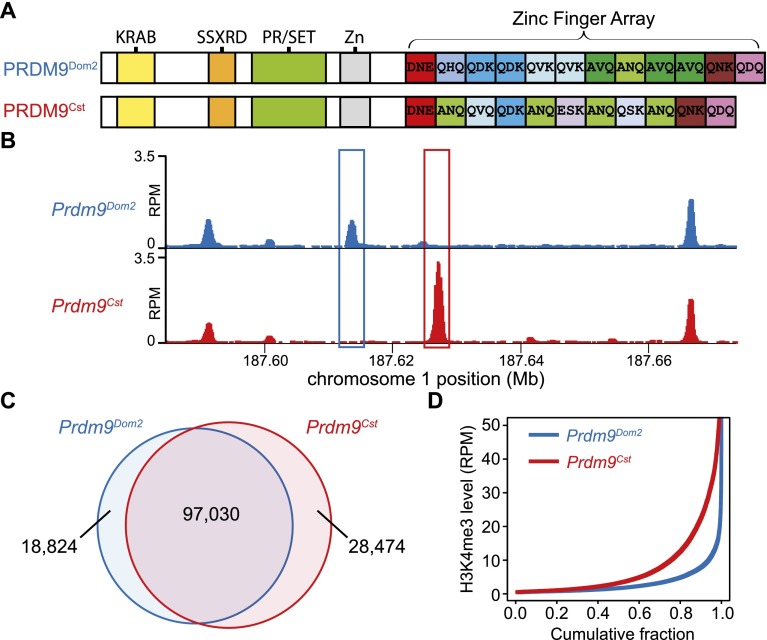
Figure 1.
H3K4me3 ChIP-seq allows detection of PRDM9-dependent sites using co-isogenic strains of mice. (A) PRDM9Dom2 and PRDM9Cst differ both in the number of zinc finger domains and sequence-specific identity of individual domains. The known domain structure of PRDM9 includes an N-terminal KRAB domain (Krüppel-associated box), an SSXRD (SSX repression domain), a PR/SET domain (methyltransferase activity), an invariant zinc finger domain, and a C-terminal zinc finger array. The three most polymorphic positions in the individual zinc finger domains are indicated by their amino acid identity. (B) H3K4me3 coverage profile from a representative 100-kb window of chromosome 1. H3K4me3 peaks can be unique or shared between the two strains of mice (in all figures, the Prdm9Cst allele originates from the CAST/EiJ strain and is represented in red, while Prdm9Dom2, the endogenous allele for C57BL/6J, is represented in blue). (C) Venn diagram showing quantity and identity of H3K4me3 peaks. Prdm9Cst results in more unique H3K4me3-marked hotspots. (D) H3K4me3 signal for strain-specific peaks. Prdm9Cst hotspots have higher methylation levels than Prdm9Dom2 in the B6 background (red: Prdm9Cst; blue: Prdm9Dom2).
Results
PRDM9-dependent chromatin modification is extensive in meiosis
We used immunoprecipitation of micrococcal nuclease (MNase)-digested chromatin from 12-d post-partum male germ cells, followed by high-throughput DNA sequencing (ChIP-seq), to generate genome-wide H3K4me3 maps from the two co-isogenic strains (Fig. 1B). Most H3K4me3-modified loci are shared between the strains, representing common features such as active promoters and enhancers. Because the only difference between strains is the zinc finger array of PRDM9, unique H3K4me3 positions represent putative allele-specific sites of modification (Fig. 1B,C). We found appreciably more PRDM9Cst-dependent H3K4me3 sites than PRDM9Dom2-dependent ones, and these PRDM9Cst sites, on average, have a greater level of trimethylation (Fig. 1C,D). One contributing factor to this difference may be that the Prdm9Cst knock-in allele, which originated in Mus musculus castaneus, has been in contact with the M. m. domesticus genome of B6 for only a few generations, insufficient time for its hotspot sequences to undergo evolutionary erosion (Boulton et al. 1997).
We estimated the number of PRDM9Dom2-activated sites in an average meiosis. Using quantitative PCR of four H3K4me3 positions that varied in activity, we established the proportionality between ChIP results and the fraction of total “input” DNA from spermatocytes that was H3K4me3-modified in the same samples (Supplemental Fig. S1). This enabled conversion of the number of ChIP-seq reads at a hotspot to the fraction of chromatids with trimethylated nucleosomes. Summing across all hotspots, after correcting for the presence of four chromatids in a meiotic cell and for the estimated representation of early meiotic prophase cells (see Methods), we estimate that there are ∼4700 ± 400 PRDM9-modified sites in the B6 background in an average meiosis. This conservative estimate of PRDM9-activated positions is ∼15 times higher than the current estimated number of DSBs per meiotic cell based on immunohistochemistry (Cole et al. 2012), raising the question of what additional factors determine the likelihood of progression from PRDM9-activation to induction of a DSB.
Hotspots contain symmetrically organized nucleosomes around a central nucleosome-depleted region
In vivo, DNA is packaged by wrapping 147–150 bp of DNA around a histone octamer to form a nucleosome, with 38–75 bp of inter-nucleosome linker DNA (Kornberg 1977; Valouev et al. 2011; Yamada and Ohta 2013). Earlier studies are contradictory regarding the question of how nucleosomes are organized within hotspots. Getun et al. (2010) reported that hotspots could contain regions >1 kb nearly devoid of nucleosomes in an arrangement that is similar for meiotic and somatic cells; however, this is difficult to accept given the much repeated observation that hotspots sequences are enriched for H3K4me3 nucleosomes. Shenkar et al. (1991) reported one hotspot that contained a DNase I hypersensitivity site overlapping the position of recombination, indicating the absence of a nucleosome there; whereas in contrast, Smagulova et al. (2011) reported that sites of DSBs typically contain a well-placed nucleosome at their center.
