Abstract
Thousands of putative enhancers are characterized in the human genome, yet few have been shown to have a functional role in cancer progression. Inhibiting oncokinases, such as EGFR, ALK, ERBB2, and BRAF, is a mainstay of current cancer therapy but is hindered by innate drug resistance mediated by up-regulation of the HGF receptor, MET. The mechanisms mediating such genomic responses to targeted therapy are unknown. Here, we identify lineage-specific enhancers at the MET locus for multiple common tumor types, including a melanoma lineage-specific enhancer 63 kb downstream from the MET TSS. This enhancer displays inducible chromatin looping with the MET promoter to up-regulate MET expression upon BRAF inhibition. Epigenomic analysis demonstrated that the melanocyte-specific transcription factor, MITF, mediates this enhancer function. Targeted genomic deletion (<7 bp) of the MITF motif within the MET enhancer suppressed inducible chromatin looping and innate drug resistance, while maintaining MITF-dependent, inhibitor-induced melanoma cell differentiation. Epigenomic analysis can thus guide functional disruption of regulatory DNA to decouple pro- and anti-oncogenic functions of a dominant transcription factor and block innate resistance to oncokinase therapy.
Personalized cancer therapy targets mutated genes of functional importance including oncogenic kinases such as BRAF in melanoma. The rapid induction of innate drug resistance in response to inhibitor treatment, however, may restrict sustained treatment responses by allowing tumor cells to survive long enough to engage additional stable genetic and epigenetic alterations that bypass inhibitor action. Innate resistance to oncogenic kinase inhibition can be mediated by microenvironment-derived hepatocyte growth factor (HGF) acting on its tumor cell-expressed receptor, MET, to reactivate the MAPK and PI3K pathways (Straussman et al. 2012; Wilson et al. 2012). Multiple cancer cell types induce MET in response to kinase inhibition (Corso and Giordano 2013); however, the mechanisms by which specific cancer cell types sense oncogene withdrawal and engage innate resistance remain to be characterized.
Oncogene expression can be controlled by enhancers that respond to cellular signaling events. MYC is a canonical example of this phenomenon, with tissue-specific enhancers capable of acting over large genomic distances to regulate its expression. GWASs of cancer risk in various tissue types led to the discovery of these enhancers (Grisanzio and Freedman 2010), and elegant mechanistic characterization demonstrated enhancer responsiveness to Wnt signaling, long-range chromatin looping with the MYC transcription start site (TSS), and regulation of MYC expression (Ghoussaini et al. 2008; Pomerantz et al. 2009; Tuupanen et al. 2009; Ahmadiyeh et al. 2010; Wasserman et al. 2010; Wright et al. 2010).
Following the precedent established by MYC gene dysregulation, we hypothesized that MET induction leading to innate drug resistance across multiple tumor types may be regulated by lineage-specific regulatory elements.
Results
To search for regulatory elements that might mediate tumor-selective MET gene induction in response to oncokinase inhibition, we analyzed the genomic landscape of the MET locus using DNase I hypersensitive sites (DHSs) derived from ENCODE data (The ENCODE Project Consortium 2012). DHSs denote regions of chromatin accessibility and are characteristic of gene regulatory DNA (Gross and Garrard 1988; Thurman et al. 2012). Unsupervised hierarchical clustering was performed with DHSs in a 2-Mb window centered on the MET TSS for 100 cell-type data sets (Fig. 1A). DHS clusters demonstrating specificity for multiple specific cell lineages (defined in Supplemental Table 1) emerged from this analysis. The largest DHS cluster was exclusive to the three cell types within the melanocyte lineage present in this data set (Fig. 1A; Supplemental Fig. 1A). These findings indicate that multiple cell lineage-specific gene regulatory sequences may exist at the MET locus.
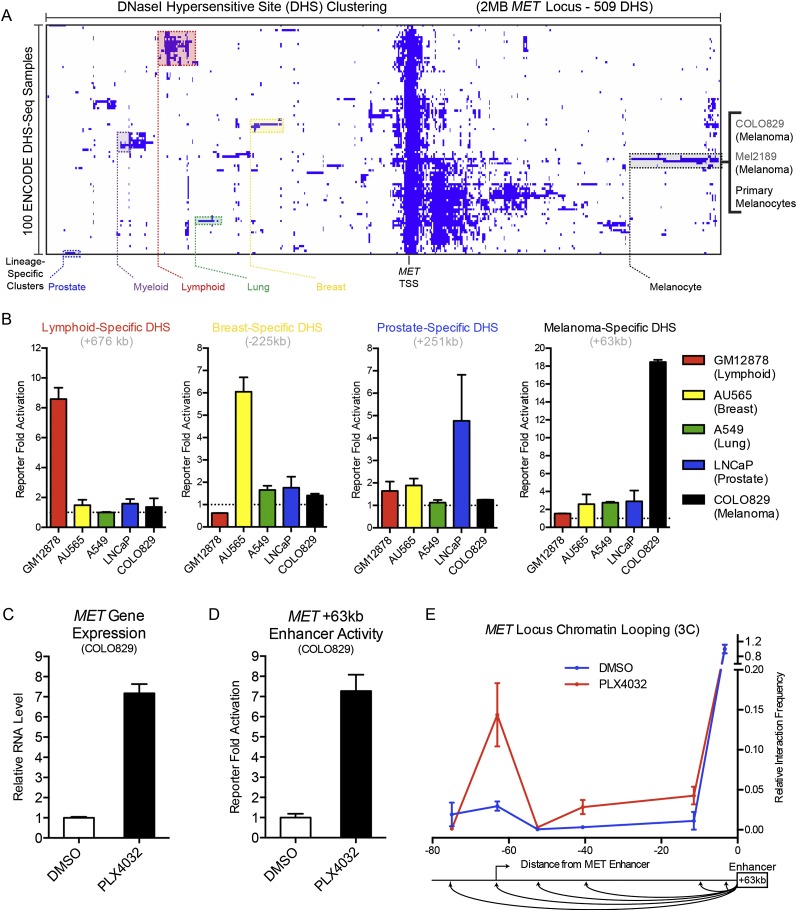
Figure 1.