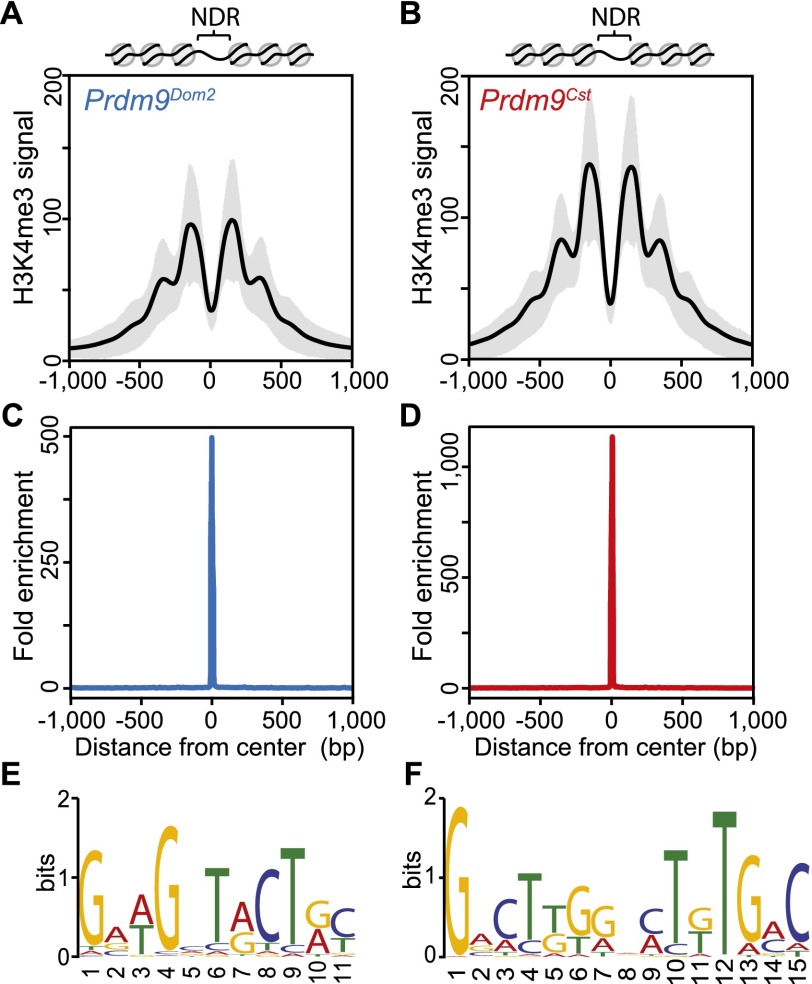
In our data, H3K4me3-modified nucleosomes at individual hotspots show a distinct, symmetrical oscillatory pattern, organized around a central nucleosome-depleted region (NDR) (Fig. 2A,B). Predicted PRDM9 DNA binding motifs for each allele are enriched at the centers of their corresponding NDRs (Fig. 2C–F). The PRDM9Dom2 motif found here matches that previously identified at B6 DSB sites (Brick et al. 2012), and the 15-bp PRDM9Cst motif derived from in vivo binding in this study closely matches the region identified by in vitro studies as most sensitive to base-pair substitution (Billings et al. 2013). Together, these data suggest that PRDM9 binds hotspots in the central NDR.
Figure 2.
H3K4me3 nucleosomes are organized symmetrically around predicted PRDM9 binding motifs, creating a central nucleosome-depleted region (NDR). (A) Aggregate plot of H3K4me3 signal across a 2-kb window from PRDM9Dom2-dependent hotspots (n = 7235 peaks; black line: mean; gray shading: SD). Predicted nucleosome positions are shown above. (B) Similar to A except for PRDM9Cst-dependent hotspots (n = 9306 peaks). (C) Fold enrichment of the PRDM9Dom2 motif over the PRDM9Cst motif at Prdm9Dom2-dependent hotspots in A. (D) Fold enrichment of the PRDM9Cst motif over the PRDM9Dom2 motif at PRDM9Cst-dependent hotspots in (B). (E) The PRDM9Dom2 motif identified in the NDRs of H3K4me3 peaks in A. (F) The PRDM9Cst motif.
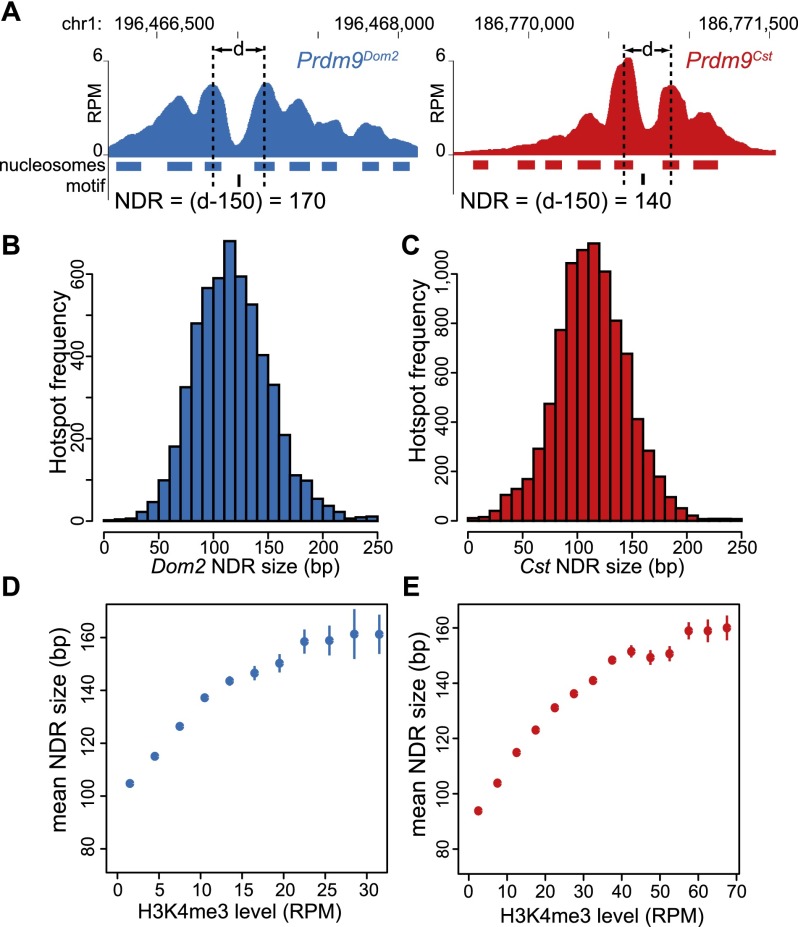
To calculate hotspot NDR lengths, we located nucleosomes, at individual hotspots, in the −1 and +1 positions relative to our identified motifs (Fig. 3A). The mean NDR length, for both alleles, is 120 bp ± 34 (Fig. 3B,C). Even though the PRDM9Cst and PRDM9Dom2 zinc finger arrays differ in length by one zinc finger domain, these data show that their average NDR lengths are similar at this level of resolution. Hotspot NDR length is not constant but instead positively correlates with hotspot strength, with higher H3K4me3 levels relating to longer NDRs (Fig. 3D,E). Hotspot NDRs are longer than the linker DNA found between the next pair of nucleosomes on either side of the NDR (66 bp ± 32) (Supplemental Fig. S2) and longer than linkers reported for mammalian cells (Valouev et al. 2011). The PRDM9 NDR is similar in size to the average 134 bp ± 25 NDR found in spermatocytes for insulator sequences bound by the highly conserved protein CTCF, another DNA binding protein containing 11 zinc fingers (Supplemental Fig. S3).
Figure 3.
NDR length is dependent on H3K4me3 level. (A) Two examples of calculating the NDR at individual hotspots, one activated by each Prdm9 allele (red: Prdm9Cst; blue: Prdm9Dom2). Hotspot nucleosome positions are indicated below the coverage profile with filled bars and PRDM9 binding motif positions in black. The NDR length was determined by subtracting 150 from the distance (d) between the centers of the −1 and +1 nucleosomes. (B) Distribution of NDR sizes for Prdm9Dom2 hotspots (n = 5538). (C) Distribution of NDR sizes for Prdm9Cst hotspots (n = 9194). (D) Relationship between H3K4me3 level and NDR length at Prdm9Dom2 hotspots (error bars: standard error of the mean NDR length for each H3K4me3 category). (E) Relationship between NDR length and H3K4me3 level at Prdm9Cst hotspots.
PRDM9 binds predicted motifs in the NDRs
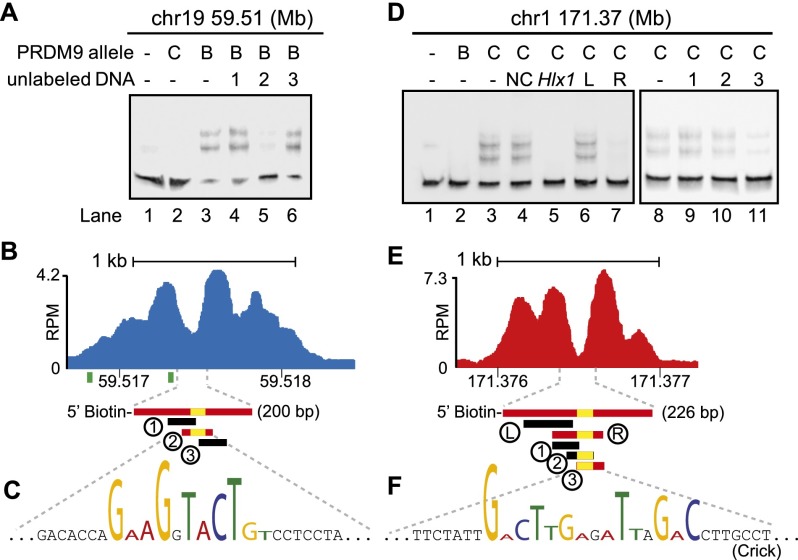
In order to confirm that the motifs found in the NDRs are the targets of PRDM9 binding, we chose four hotspots as models, two for each allele, and tested their binding specificity using an in vitro mobility shift assay. We found that only the allele that activates a hotspot in vivo binds DNA from that NDR in vitro, and the minimum DNA fragment required for binding always contains a close match to the consensus motif (Fig. 4; Supplemental Figs. S4, S5).
Figure 4.
PRDM9 binds hotspots at the NDR. (A) Mapping of a PRDM9Dom2 binding site. EMSA shows specific binding of PRDM9Dom2 at the PRDM9Dom2-activated hotspot located on chromosome 19 at Mb 59.51. Only unlabeled DNA oligo 2, containing the consensus motif, can compete for binding (Lane 5). All lanes contain labeled DNA; otherwise, the composition of the binding reactions is shown above the blot (B: PRDM9Dom2; C: PRDM9Cst). (B) ChIP-seq coverage profile from Prdm9Dom2 showing H3K4me3 nucleosome positions, the NDR, and DNA sequences used for the EMSA in A (red bars indicate DNA that binds PRDM9Dom2; black bars indicate nonbinding sequences; yellow bars show the position of the PRDM9Dom2 motif; green squares under the graph show the position of the predicted binding site from Getun et al. [2012]). (C) The partial DNA sequence of oligo 2. Nucleotides matching the PRDM9Dom2 consensus motif in Figure 2E are formatted to indicate similarity. (D–F) Similar to A–C except showing results for PRDM9Cst. (D) PRDM9Cst binds hotspot chr1 171.36 Mb at the NDR ([NC] Noncompeting oligo 2 from chr19 59.51 in panel A. [Hlx1] Binding site for Hlx1 [chr1 186.44 Mb] from Billings et al. [2013]). (E) ChIP-seq coverage profile at hotspot chr1 171.37 from B6-Prdm9CAST-KI. (F) Partial DNA sequence of oligo 3 indicating nucleotides that match the consensus in Figure 2F.
These binding experiments also revealed several properties of PRDM9-DNA interactions. First, competition for PRDM9 binding in vitro is allele-specific. DNAs from different hotspots activated by the same allele compete with each other for binding (Fig. 4D, lanes 3,5), consistent with previous results (Billings et al. 2013). The hotspot at chr1 171.37 Mb is specifically bound by PRDM9Cst, and this binding can be inhibited by addition of excess unlabeled DNA from the PRDM9Cst-activated hotspot Hlx1 (found at chr1 186.44 Mb). However, DNAs from hotspots activated by different alleles do not compete for binding (Fig. 4D, lanes 3,4). The PRDM9Dom2-activated hotspot chr19 59.51 Mb (labeled NC in Fig. 4D) does not inhibit PRDM9Cst binding to the PRDM9Cst-activated hotspot at chr1 171.37 Mb.
We also find that the consensus motif alone is not always sufficient for binding (Fig. 4F, oligo 2). This last observation indicates that other DNA features surrounding the motif also influence binding. PRDM9Cst has 11 zinc fingers in its array and requires ∼3 bp for each zinc finger for complete binding (Billings et al. 2013). Because the PRDM9Cst motif identified in the NDRs here aligns to zinc fingers 4–8, if the motif occurs near the edge of short DNA fragments (such as oligos 2 and 3 in Fig. 4E), there may be insufficient flanking DNA for full binding.
Finally, in silico predictions of PRDM9 binding sites based on the structure of zinc finger arrays can be a poor predictor of the true in vivo position. As an example, the actual in vivo binding site and NDR at hotspot chr19 59.51 Mb is not covered by sites previously predicted in silico (Fig. 4B, green bars; Getun et al. 2012), highlighting the importance of experimentally confirming protein-DNA interactions.
Combined, these results confirm that the motifs identified here and previously are the in vivo targets for PRDM9 binding.
PRDM9 binding organizes the nucleosomes surrounding meiotic hotspots
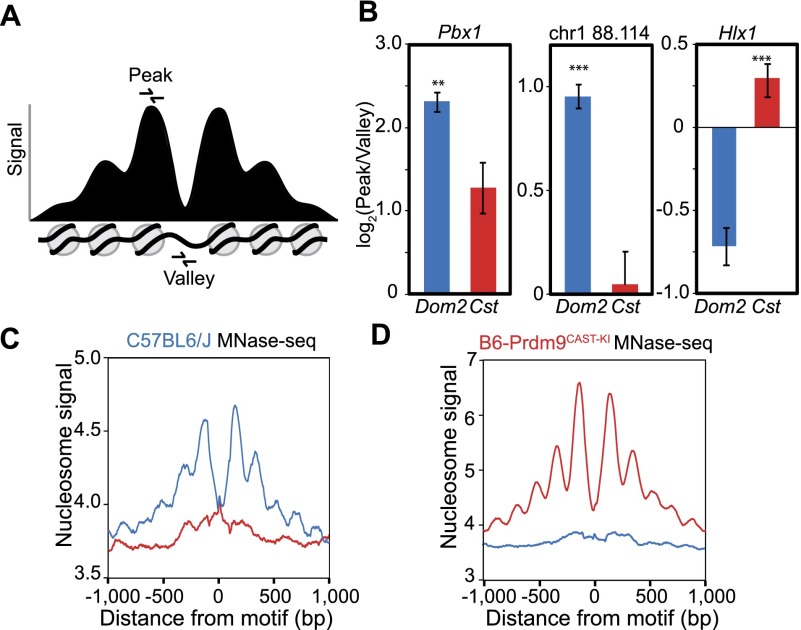
To determine whether nucleosome organization at hotspots precedes or is a consequence of PRDM9 binding, we first identified three hotspots with clear nucleosome peaks and an obvious NDR and compared nucleosome positions in our co-isogenic strains (Fig. 5A,B). Using qPCR, we quantified DNA from MNase-digested spermatocytes both at the nucleosome peak and within the NDR to calculate a peak-to-valley ratio. Nucleosomes should protect DNA from MNase digestion while the NDR should not, resulting in a peak-to-valley ratio >1. If PRDM9 binding leads to reorganization of nucleosomes and establishment of the NDR, the peak-to-valley ratio should be higher in the strain that contains the activating allele for a given hotspot. Nucleosome positions in the absence of an activating Prdm9 allele varied from one hotspot to another; most obviously, hotspot Pbx1 appears to lack a nucleosome at its binding site while hotspot Hlx1 has one (Fig. 5B). However, the peak-to-valley ratio at all three hotspots was significantly increased when an activating Prdm9 allele was present. Taking into account that not all copies of a hotspot are activated in every cell, this suggests that nucleosomes are remodeled in coordination with PRDM9 binding, reinforcing the position of the −1 or +1 nucleosome and granting greater MNase access to the NDR.