Discovery of a lineage-specific, kinase inhibitor-responsive MET enhancer. (A) DNase I hypersensitive site (DHS) clustering at the MET gene locus. Lineage-specific clusters are noted in color, with individual cell types within the melanocyte lineage enumerated at right. (B) Enhancer reporter activation levels of selected lineage-specific DHS over an empty vector background in lymphoid, breast, lung, prostate, and melanocyte cell types. (C) MET gene expression in human melanoma cell line COLO829 after a 24-h treatment with DMSO or a selective oncogenic BRAF inhibitor PLX4032. (D) Enhancer reporter activation levels of the melanoma lineage-specific MET +63-kb enhancer in COLO829 cells after a 24-h treatment with DMSO or PLX4032. (E) Chromosome conformation capture (3C) at the MET locus as a function of BRAF inhibition with the +63-kb enhancer as an anchor region. Schematic of the assayed genomic locations below.
To investigate the potential gene regulatory activity of these regions, selected DHSs were cloned into lentiviral enhancer reporter constructs. Five different cell lines from selected lineages were then infected with these constructs. Enhancer activity significantly enriched in a single cell type over all other cell types assayed was observed for lymphoid, breast, prostate, and melanoma lineages (Fig. 1B), experimentally confirming the prediction of lineage-specific enhancers in the MET locus. Interestingly, the melanoma lineage-specific enhancer 63 kb downstream from the MET TSS (MET +63 kb) also demonstrated activation in primary human melanocytes—the cell of origin for melanoma—but not primary fibroblasts or keratinocytes (Supplemental Fig. 1B), indicating that this enhancer demonstrates specificity for the melanocyte lineage in both primary and cancer cells.
Inhibition of oncogenic kinases can elicit MET gene induction, resulting in a process of innate drug resistance that enables tumor cell survival (Corso and Giordano 2013). We hypothesized that lineage-specific enhancers at the MET locus may facilitate MET up-regulation, and thus these enhancers would also be responsive to oncogene withdrawal. To test this, we utilized the well-characterized (Pleasance et al. 2010) melanoma cell line COLO829 that harbors a constitutively active BRAFV600E kinase mutant, and the selective mutant BRAF kinase inhibitor PLX4032 (Bollag et al. 2010). Upon PLX4032 treatment for 24 h, MET gene expression was induced approximately sevenfold (Fig. 1C), and the MET +63-kb enhancer reporter also demonstrated approximately sevenfold activation (Fig. 1D), suggesting a common regulatory mechanism and possible functional link between this enhancer and the MET gene.
Enhancers may act over large genomic distances, “skipping over” multiple proximal genes to regulate targets at a distal location (Sanyal et al. 2012). As this potentially confounds the assignment of an enhancer to its gene regulatory target (Pennacchio et al. 2013), we sought to establish a physical interaction between the MET TSS and the melanocytic lineage-specific enhancer element located 63 kb downstream. Chromosome conformation capture (3C) (Dekker et al. 2002) was performed in COLO829 cells in the presence or absence of PLX4032 using the MET +63-kb enhancer as an anchor region (Fig. 1E; Supplemental Table 2). We observed that BRAF inhibition strikingly increased the interaction between the MET +63-kb enhancer and the MET TSS, demonstrating that this lineage-specific enhancer undergoes inducible chromatin looping in response to oncogene withdrawal in melanoma cells.
To identify potential mediators of lineage-specific enhancer function, we developed a new analysis tool, here termed FOCIS (feature overlapper for chromosomal interval subsets). FOCIS performs an interval-based screen of a genomic feature database—containing ChIP-seq peaks, motif matches, and others (Supplemental Table 3)—for overlap enrichment at a specific subset of genomic regions relative to a data set matched background (Fig. 2A). FOCIS analysis was performed on lineage-specific DHS clusters and a motif for the transcription factor MITF was identified as significantly enriched in melanocyte lineage-specific DHSs relative to all other DHSs at the MET locus (Fig. 2B). While MYC ChIP-seq peaks were the most enriched feature for this analysis, we prioritized MITF due to its role as a master regulator (Steingrímsson et al. 2004; Levy et al. 2006), lineage-specific oncogene (Garraway et al. 2005; Tsao et al. 2012), and its association with MET regulation (McGill et al. 2006; Beuret et al. 2007) and enhancer function (Gorkin et al. 2012). These results raised the possibility that MITF may utilize lineage-specific regulatory elements to control MET expression in melanoma downstream from BRAF inhibition.