Figure 5.
Nucleosome positions at hotspots are variable prior to PRDM9 binding. (A) Schematic of an idealized PRDM9-modified hotspot showing signal intensity representing nucleosome positions. Primers for qPCR were designed to amplify DNA from either the −1 or +1 nucleosomes flanking the NDR (peak) or from within the NDR (valley). (B) Ratio of nucleosome peak vs. nucleosome-depleted valley signal in two genetic backgrounds. qPCR was performed on MNase-digested spermatocyte chromatin DNA from both strains (blue: DNA from Prdm9Dom2; red: Prdm9Cst). Error bars represent standard deviation of triplicate measurements from three biological replicates. (**) P < 0.01, (***) P < 0.001; one-sided t-test. Hotspots Pbx1 and chr1 88.114 Mb are PRDM9Dom2-activated; hotspot Hlx1 is PRDM9Cst-activated. (C) Aggregation plot showing nucleosome positions at hotspots inferred from MNase-seq of C57BL/6J spermatocytes. Only hotspots containing a motif that is predicted to be recognized by the endogenous Prdm9 allele show organized nucleosomes (blue: aggregation of Prdm9Dom2 sites; red: Prdm9Cst sites). (D) Aggregation plot of Prdm9Dom2 and Prdm9Cst hotspots from sequencing of mononucleosome DNA prepared from B6-Prdm9CAST-KI spermatocytes.
To characterize average hotspot nucleosome positioning for both Prdm9 alleles, this analysis was extended to the whole genome by sequencing all nucleosome-associated DNA in spermatocytes (MNase-seq). These data show that in C57BL/6J spermatocytes, nucleosomes are organized in a symmetrical pattern only within hotspots containing the DNA binding motif recognized by PRDM9Dom2 (Fig. 5C, blue line) and not at PRDM9Cst-activated hotspots (Fig. 5C, red line). The reverse is true in the B6-Prdm9CAST-KI strain, where hotspots containing the PRDM9Cst motif only show nucleosome organization (Fig. 5D, red line). Because the knock-in strain was created on the B6 background, hotspot sequences are identical between the two strains and would not contribute to a difference in nucleosome positioning (cis-control); the only difference is the identity of the Prdm9 allele (trans-control). In contrast, nucleosome organization at CTCF binding sites is similar in these two co-isogenic strains, highlighting the allele-specific nature of PRDM9 sites (Supplemental Fig. S3C,D). Collectively, these data show that PRDM9 binding actively reorganizes nucleosomes at hotspots.
PRDM9-dependent modification limits the branch migration of Holliday junctions
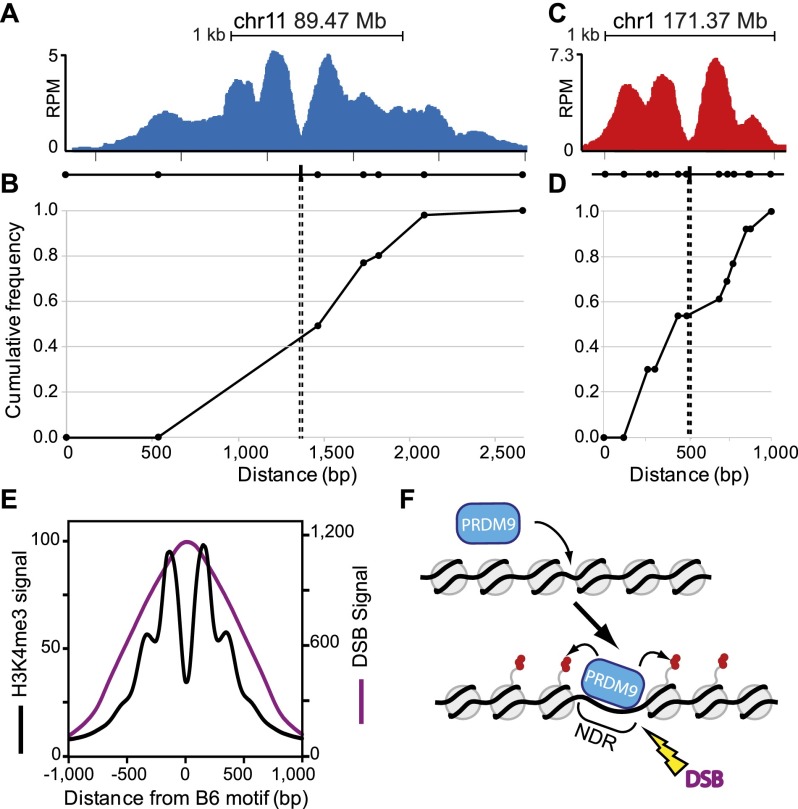
We combined H3K4me3 ChIP-seq data with genetic recombination maps from crosses between B6 and CAST mice (Paigen et al. 2008; Billings et al. 2010) to test whether the locations of H3K4me3 nucleosomes influence sites of genetic recombination. Initially, we chose two hotspots (Fig. 6A,C) containing SNPs distributed across the H3K4me3 interval and expanded the mapping of all crossovers at these hotspots using the repertoire of available SNPs. At both hotspots, the NDR and PRDM9 binding sites reside at the midpoint of all recombination events, and crossovers are limited to the region of H3K4me3 modification (Fig. 6B,D). These observations were confirmed for previously mapped recombinants at the Hlx1 hotspot activated by PRDM9Cst (Parvanov et al. 2009) and the A3 hotspot activated by PRDM9Dom2 (Supplemental Fig. S6; Kelmenson et al. 2005; Cole et al. 2010b).
Figure 6.