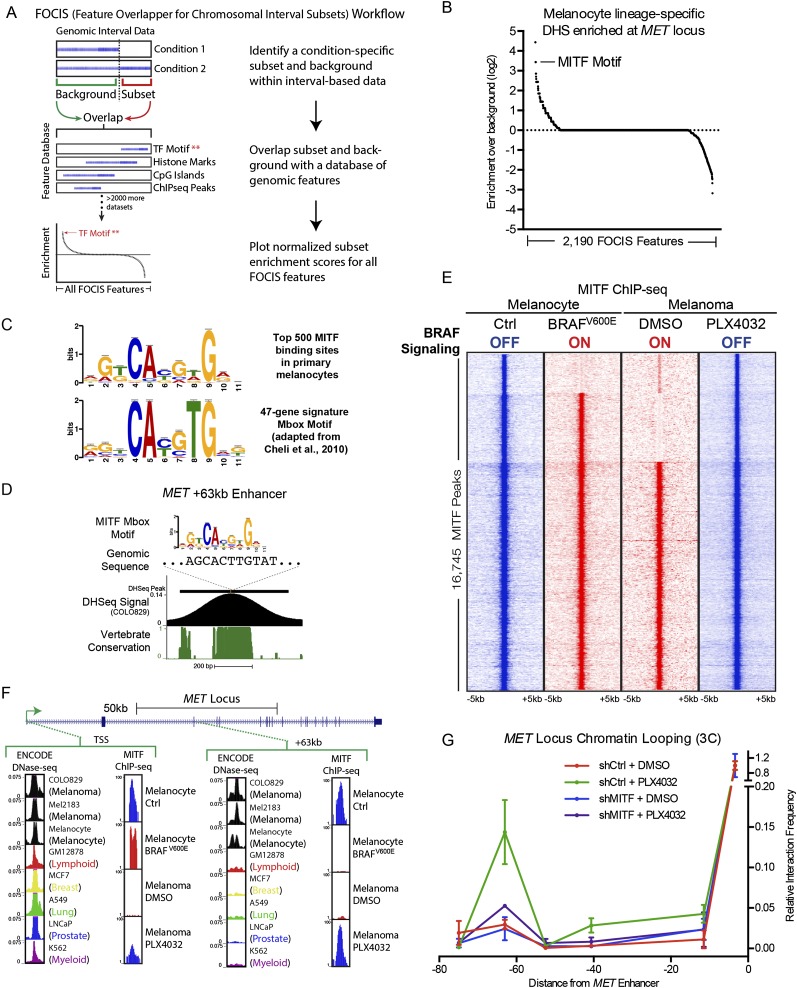
Figure 2.
Integrative epigenomic analysis identifies MITF as a regulator of MET enhancer function. (A) Schematic workflow for the feature overlapper for the chromosomal interval subsets (FOCIS) algorithm. (B) FOCIS feature enrichment for melanocyte lineage-specific DHS at the MET locus. Enrichment for the transcription factor MITF is noted. (C) MITF “Mbox” motif derived from the top 500 MITF-binding sites in ChIP-seq data from primary melanocytes (top, the present study) and from the previously published 47-gene signature (bottom). (D) Characteristics of the lineage-specific enhancer 63 kb downstream from the MET TSS, including the MITF Mbox motif sequence, COLO829 DNase-seq signal, and phastCons vertebrate conservation. (E) Peak-centered heatmap of MITF ChIP-seq signal in primary melanocytes and melanoma cells in the context of inactive (blue) and active (red) BRAF signaling. (F) DNase-seq and MITF ChIP-seq signal at the MET gene TSS and the +63-kb melanocyte lineage-specific enhancer. Normalized ChIP-seq signal is shown on an arbitrary 1–100 scale; ENCODE DNase-seq data shown as F-seq density signal. (G) 3C at the MET locus as a function of BRAF inhibition and MITF depletion in COLO829 melanoma cells. Matched control 3C data from Figure 1E are also shown in this figure for reference.
Overall MITF levels can be regulated by MAPK signaling (Price et al. 1998; Wu et al. 2000), including by oncogenic BRAF (Wellbrock et al. 2008); however, the extent to which these signaling events affect global MITF genomic occupancy is unknown. To investigate the role of MITF in mediating the response to BRAF inhibition, ChIP-seq of endogenous MITF was performed in primary human melanocytes and melanoma cells in the context of either active or inactive oncogenic BRAF signaling. These conditions were achieved in primary cells by enforced expression of BRAFV600E versus marker gene control and in COLO829 by PLX4032-mediated BRAF inhibition versus DMSO vehicle control (Supplemental Fig. 2A,B). As endogenous MITF ChIP-seq had not been previously reported in human melanocytes or melanoma, we performed a global analysis of potential target genes using the GREAT algorithm (McLean et al. 2010) and found enrichment for GO terms such as “melanosome” and “pigmentation” (Supplemental Fig. 2C) known to be associated with MITF function in melanocytes, supporting the validity of these genome-wide results. We next analyzed MITF ChIP-seq peaks for a consensus motif. The observed motif derived from ChIP-seq data is highly similar to a previously described consensus MITF motif, or “Mbox,” derived from analysis of 47 MITF target genes (Fig. 2C; Cheli et al. 2010). Interestingly, this motif was found at the apex of the melanoma lineage-specific MET +63-kb DHS peak in COLO829 cells and demonstrated strong conservation throughout vertebrate evolution (Fig. 2D).
MITF global binding profiles were then compared to evaluate differences as a function of BRAFV600E, revealing dynamic occupancy at a subset of MITF-binding sites (Fig. 2E; Supplemental Table 4). This dynamic occupancy followed a reciprocal pattern in melanocytes and melanoma cells, with introduction of oncogenic BRAF expelling MITF, while BRAF inhibition allowed re-occupancy. By integrating ChIP-seq data with ENCODE DNase-seq data, we observed this pattern at the MET +63-kb lineage-specific enhancer. In contrast, while the MET TSS also demonstrates dynamic MITF localization, this regulatory element is DNase I hypersensitive in 99/100 analyzed cell types (Fig. 2F). These results suggest that the MET +63-kb enhancer may operate as a unique “handle” on MET expression that is lineage specific and drug responsive, and that MITF may regulate these enhancer functions.
To test the necessity of MITF for inducible chromatin looping between the MET +63-kb enhancer and MET TSS in response to BRAF inhibition, 3C was performed in COLO829 cells in the presence or absence of PLX4032 and MITF depletion. We observed that the inducible enhancer–promoter interaction demonstrated a marked decrease with MITF depletion, establishing that MITF is essential for the interaction between the MET enhancer and TSS (Fig. 2G). To explore the global gene regulatory role of MITF in response to oncogenic kinase inhibition, microarray profiling was performed on mRNA from COLO829 cells treated with PLX4032 for 24 h with or without MITF depletion. Loss of MITF disables 20.2% of the transcriptional response to acute BRAF inhibition (Fig. 3A; Supplemental Table 5). Gene Ontology analysis of this MITF-dependent gene set (Fig. 3B) was enriched for the GO terms “melanocyte differentiation,” which includes a melanocyte antigen recognized by T-cells (MLANA [also known as MART1]) among many other melanocyte differentiation genes, and “cell migration,” which includes the innate resistance mediator MET (Fig. 3C; Supplemental Table 5).
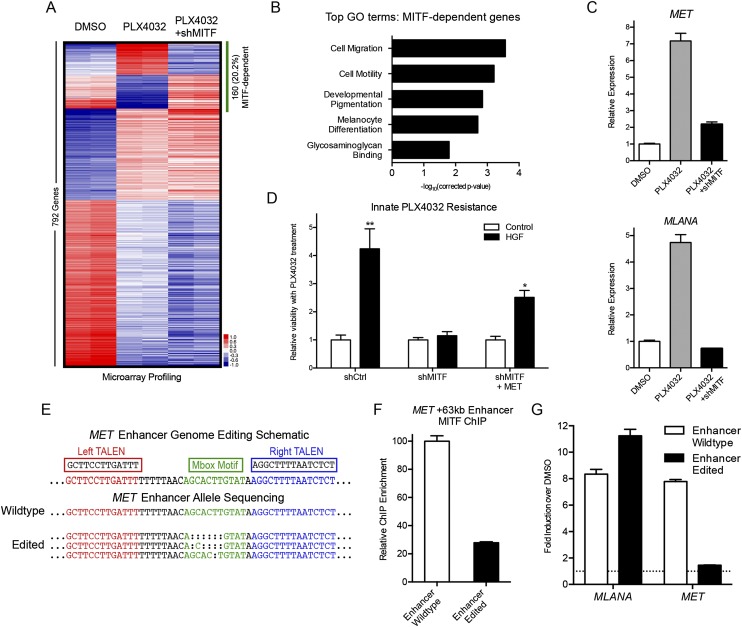
Figure 3.
Enhancer genome editing decouples BRAF inhibitor-driven induction of MET and melanoma differentiation. (A) Mean-centered heatmap of significantly changed genes in COLO829 melanoma cells treated with vehicle control (DMSO), PLX4032 BRAF inhibitor, and PLX4032 plus shRNA to MITF. MITF-dependent gene expression changes are highlighted by the green bar. (B) Top Gene Ontology (GO) terms from MITF-dependent genes in A. (C) Expression of MET (top) and the melanocyte differentiation antigen MLANA (bottom) in COLO829 melanoma cells as a function of PLX4032 and MITF depletion. (D) Viability of COLO829 melanoma cells in response to PLX4032 in the presence or absence of exogenous HGF, MITF depletion, and MET overexpression. (**) P = 0.0043; (*) P = 0.0152. (E) Schematic of the MET +63-kb enhancer TALEN targeting strategy; the Mbox sequence motif is shown in green. The wild-type (WT) normal allele found in COLO829 and the enhancer-edited (EE) clone alleles are shown. Deep sequencing validated that no WT alleles remained in the EE clone. (F) MITF ChIP-qPCR at the MET +63-kb enhancer of WT versus EE cells. (G) Induction of MLANA and MET expression by PLX4032 in WT and EE cells.
To determine whether MITF is necessary for innate resistance to BRAF inhibition, we assayed melanoma cell viability in the presence or absence of exogenous HGF in an approach that has been successfully used to study the effect of paracrine signaling from the stroma to elicit innate drug resistance in tumor cells (Straussman et al. 2012; Wilson et al. 2012). Exogenous HGF significantly enhanced viability of COLO829 melanoma cells after acute PLX4032 exposure, consistent with innate drug resistance. MITF depletion prevented this innate resistance, which was partially reverted by forced expression of MET (Fig. 3D; Supplemental Fig. 3). These results are consistent with other work implicating MITF in melanoma survival (Haq et al. 2013) and establish a role for MITF in mediating innate resistance to BRAF kinase inhibition by enabling HGF-MET signaling. MITF therefore regulates two contrary effects of BRAF inhibition, namely, induction of melanocyte differentiation, which is an anti-tumorigenic effect important for immune cell recognition of melanoma (Kono et al. 2006; Boni et al. 2010; Liu et al. 2013), and the pro-tumorigenic effect of engaging innate drug resistance to BRAF inhibition.
While global MITF loss has pleiotropic effects downstream from BRAF inhibition, we sought to specifically eliminate the undesirable MET induction component of MITF function, while maintaining desired MITF-driven melanoma cell differentiation. We hypothesized that disrupting MITF functionality at the MET +63-kb enhancer may achieve this selective effect. To disrupt MITF binding at the MET enhancer in its native context, targeted genome editing by transcription activator-like effector nucleases (TALENs) (Bogdanove and Voytas 2011; Miller et al. 2011) was performed in COLO829 melanoma cells to remove the Mbox motif. Clonal selection and deep targeted resequencing of the MET enhancer locus confirmed homozygous disruption of the Mbox (Fig. 3E), while targeted sequencing of the four most highly homologous sites showed no mutations indicative of off-target effects of TALEN treatment. Genome editing of the +63-kb enhancer within the third intron of MET did not diminish the capacity for full-length MET protein production (Supplemental Fig. 4). Three specific edited alleles were observed consistent with the known aneuploidy of these melanoma cells. ChIP-qPCR analysis at the MET +63-kb enhancer demonstrated that Mbox deletion significantly reduced MITF occupancy (Fig. 3F), though residual MITF binding was still observed at ∼15-fold greater than the IgG control. Strikingly, MET gene induction upon BRAF inhibition was prevented, whereas induction of differentiation genes including melanocyte antigen MLANA remained intact (Fig. 3G; Supplemental Fig. 4). These results were replicated with a second, independently derived homozygous Mbox-deleted TALEN-treated clone with distinct genetic lesions at the MET enhancer (Supplemental Fig. 5). Thus, genetically disabling the function of a single MET enhancer decoupled pro- and anti-oncogenic gene regulatory impacts of MITF initiated by BRAF inhibition.