Location of meiotic crossing-over is constrained by PRDM9-dependent chromatin modification. (A) ChIP-seq coverage profile for the PRDM9Dom2-activated hotspot at chr11 89.47 Mb showing nucleosome peaks and central NDR. The black line underneath the nucleosome profile indicates the positions of SNPs (circles) between the B6 and CAST strains used to map recombination and the PRDM9Dom2 binding site from Supplemental Figure S5 (black box). (B) Cumulative frequency of recombination in B6 × CAST F1 progeny plotted against the scaled distance across the H3K4me3 signal track (n = 56 recombinant mice). (C,D) Similar to A and B except for the PRDM9Cst-activated hotspot at chr1 171.37 Mb. (C) ChIP-seq coverage profile showing nucleosome peaks and central NDR. SNPs assayed for recombination between B6 and CAST strains are indicated below as circles and PRDM9Cst binding site from Figure 4F is shown (black box). (D) Cumulative frequency of recombination at this hotspot (n = 13). (E) Aggregation plot of Prdm9Dom2 hotspots for mean H3K4me3 signal (black line) and mean DMC1 ChIP-seq signal representing the position of meiotic DSBs (purple line, n = 7235 peaks, DSB data from Brick et al. [2012]). (F) Model of PRDM9 activation of hotspots. Prior to PRDM9 binding, hotspot nucleosome positions can be variable. Upon binding, PRDM9 modifies surrounding nucleosomes (red circles), which are remodeled to create a central nucleosome-depleted region (NDR). Later, DNA double-strand breaks (DSBs) are introduced at the NDR.
DSBs are located at the center of the NDR
To determine the locations of DSBs in relation to the PRDM9-dependent NDRs, we combined our present data with the DSB data generated from DMC1 ChIP-seq in testis from B6 mice (Brick et al. 2012). Aggregating the genome-wide DSB signals in relation to the center of the NDR, we find that the peak of the mean DMC1 tag distribution signal is precisely aligned with the center of the PRDM9 NDR (Fig. 6E). The shape of the DSB distribution curve is expected if DSBs are located at the center of the NDR, given the 100- to 200-bp size of the DNA fragments from which sequences were obtained (Brick et al. 2012) and the ∼1-kb length of DMC1 coated filaments (Neale and Keeney 2006). Previous estimates found that, for a particular hotspot, the median of the DMC1 tag distribution could predict the location of meiotic DSBs with a precision of ∼180 nt (Smagulova et al. 2011); we now show that the peak of the average signal aligns with the center of PRDM9-dependent NDRs. These data support the idea that meiotic DSBs occur within NDRs and fit well with the above observation that crossing-over occurs symmetrically around the PRDM9 binding site.
Discussion
We have characterized the genome-wide positions of PRDM9-dependent chromatin modifications by comparing two inbred mouse strains that only differ in the zinc finger array of PRDM9. Because PRDM9 is an H3K4 methyltransferase, our fine-scale mapping of H3K4me3-modified nucleosomes reflects nucleosome positions after PRDM9 binding. These post-PRDM9 binding snapshots of nucleosome positions reveal several previously unknown features of hotspot chromatin architecture and provide mechanistic insight into how PRDM9 activates hotspots (Fig. 6E).
Hotspots contain a central nucleosome-depleted region that, on average, covers ∼120 bp (Fig. 3). PRDM9 can trimethylate 1–2 nucleosomes beyond the −1 and +1 nucleosomes flanking the binding sites. The ability of the catalytic domain of PRDM9 to reach this far may depend on supercoiling of chromatin to bring nucleosomes into closer proximity to PRDM9 in three-dimensional space. Previously, Valouev et al. (2011) reported a 130-bp NDR for the zinc finger protein CTCF and a 93-bp NDR for the REST (also known as NRSF) repressor in human cell lines. We have found a 134-bp NDR for mouse CTCF in spermatocytes (Supplemental Fig. S3), indicating that our MNase-seq methodology is robust.
Nucleosomes surrounding the PRDM9 NDR are organized in an oscillatory pattern, with increasing variability as a function of distance from the binding site, similar to CTCF. This symmetrical distribution of nucleosomes around an NDR fits with the previously established barrier model for nucleosome organization first proposed in yeast (Mavrich et al. 2008) and subsequently confirmed in mammalian cells (Valouev et al. 2011), which proposes that nucleosomes are packed against a well-positioned barrier whose location depends on either primary DNA sequence or the presence of a sequence-dependent DNA-binding factor. NDRs, such as the one reported for PRDM9 here, are likely general features of sequence specific DNA-binding factors, with the length of the depleted region being dependent on the protein complex examined.
We also find that the PRDM9-dependent H3K4me3 level is positively correlated with NDR length. If H3K4me3 level is a measure of the frequency with which a hotspot binds PRDM9 and becomes trimethylated, which in turn depends on the affinity of PRDM9 for its binding site in that hotspot, it appears that the extent to which PRDM9 reorganizes nucleosomes at a hotspot depends on its affinity for the PRDM9 binding site in that hotspot. That is, the stronger the binding, the greater the effect on nucleosome positioning.
PRDM9 binding alters hotspot nucleosomes organization. Nucleosomes are only symmetrically phased around PRDM9 binding sites when the appropriate Prdm9 allele is present (Fig. 5). This process probably requires the activity of the SWI/SNF family of ATP-dependent remodeling enzymes (Narlikar et al. 2013), which are often recruited to reposition or remove existing nucleosomes. Currently, it is unclear whether accessibility of PRDM9 to its binding site prior to nucleosome remodeling ultimately influences the recombination rate. Two hotspots assayed here, Pbx1 and Hlx1, are among the most highly active for recombination in male mice on chromosome 1 (2.6 and 3.0 cM, respectively) (Parvanov et al. 2009; Billings et al. 2013). However, in the absence of PRDM9 binding, the two hotspots do differ in chromatin configuration; the Pbx1 binding site is more susceptible to MNase digestion than the Hlx1 binding site, suggesting differing chromatin states and therefore accessibility to trans-acting factors. To fully answer this question, nucleosome positions need to be resolved in the absence of PRDM9 binding at the individual hotspot level and genome-wide hotspot recombination rates measured to correlate the two.
The position of crossing-over is restricted to the area modified by PRDM9 (Fig. 6). This suggests that not only does PRDM9 direct the DSB machinery to its genomic binding locations (Brick et al. 2012), but that it is also necessary for creating an environment permissive to branch migration. This limiting of branch migration to the area of trimethylated nucleosomes was seen when the span of H3K4me3 nucleosomes among hotspots varied from 1000 to 2500 bp (Fig. 6A,C). Since DSBs are introduced in close proximity to PRDM9 binding sites (Fig. 6E), we conclude that Holliday junctions can migrate equally well in either direction but only within the space defined by trimethylated nucleosomes. Whether other factors help limit the locations of Holliday junctions is not known, but it is clear that branch migration does not extend beyond the chromatin environment created by PRDM9-dependent modification.
While our data show a clear relationship between H3K4me3 track length and crossover track length, most meiotic DSBs are repaired as noncrossover (NCO) gene conversions. Previous work has shown that gene conversion tracks are generally short (∼10 to 120 bp), found within hotspots, and are limited to a narrower window than crossovers (Ng et al. 2008; Paigen et al. 2008; Cole et al. 2010b). It has been proposed that the position of noncrossovers may correlate with the position of DSBs (Cole et al. 2010b). If this is true, they should cluster within the NDR at hotspots. Indeed, for the hotspots Hlx1 and A3, where NCOs have been extensively mapped, the majority of noncrossovers are concentrated at the center of hotspots near the NDR, although they can still be found outside of the NDR (Ng et al. 2008; Cole et al. 2010b). It is possible that some DSBs occur outside the NDR, given the DMC1 distribution (Fig. 6E); alternatively, NCOs outside the NDR could result from extended single-strand resection, followed by limited repair synthesis. Co-conversion events, when multiple SNPs within a hotspot are converted together in the same meiotic cell, are rare. Co-conversions at the Hlx1 hotspot were only found within the PRDM9Cst binding site, and 92% of co-conversions found at the A3 hotspot are in a ∼100-bp window within the A3 NDR, which includes the predicted PRDM9Dom2 binding site (Supplemental Fig. S6B; Cole et al. 2010b).