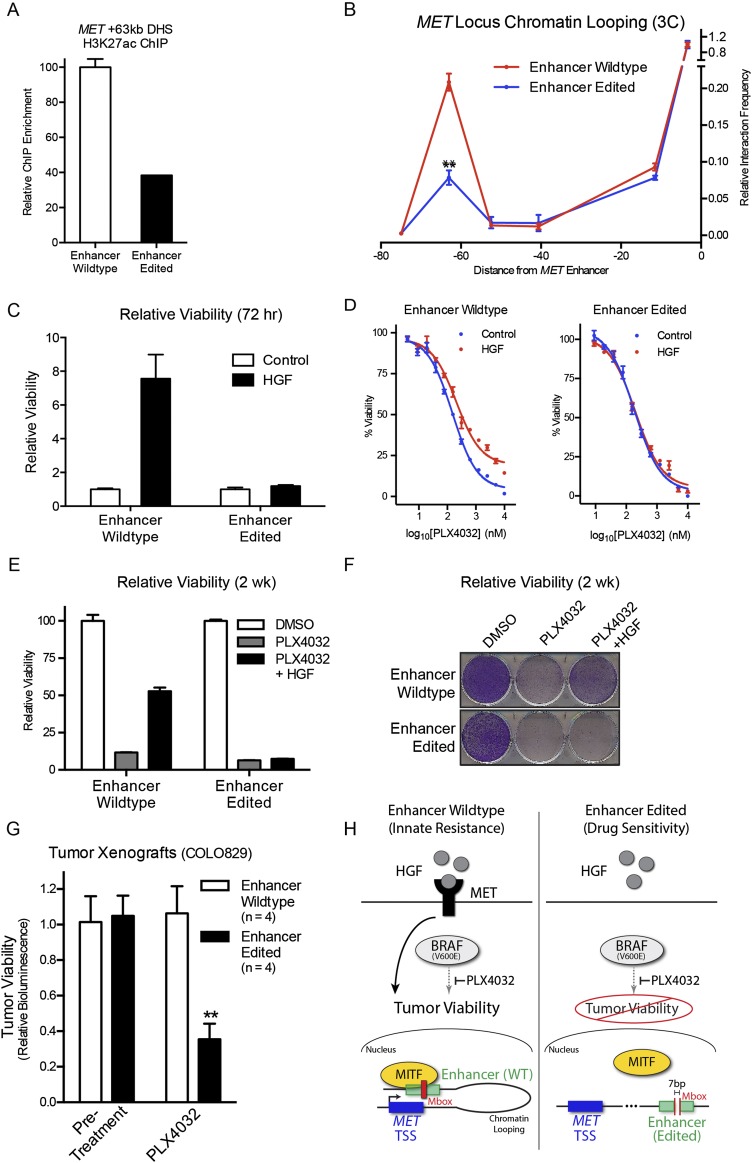
Figure 4.
Targeted MET enhancer disruption suppresses innate resistance to inhibition of the BRAF oncokinase. (A) H3K27ac ChIP-qPCR at the MET +63-kb enhancer of wild-type (WT) versus enhancer-edited (EE) cells. (B) 3C of WT versus EE cells at the MET locus. (**) P = 1.00 × 10−3. (C) Viability quantification at 72 h of D, WT and EE cells in PLX4032 dose-response curve ± exogenous HGF. (E) Viability quantification at 2 wk in vitro of WT and EE cells in PLX4032 ± exogenous HGF. (F) Cresyl violet staining of cell populations in E. (G) Tumor cell viability in vivo assessed by bioluminescent signal in COLO829 tumor xenografts before and after treatment with BRAF inhibitor PLX4032. (**) P = 3.49 × 10−3; n = 4 mice/group. (H) Hypothetical working model for MITF-dependent induction of HGF/MET-mediated innate resistance to BRAF (left), and the disruption of this process by targeted genomic editing of the MET +63-kb enhancer (right).
To explore a mechanistic basis for the observed impacts of enhancer genome editing on MET expression, ChIP and 3C analysis were performed on wild-type or enhancer-edited cells treated with PLX4032. A significant decrease in both H3K27 acetylation (Fig. 4A; Supplemental Fig. 5C), a histone modification associated with active enhancer state (Creyghton et al. 2010), and enhancer–promoter chromatin looping (Fig. 4B; Supplemental Fig. 5D) was observed in enhancer-edited cells. These results indicate that genome editing of the MET +63-kb enhancer prevents PLX4032-induced MET induction by suppressing MITF binding, enhancer activation, and chromatin looping interactions with the MET TSS.
Given its impact on MET regulation, we hypothesized that MET enhancer function would be necessary for innate resistance to BRAF inhibition in melanoma cells. To test this, wild-type and MET enhancer-edited clones lacking an intact Mbox were assayed for viability in response to increasing doses of PLX4032 in vitro. Targeted disruption of the MET enhancer prevented innate resistance to acute BRAF kinase inhibition in the presence of HGF in vitro, in both MET enhancer-edited clones (Fig. 4C,D; Supplemental Fig. 5F) along with a significant decrease in longer-term growth of melanoma cells under continuous drug treatment (Fig. 4E,F; Supplemental Fig. 5F).
To test the effect of MET enhancer disruption in an in vivo setting, wild-type and enhancer-edited COLO829 melanoma cells were used to generate subcutaneous tumors in immune-deficient mice. Within 24 h of systemic PLX4032 treatment, enhancer-edited tumors showed a significant decrease in tumor viability signal compared with wild-type (Fig. 4G), consistent with a failure to engage innate oncokinase resistance in vivo. These results present multiple lines of evidence that genome editing of the MET enhancer functionally suppresses innate drug resistance to oncogenic kinase inhibition. Our findings suggest a hypothetical working model (Fig. 4H) in which oncogenic BRAF inhibition leads to MITF binding to the +63-kb MET enhancer and mediates an inducible chromatin looping interaction with the MET TSS to increase MET expression and engage innate drug resistance pathways in melanoma.
Discussion
Taken together, these data provide a framework for making precise alterations in regulatory elements to influence the genomic response to upstream signaling events. Using genome editing and chromosome conformation guided by integration of multiple epigenomics data sets, we identified a novel, lineage-specific MET enhancer and characterized its functional role in regulating innate tumor drug resistance. This approach facilitated prevention of an adverse effect of oncogenic kinase inhibition by precise deletion of <7 bp of regulatory DNA, and demonstrated the ability to decouple adverse and beneficial functions of a lineage-specific oncogenic transcription factor.
It is notable that disruption of a transcription factor motif within a single enhancer was sufficient to significantly alter gene regulation. It remains to be seen whether this example is more generally applicable or whether regulatory redundancy could mask the effect of single enhancer targeting in other contexts. The recent development of genome editing proteins fused to chromatin remodeling enzymes should allow for combinatorial targeting of multiple regulatory elements without the necessity of genetic deletion and clonal selection.
As full genome sequencing of multiple tumor cell types becomes more common, it is likely that recurrent somatic mutations will facilitate the identification of additional noncoding regulatory elements that control biological processes relevant to tumor progression. The discovery and characterization of “cancer enhancers” by integrative epigenomics analysis as well as the potential for their disruption by targeted agents, represents a promising avenue for further study.
Methods
DHS clustering analysis
ENCODE DNase-seq peaks (DHS) were downloaded from the UCSC Genome Browser (Rosenbloom et al. 2013) (Duke DNase I HS July 2012 freeze) for human genome build hg19. DHS intervals were merged using BEDTools (Quinlan and Hall 2010) (v2.17) and those intersecting a 2-Mb window centered on the MET TSS were clustered and visualized with TreeView (Eisen et al. 1998). DHS exclusive to one or more samples within a defined lineage (Supplemental Table 1) were classified as lineage specific.
Lentiviral reporter constructs and reporter assay
All reporter constructs were derived from the pGreenFire Lenti-Reporter (Puro) system (System Biosciences). COLO829 genomic DNA sequences from the MET +63-kb DHS were cloned into the empty vector using primers listed in Supplemental Table 6. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) following the manufacturer's instructions. Signal was normalized as described previously (Sen et al. 2012).
Chromosome conformation capture (3C) assay
The 3C-qPCR assay was performed as described (Hagège et al. 2007) with the following modifications (Court et al. 2011). The 3 × 107 COLO829 cells that were stably transduced with control or MITF knockdown vectors, or transient TALEN treatment, were cross-linked with 1% formaldehyde at room temperature for 10 min, followed by glycine quenching and cell lysis. Cells were then divided into three 1 × 107 cell aliquots, creating triplicate samples for each condition. Nuclei were resuspended in 500 µL NEB buffer 3, then lysed with 0.2% SDS, followed by SDS sequestration with 1.2% Triton X-100 resuspended in 50 µL 1× Fermentas ligation buffer containing 5 mM ATP. Lysates were digested overnight at 37°C with ApoI (NEB), and the restriction enzyme then inactivated for 1 h at 37°C with 1.6% SDS. Ligation was performed with NEB T4 ligase in 1.15X T4 Ligase buffer at 16°C for 4 h. 3C ligation products were quantitated with eight replicates by SYBR Green qPCR. The 3C signals at the MET locus were normalized to an undigested region to correct for differences in input DNA, then further normalized to an interacting region in close proximity to the anchor region or to a high-frequency GAPDH locus interaction, both producing similar results. Primers were designed using Primer3 Plus online software (Untergasser et al. 2007) and tested against an ApoI-digested, ligated MET locus BAC template by serial dilution and melt-curve analysis to ensure specific and linear amplification. Digestion efficiencies were determined with SYBR Green qPCR by comparison of aliquots taken pre- and post-digestion using primer pairs that amplified a region in each interrogated fragment spanning an ApoI digestion site. Primer sequences are listed in the Supplemental Table 6.
FOCIS analysis
Melanocyte lineage-specific DHS intervals were defined as the subset, with all other DHS at the 2-Mb MET locus defined as the background. FOCIS analysis was performed using the algorithm and feature database described in Supplemental Table 2. A minimum overlap cutoff of 1% was used. The subset enrichment score was plotted in Prism (ABI) for all features. FOCIS source code is freely available on SourceForge at http://sourceforge.net/projects/focis/ and at http://khavarilab.stanford.edu/resources.html.
Transcriptome profiling
RNA isolation was performed with Qiagen RNeasy Plus kit following the manufacturer's instructions. These samples were profiled with Affymetrix GeneChip Human Gene 1.0 ST arrays. Microarray analysis was performed as described previously (Kretz et al. 2012).
Cell culture, drug treatment, and viability assay
COLO829, A549, AU565, LNCaP, and GM12878 cells were obtained (ATCC) and cultured according to the provider's instructions in their recommended media. Primary human melanocytes, fibroblasts, and keratinocytes were obtained through an Institutional Review Board–approved protocol and cultured as described previously (Flockhart et al. 2012). The mutant BRAF inhibitor PLX4032 (Cellagen) was resuspended in DMSO and used at a concentration of 1 μM for all experiments excluding the dose-response viability assay. A range of PLX4032 concentrations in a twofold dilution series beginning at 10 μM was used for dose-response viability measurements. HGF (Peprotech) was resuspended in water and used at a concentration of 50 ng/mL. Quadruplicate viability measurements were taken at 72-h post drug addition in 96-well plates using Cell Titer Blue (Promega) following the manufacturer's instructions. Relative viability was measured by fitting survival curves for all replicates in ABI Prism software and calculating IC20 values. Longer-term viability was measured in the same way in 6-well plates after 2 wk, and these cells were stained for visualization with cresol violet.