Our findings relate to the question of what determines the likelihood that a methylated hotspot will transition to DSB formation in mammals. In yeast, a species that lacks Prdm9, Pan et al. (2011) found that Spo11 preferentially cuts DNA within the NDRs found near promoters. In addition, two recent studies demonstrated the molecular link between H3K4me3-dependent binding of the yeast protein Spp1 in proximity to promoter NDRs and the meiotic chromosome axis containing DSB machinery, including Spo11 (Acquaviva et al. 2013; Sommermeyer et al. 2013). Similar to yeast, the DSBs found in mutant mice lacking Prdm9 are frequently located at H3K4me3 sites near promoters (Brick et al. 2012). Here we show that PRDM9 binding actively creates an NDR that overlaps with the position of DSBs in mice, thereby potentially providing SPO11 access to DNA.
Taken together, the data presented here demonstrate that PRDM9 has a large influence over the state of hotspot chromatin. This influence is most apparent at the initiation of recombination, when it reorganizes hotspot nucleosomes, and later in its ability to constrain the migration of Holliday junctions and the location of genetic crossing-over.
Methods
Mouse strains and genotyping
C57BL/6J (stock number 000664) and CAST/EiJ (stock number 000928) mice were obtained from The Jackson Laboratory. All animal experiments were approved by the Animal Care and Use Committee of The Jackson Laboratory (Summary #04008).
B6-Prdm9CAST-KI/Kpgn was generated by targeted replacement of exon 12 of B6 Prdm9Dom2. The targeting construct was assembled by recombineering technology (Liu et al. 2003) such that exon 12 coding sequencing of the Prdm9Dom2 from the C57BL/6J strain (1,388 bp), along with the 5′ flanking sequence from intron 11 (55 bp) and the 3′ flanking sequence from the 3′ UTR sequence of exon 12 (106 bp), were replaced by the corresponding Prdm9Cst sequence from CAST/EiJ. In addition, a neomycin-resistance cassette flanked by the FRT sites was inserted 621 bp downstream from the end of exon 12. The targeting construct was electroporated into an embryonic stem cell line derived from B6(Cg)-Tyrc-2J/J (albino B6, JAX stock number 000058), and clones surviving G418 and ganciclovir were screened for the successfully recombined allele that had retained Prdm9Cst exon 12. Targeted clones with normal chromosome count were injected into C57BL/6J, and male chimeras were bred with B6(Cg)-Tyrc-2J/J female mice to transmit the targeted allele. Subsequent mice were backcrossed with B6.Cg-Tg(ACTFLPe)9205Dym/J (JAX stock number 005703) to excise the neomycin-resistance cassette. Mice were bred to homozygosity for the knock-in allele for this study.
For fine-mapping hotspots genetically, we used a library of DNA samples stored at −80°C taken from progeny of F1 B6 × CAST backcrossed to B6 (Paigen et al. 2008; Billings et al. 2010). SNPs between B6 and CAST were selected from publicly available databases for further genotyping crossovers. Recombination was detected as a transition from homozygous to heterozygous genotype using PCR-based KASP genotyping reagents (LGC Genomics) to narrow the interval to hotspot resolution. The final position of crossovers was confirmed by DNA sequencing of the entire hotspot.
Spermatocyte isolation and crosslinking
Crude isolation of germ cells was conducted from male mice at 12 d post-partum (dpp). Testes were isolated from six mice and the tunica albuginea removed. Tubules were placed in 20 mL DMEM (Life Technologies) with 0.5 mg of Liberase TM added (Roche) and incubated with shaking at 32°C for 15 min. Tubules were washed with 20 mL DMEM and resuspended in the same volume with 0.5 mg Liberase and 100 units (U) DNase I (Affymetrix). Tubules were disrupted by pipetting several times and incubated another 15 min at 32°C. After final incubation, tubules were disrupted by gentle pipetting and passed through an 80-µm Nitex nylon mesh filter (Sefar America). The resulting cell solution was pelleted by centrifugation at 1500 rpm. The cell pellet was washed with 10 mL DMEM supplemented with 25 U DNase I by gentle pipetting and spun again. The cell pellet was resuspended in 5 mL DMEM and passed through the nylon filter a second time, and the number of cells was counted by hemocytometer. Typical yield of crude spermatocytes after isolation was 1 million cells per mouse.
For crosslinking, cells were pelleted and resuspended in PBS at 1 million cells per mL. Formaldehyde (Sigma-Aldrich) was added to a final concentration of 1%, and the cells were incubated with rotation at room temperature for 10 min. Crosslinking was stopped by drop-wise addition of glycine to 125 mM, followed by incubation for 5 min at room temperature with rotational mixing. The cells were pelleted, washed once with PBS, and snap-frozen in liquid nitrogen for storage at −80°C until use.
Chromatin immunoprecipitation
ChIP was performed using antibody against H3K4me3 (Millipore, #07-473) as previously described (Billings et al. 2013). To obtain sufficient material for high-throughput sequencing, four ChIP reactions from a single preparation of spermatocyte chromatin were pooled after final DNA elution. ChIP DNA was concentrated using Agencourt AMPure XP beads (Beckman Coulter) following the manufacturer’s protocol. Quantitation of ChIP and input DNA was measured using the Qubit dsDNA HS assay (Life Technologies). Typically, 7–10 ng of ChIP DNA was obtained after concentration and used for the sequencing library preparation. For all samples, an equal amount of MNase-treated input DNA was sequenced as a control. Biological replicates for each genotype were prepared independently from different litters of mice. Libraries were prepared for sequencing using Bioo Scientific’s NEXTflex ChIP-Seq Kit (protocol version V11.11) without size selection. Amplification of the libraries was done with 20 µL ligation product, 16 µL water, 12 µL NEXTflex ChIP PCR master mix, 2 µL NEXTflex ChIP primer mix, and 14–18 cycles of PCR.
High-throughput sequencing and data processing
High-throughput sequencing was performed using the Illumina HiSeq 2000 platform. Raw sequences were aligned to the mouse genome NCBI Build 37 (mm9) using BWA (v.1.2.3) (Li and Durbin 2009) with default settings. The alignments were subsequently filtered to retain only uniquely mapped reads. Peak calling was performed using MACS (v.1.4.2) (Zhang et al. 2008a) with input DNA for control and ChIP samples as treatment, setting the P-value to 0.01, and nodup = “all.” Because nucleosomes are protected from micrococcal nuclease digestion, and nucleosomes appear to be sharply positioned at hotspots, we chose to keep all mapped reads with the same 5′ starting position in the final coverage profiles and when quantifying ChIP-seq signal. All coverage profiles presented in figures are from MACS bedgraph output after tag shifting.
The only difference between the co-isogenic strains created here was the identity of the PRDM9 zinc finger array. Therefore, we were able to identify PRDM9-dependent H3K4me3 positions by categorizing peaks as unique to one strain. Biological replicates for both B6 and B6-Prdm9CAST-KI were highly reproducible (Pearson’s r = 0.956 and 0.946, respectively), so data sets were pooled prior to final peak-calling. In our samples, MACS tended to identify more H3K4me3 peaks with low activity in the Prdm9Dom2 samples over Prdm9Cst samples. By inspecting the bedgraph coverage profiles manually, it was clear that many of the corresponding PRDM9Cst-dependent regions contained visually clear uncalled peaks, thus creating many false positive unique PRDM9Dom2 positions. To overcome this, we implemented a conservative bioinformatics strategy to maximize identification of common peaks between strains by performing two rounds of peak calling. The first round of peak calling was performed using MACS with standard treatment and control samples with a P-value 0.01. After identifying common peaks between the two genotypes, all reads falling within those intervals were subtracted from both treatment and control alignment files and MACS run a second time to identify peaks with the same P-value. Common peaks between Prdm9Dom2 and Prdm9Cst from the second run were identified and pooled with common peaks from the first run. We then used this pooled common set of peaks to filter the original MACS results, removing common peaks found in both rounds, leaving peaks unique to the Prdm9 genotype. This had the effect of enriching the sample for potential PRDM9 signals by eliminating all of the common H3K4me3 sites, even if they were low activity. While this strategy may miss potential quantitative differences in H3K4me3 strength due to PRDM9, such as sites that are coincident with other H3K4me3 sites, this strategy did satisfy our primary concern of identifying where in the genome PRDM9 modifies chromatin with the fewest false positive positions.
Electrophoretic mobility shift assay
EMSA was performed as previously described (Billings et al. 2013). DNA was 5′-labeled for EMSA using biotinylated primers (Eurofins MWG Operon) for PCR of genomic DNA. For the competition assay, synthetic DNA oligos from both strands were annealed to create unlabeled dsDNA and used at 20× concentration of labeled DNA. All oligo sequences are listed in Supplemental Table S1.
Quantitative PCR
Quantitative PCR was carried out using the Eppendorf Realplex cycler and the QuantiFast SYBR Green PCR kit (Qiagen) following the manufacturer’s protocols, except that primers were used at a final concentration of 0.5 µM to help eliminate primer dimers in later cycles. qPCR primers were designed using OligoPerfect (Life Technologies) with 40%–60%GC and 80- to 120-bp product size (primer sequences are listed in Supplemental Table S1). PCR was carried out for 40 cycles followed by melting curve analysis, and all samples were run in triplicate. Cycle number was determined using the automated threshold analysis.
For estimating the total number of PRDM9-dependent hotspots per meiosis, an equal volume of MNase-digested input DNA was analyzed alongside the ChIP DNA. Cycle threshold (Ct) values were calculated by averaging the technical replicates. Percent input was calculated by using the formula: % Input = 2(Ct Input−Ct ChIP)*100.
To determine peak-to-valley ratios, the peak and valley primer pairs were assayed using five 10-fold dilutions of genomic DNA to compare amplification efficiencies. For each hotspot, after qPCR the Ct values for peaks and valleys were subtracted at each dilution and plotted against the template amount. Only peak and valley primer sets that had a delta-Ct slope ∼ 0.1 or less were used for subsequent analysis of the peak-to-valley ratio.
Estimating the number of PRDM9-dependent H3K4me3 sites per meiosis
qPCR of ChIP DNA allows us to estimate the average number of modified chromatids per meiosis. Using the hotspot Pbx1 as an example, we measured that 25% of the total input DNA had H3K4me3 at this hotspot (Supplemental Fig. S1). Given that there are four chromatids per meiotic cell, we would estimate that one chromatid in four is bound by PRDM9 and modified. However, the actual fraction of early prophase I meiotic cells in 12-dpp mouse testis is ∼46% of the total cell population (Bellve et al. 1977). Therefore, we adjust the estimated number of H3K4me3 sites per meiosis by the fraction of cells in leptotene/zygotene. From this we conclude that ∼2.2 chromatids are modified at Pbx1 per average meiosis. To estimate the genome-wide number of sites, we extended the principle of this analysis to all hotspots.
The proportionality established between qPCR measurement of the fraction of modified loci (Supplemental Fig. S1) and ChIP-seq reads was applied to all PRDM9-dependent H3K4me3 positions from B6 mice. These converted values were summed, multiplied by four to correct for the number of chromatids per cell, and divided by 0.46, the estimated fraction of cells in early prophase I. The standard error for the slope of the regression in Supplemental Figure S1 is 0.00039, or ∼8%, equaling ∼400 hotspots. We believe this is a conservative estimate. If our crude isolation of germ cells resulted in 100% efficiency for recovering only early meiotic prophase I cells, then, at most, our estimate here is twice the actual value, still nearly 10 times the number of DSBs seen by immunofluorescence. However, if our estimate of early prophase I meiotic cells is too high, and our actual recovery of germ cells is much lower, a more likely scenario, then we would have underestimated the number of PRDM9-dependent modifications per meiosis.
Motif identification and aggregation plot
We used the MEME Suite (v4.9.0, accessed online) for motif discovery and sequence searching (Bailey et al. 2009). MEME was used to identify allele-specific motifs with default parameters. Reasoning that PRDM9 would have the most access to DNA in the NDR, we narrowed the search space by restricting the DNA sequence to 500 bp surrounding the local minimum of ChIP-seq tag density near the center of H3K4me3 tracks. Only a single significant motif was identified for each Prdm9 allele. Once the motifs were identified using this narrow search space, FIMO was subsequently used to scan the entire 2 kb surrounding each NDR to determine if the motifs (1) were found outside of the NDR, and (2) showed allele-specific enrichment. To calculate fold enrichment, the number of 5′ start positions for each motif was counted for a sliding 20-bp window with steps of 1 bp. The number of occurrences of the active motif was divided by the number of occurrences of the inactive motif at each bp position. MEME was also used to identify the de novo CTCF motif in Supplemental Figure S3. CTCF binding sites in adult mouse testis were obtained from the Mouse ENCODE Project and are publicly available at The University of California, Santa Cruz Genome Browser or at GEO GSM918711 (The Mouse ENCODE Project Consortium et al. 2012).
To determine the nucleosome profile at hotspots, the ChIP-seq tag densities were aggregated using the agg-py script from the Aggregation and Correlation Toolbox (Jee et al. 2011) with nbins = 200 and radius = 1000. The first aggregation plots were anchored at the local minimum of H3K4me3 signal found within the NDR. Once the position of the allele-specific motifs were located using FIMO, aggregation plots were anchored using the 5′ starting position and strand orientation of the motif to center each hotspot. These motif-center aggregation plots are shown in Figure 2. Aggregation of DMC1 ChIP-seq was performed using the PRDM9Dom2 motif positions. Sequencing data for DMC1 ChIP-seq were previously published (Brick et al. 2012) and can be found at NCBI GEO GSE35498.