Lentiviral overexpression and knockdown constructs
Overexpression of BRAFV600E and a control vector expressing RFP was performed as described previously (Flockhart et al. 2012). The MET CDS was cloned out of pLenti-MetGFP (Addgene) and cloned into the pLEX-MCS (Puro) overexpression vector (Open Biosystems) using primers listed in Supplemental Table 6. Knockdown of MITF was achieved by cloning a custom sequence, TTCGCACAAACTGCTTCCTTTC, targeting the 3′UTR of MITF into the pGIPZ (Puro) lentiviral knockdown system (Open Biosystems). A nontargeting hairpin vector available from Open Biosystems was used as a control. Reagent specificity was confirmed by rescue of the MITF knockdown proliferation phenotype by overexpression of MITF (data not shown). Lentiviral supernatant was concentrated with Lenti-X concentrator (Clontech), resuspended at a 50x concentration in PBS followed by freezing at −80°C. Cells were infected once (overexpression) or twice (knockdown) at 24-h intervals with high-titer lentivirus and selected for 2 d with 1 μg/mL puromycin.
qRT-PCR and Western blotting
RNA was isolated with Qiagen RNeasy kits and reverse transcribed with iScript cDNA synthesis kit (Bio-Rad) following the manufacturer's instructions. qPCR was performed as described previously (Kretz et al. 2013) with the Roche LightCycler using primers listed in Supplemental Table 6. For Western blotting, 20 μg of cell lysates were used for immunoblotting and resolved on a 4%–12% gradient SDS-PAGE and transferred to PVDF membranes. Membranes were incubated in primary and secondary antibodies for 1 h each at room temperature. Primary antibodies used are: c-MET (Cell Signaling #8198) at 1:500 and beta-actin (Sigma A5441) at 1:5000. Donkey anti-rabbit (GE Amersham) or Goat anti-mouse (Santa Cruz Biotechnology) secondary antibodies were used at 1:5000.
Chromatin immunoprecipitation
For ChIP followed by qPCR, 2–5 million cells were used as starting material; for ChIP-seq, 20–50 million cells were used. Cells were trypsinized, washed, and cross-linked as described previously (Sen et al. 2008). Lysis with a modified RIPA buffer (1% NP40/Igepal, 0.5% Sodium Deoxycholate, 0.1% SDS, 100 uM EDTA in PBS), nuclei were isolated and overnight pulldown was performed with an antibody to MITF (Sigma HPA003259), H3K27ac (Abcam ab4729), H3K4me1 (Abcam ab8895), or Rabbit IgG control (Abcam ab37415). Staph A cells were used for pulldown and washes were performed as described previously (Sen et al. 2008). Isolated DNA was subject to qPCR with primers in Supplemental Table 6. High-throughput ChIP-sequencing libraries were prepared with a modified version of NEBNext ChIP-seq library prep kit (NEB) using AMPure beads (Agencourt) for purification. Barcodes were introduced and these libraries were sequenced with 1 × 50-bp reads on the Illumina HiSeq platform.
ChIP-seq data analysis
MITF ChIP-seq data was mapped to the hg19 genome assembly by Bowtie (Langmead et al. 2009). Peaks were called using MACS (Feng et al. 2012) with a P-value cutoff of 1 × 10−10. For comparative analysis, MITF peaks were selected that were present in both primary melanocytes and COLO829 cells, but the called peak did not need to be present in both data sets for a given cell type. For visualization in TreeView, signal was normalized to the total number of mapped reads, and mean was normalized based on a 5-kb sliding window around called peaks similar to the process described in Taslim et al. (2009). Gene Ontology analysis for all MITF peaks was performed using GREAT v1.8.2 (McLean et al. 2010), with basal plus 100 kb maximal extension. GO terms enriched by both binomial and hypergeometric tests with an FDR less than 0.05 were considered significant. Motif analysis was performed with MEME (Bailey and Elkan 1994) for the 47 sequences identified to be MITF-binding sites from previous literature (Cheli et al. 2010) or from the top 500 MITF-binding sites in primary melanocytes, as measured by read counts.
Genome editing with TALENs
TALE nucleases were designed flanking the MITF motif generated from TAL plasmids following a previously described protocol (Cermak et al. 2011) and then moved to a mammalian expression vector provided as a generous gift from Dr. M. Porteus (Stanford University). A total of 3 μg of the left and right TALEN plasmids and a plasmid encoding the T antigen were nucleofected (Lonza) into COLO829 cells following the manufacturer's instructions. The nucleofected cells were transiently selected with NEO for 3 d. These cells received two additional nucleofections and the population was clonally selected from single cells. To verify loss of the wild-type allele, gDNA was isolated from clonal TALEN-treated cell populations and the parental COLO829 cells. The MET +63-kb DHS region was amplified by PCR, then sheared and cloned into a barcoded, high-throughput sequencing library. Libraries were sequenced with 2 × 100-bp reads on the Illumina MiSeq machine. WT alleles in the genome-edited lines were present at <2.5% of the analyzed reads. Alleles observed at <10% allele frequency from the pure cell population were considered artifactual and not included for downstream analysis.
Targeted Sanger sequencing to an average depth of 13-fold coverage was performed on the four sites most homologous to the MET enhancer region targeted by the TALEN design (chr17:54087425-54087463, chr6:49899544-49899563, chr7:79288419-79288439, chr10:89915164-89915183) in wild-type and enhancer-edited cell line genomic DNA.
Xenografts and bioluminescence imaging
Enhancer wild-type and enhancer-edited COLO829 melanoma cells were stably infected with a hygromycin-selectable retroviral vector containing firefly luciferase. Ten million cells were suspended in a 1:1 solution of PBS and Matrigel (BD Biosciences) and injected into the subcutaneous space of eight hairless 6-wk-old severe combined immunodeficient mice (SHO stock, Charles Rivers). Xenografts were allowed to grow untreated for 1 wk, then mice were treated by intraperitoneal injection with 25 mg/kg PLX4032 dissolved in DMSO. Tumor viability was measured by total flux of bioluminescent signal in a fixed region of interest around all tumors with the In Vivo Imaging System-200 (Xenogen) at 3-d post-subcutaneous injection and 24 h after PLX4032 treatment.