NDR length calculation
The Aggregation and Correlation Toolbox script generates the aggregate profile for the PRDM9-dependent H3K4me3 sites. This only allows a single calculation of the distance between nucleosome peaks and is biased toward higher active hotspots. To examine the distribution of NDRs at hotspots in relation to H3K4me3 signal strength, nucleosome positions at individual hotspots were identified using NPS (v. 1.3.2) with default settings (Zhang et al. 2008b). The centers of the resulting genomic intervals align with the subpeaks in the coverage profile within hotspots (Fig. 3A). Hotspots with nucleosomes surrounding the identified motifs further than 500 bp apart were discarded, because manual spot inspection showed that these cases were skewed, having only one nucleosome per interval. NDR lengths at each hotspot were calculated by subtracting 150 from the distance between the centers of the −1 and +1 nucleosomes flanking identified motifs. This analysis was extended to the next pair of nucleosomes to find the linker distances to the left and right of the NDR.
Data access
Raw sequencing data, bedgraph files, and unique PRDM9 intervals generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE52628.
Acknowledgments
We thank members of the laboratory, Mary Ann Handel, and Greg Carter for their discussion of the data and manuscript. We thank Tim Billings for his assistance and reagents for the EMSA assays. We also thank the Mouse ENCODE Project Consortium for making the CTCF ChIP-seq data publicly available. This work was assisted by JAX scientific services, which is supported through NIH Cancer Core grant CA34196. Funding for the work was provided by NIH T32 HD007065-32 and NIGMS F32 GM101736 to C.L.B. S.K. was supported by the Japan Society for the Promotion of Science (JSPS) through its Strategic Young Researcher Overseas Visits Program for Accelerating Brain Circulation, NIH P50 GM076468 to Gary Churchill, R01 GM078452 to P.M.P., and P01 GM099640 and R01 GM078643 to K.P.
Author contributions: C.L.B. and S.K. performed all experiments. C.L.B. and M.W. performed computational data analyses. C.L.B., P.M.P., and K.P. contributed to the experimental design. K.P. supervised the study. C.L.B. and K.P. wrote the manuscript. All authors discussed the results and interpretation, read the paper, and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.170167.113.
References
- Acquaviva L, Szekvolgyi L, Dichtl B, Dichtl BS, de La Roche Saint Andre C, Nicolas A, Geli V 2013. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339: 215–218 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellve AR, Cavicchia JC, Millette CF, O’Brien DA, Bhatnagar YM, Dym M 1977. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol 74: 68–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, May CA, Jeffreys AJ 2010. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet 42: 859–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IL, Neumann R, Sarbajna S, Odenthal-Hesse L, Butler NJ, Jeffreys AJ 2011. Variants of the protein PRDM9 differentially regulate a set of human meiotic recombination hotspots highly active in African populations. Proc Natl Acad Sci 108: 12378–12383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings T, Sargent EE, Szatkiewicz JP, Leahy N, Kwak IY, Bektassova N, Walker M, Hassold T, Graber JH, Broman KW, et al. 2010. Patterns of recombination activity on mouse chromosome 11 revealed by high resolution mapping. PLoS ONE 5: e15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings T, Parvanov ED, Baker CL, Walker M, Paigen K, Petkov PM 2013. DNA binding specificities of the long zinc-finger recombination protein PRDM9. Genome Biol 14: R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton A, Myers RS, Redfield RJ 1997. The hotspot conversion paradox and the evolution of meiotic recombination. Proc Natl Acad Sci 94: 8058–8063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485: 642–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J, Barthes P, Grey C, de Massy B 2009. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J 28: 2616–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J, Rivals E, Dunoyer de Segonzac D, Garres C, Caminade P, de Massy B, Boursot P 2014. Diversity of Prdm9 zinc finger array in wild mice unravels new facets of the evolutionary turnover of this coding minisatellite. PLoS ONE 9: e85021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F, Keeney S, Jasin M 2010a. Evolutionary conservation of meiotic DSB proteins: more than just Spo11. Genes Dev 24: 1201–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F, Keeney S, Jasin M 2010b. Comprehensive, fine-scale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Mol Cell 39: 700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F, Kauppi L, Lange J, Roig I, Wang R, Keeney S, Jasin M 2012. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol 14: 424–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getun IV, Wu ZK, Khalil AM, Bois PR 2010. Nucleosome occupancy landscape and dynamics at mouse recombination hotspots. EMBO Rep 11: 555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getun IV, Wu ZK, Bois PR 2012. Organization and roles of nucleosomes at mouse meiotic recombination hotspots. Nucleus 3: 244–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey C, Barthes P, Chauveau-Le Friec G, Langa F, Baudat F, de Massy B 2011. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol 9: e1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel MA, Schimenti JC 2010. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 11: 124–136 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y 2005. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438: 374–378 [DOI] [PubMed] [Google Scholar]
- Jee J, Rozowsky J, Yip KY, Lochovsky L, Bjornson R, Zhong G, Zhang Z, Fu Y, Wang J, Weng Z, et al. 2011. ACT: aggregation and correlation toolbox for analyses of genome tracks. Bioinformatics 27: 1152–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S 2008. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn Stab 2: 81–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelmenson PM, Petkov P, Wang X, Higgins DC, Paigen BJ, Paigen K 2005. A torrid zone on mouse chromosome 1 containing a cluster of recombinational hotspots. Genetics 169: 833–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl KP, Sekelsky J 2013. Meiotic and mitotic recombination in meiosis. Genetics 194: 327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono H, Tamura M, Osada N, Suzuki H, Abe K, Moriwaki K, Ohta K, Shiroishi T 2014. Prdm9 polymorphism unveils mouse evolutionary tracks. DNA Res. doi: 10.1093/dnares/dst059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD 1977. Structure of chromatin. Annu Rev Biochem 46: 931–954 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res 13: 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Mouse ENCODE Project Consortium, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, et al. 2012. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Sundaramoorthy R, Owen-Hughes T 2013. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154: 490–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Keeney S 2006. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SH, Parvanov E, Petkov PM, Paigen K 2008. A quantitative assay for crossover and noncrossover molecular events at individual recombination hotspots in both male and female gametes. Genomics 92: 204–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Petkov P 2010. Mammalian recombination hot spots: properties, control and evolution. Nat Rev Genet 11: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Szatkiewicz JP, Sawyer K, Leahy N, Parvanov ED, Ng SH, Graber JH, Broman KW, Petkov PM 2008. The recombinational anatomy of a mouse chromosome. PLoS Genet 4: e1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, et al. 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov ED, Ng SH, Petkov PM, Paigen K 2009. Trans-regulation of mouse meiotic recombination hotspots by Rcr1. PLoS Biol 7: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenkar R, Shen MH, Arnheim N 1991. DNase I-hypersensitive sites and transcription factor-binding motifs within the mouse E β meiotic recombination hot spot. Mol Cell Biol 11: 1813–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermeyer V, Beneut C, Chaplais E, Serrentino ME, Borde V 2013. Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell 49: 43–54 [DOI] [PubMed] [Google Scholar]
- Thomas JH, Emerson RO, Shendure J 2009. Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS ONE 4: e8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A 2011. Determinants of nucleosome organization in primary human cells. Nature 474: 516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Ohta K 2013. Initiation of meiotic recombination in chromatin structure. J Biochem 154: 107–114 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008a. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shin H, Song JS, Lei Y, Liu XS 2008b. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics 9: 537. [DOI] [PMC free article] [PubMed] [Google Scholar]