Data access
The sequence and array data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE50686.
Acknowledgments
This work was supported by the U.S. Veterans Affairs Office of Research and Development, NIH/NCI grant CA142635 to P.A.K. We thank J. Wysocka, S. Artandi, M. Snyder, J. Ferrell, H. Chang, A. Oro, and G. Crabtree for pre-submission review and G. Sen, M. Kretz, J. Reuter, A. Ungewickell, R. Spitale, P. Giresi, R. Corces-Zimmerman, R. Chen, J. Ronan, C. Webster, and M. Zeman for helpful comments and discussions.
Author contributions: D.E.W. designed and executed the experiments, generated informatics algorithms, analyzed the data, and wrote the manuscript. D.E.W., B.B., R.T.B., K.J.Y., P.H.N., R.J.F., J.K., and A.Z. executed the experiments, analyzed the data, and contributed to the design of experimentation. P.A.K. designed the experiments, analyzed the data, and wrote the manuscript. Correspondence and requests for materials should be addressed to khavari@stanford.edu.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.166231.113.
References
- Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, Jia L, Almendro V, He HH, Brown M, Liu XS, Davis M, et al. 2010. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci 107: 9742–9746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Beuret L, Flori E, Denoyelle C, Bille K, Busca R, Picardo M, Bertolotto C, Ballotti R 2007. Up-regulation of MET expression by α-melanocyte-stimulating hormone and MITF allows hepatocyte growth factor to protect melanocytes and melanoma cells from apoptosis. J Biol Chem 282: 14140–14147 [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF 2011. TAL effectors: customizable proteins for DNA targeting. Science 333: 1843–1846 [DOI] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, et al. 2010. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467: 596–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni A, Cogdill AP, Dang P, Udayakumar D, Njauw CNJ, Sloss CM, Ferrone CR, Flaherty KT, Lawrence DP, Fisher DE, et al. 2010. Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res 70: 5213–5219 [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheli Y, Ohanna M, Ballotti R, Bertolotto C 2010. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res 23: 27–40 [DOI] [PubMed] [Google Scholar]
- Corso S, Giordano S 2013. Cell-autonomous and non-cell-autonomous mechanisms of HGF/MET-driven resistance to targeted therapies: from basic research to a clinical perspective. Cancer Discov 3: 978–992 [DOI] [PubMed] [Google Scholar]
- Court F, Miro J, Braem C, Lelay-Taha M-N, Brisebarre A, Atger F, Gostan T, Weber M, Cathala G, Forné T 2011. Modulated contact frequencies at gene-rich loci support a statistical helix model for mammalian chromatin organization. Genome Biol 12: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N 2002. Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7: 1728–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, Khavari PA 2012. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res 22: 1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. 2005. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436: 117–122 [DOI] [PubMed] [Google Scholar]
- Ghoussaini M, Song H, Koessler T, Olama Al AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall E, et al. 2008. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst 100: 962–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorkin DU, Lee D, Reed X, Fletez-Brant C, Bessling SL, Loftus SK, Beer MA, Pavan WJ, McCallion AS 2012. Integration of ChIP-seq and machine learning reveals enhancers and a predictive regulatory sequence vocabulary in melanocytes. Genome Res 22: 2290–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisanzio C, Freedman ML 2010. Chromosome 8q24-associated cancers and MYC. Genes Cancer 1: 555–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DS, Garrard WT 1988. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem 57: 159–197 [DOI] [PubMed] [Google Scholar]
- Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forné T 2007. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2: 1722–1733 [DOI] [PubMed] [Google Scholar]
- Haq R, Yokoyama S, Hawryluk EB, Jönsson GB, Frederick DT, McHenry K, Porter D, Tran T-N, Love KT, Langer R, et al. 2013. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc Natl Acad Sci 110: 4321–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentsis A, Reed C, Rice KL, Sanda T, Rodig SJ, Tholouli E, Christie A, Valk PJM, Delwel R, Ngo V, et al. 2012. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat Med 18: 1118–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono M, Dunn IS, Durda PJ, Butera D, Rose LB, Haggerty TJ, Benson EM, Kurnick JT 2006. Role of the mitogen-activated protein kinase signaling pathway in the regulation of human melanocytic antigen expression. Mol Cancer Res 4: 779–792 [DOI] [PubMed] [Google Scholar]
- Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GXY, Chow J, Kim GE, et al. 2012. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev 26: 338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. 2013. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493: 231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE 2006. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 12: 406–414 [DOI] [PubMed] [Google Scholar]
- Liu C, Peng W, Xu C, Lou Y, Zhang M, Wargo JA, Chen JQ, Li HS, Watowich SS, Yang Y, et al. 2013. BRAF inhibition increases tumor infiltration by T cells and enhances the antitumor activity of adoptive immunotherapy in mice. Clin Cancer Res 19: 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill G, Haq R, Nishimura E, Fisher D 2006. c-Met expression is regulated by Mitf in the melanocyte lineage. J Biol Chem 281: 10365–10373 [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. 2011. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148 [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G 2013. Enhancers: five essential questions. Nat Rev Genet 14: 288–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin M-L, Ordóñez GR, Bignell GR, et al. 2010. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. 2009. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet 41: 882–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ER, Ding HF, Badalian T, Bhattacharya S, Takemoto C, Yao TP, Hemesath TJ, Fisher DE 1998. Lineage-specific signaling in melanocytes. C-kit stimulation recruits p300/CBP to microphthalmia. J Biol Chem 273: 17983–17986 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, et al. 2013. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res 41: D56–D63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J 2012. The long-range interaction landscape of gene promoters. Nature 489: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA 2008. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev 22: 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, Johnston D, Siprashvili Z, Khavari PA 2012. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev Cell 22: 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrímsson E, Copeland NG, Jenkins NA 2004. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet 38: 365–411 [DOI] [PubMed] [Google Scholar]
- Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, et al. 2012. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487: 500–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taslim C, Wu J, Yan P, Singer G, Parvin J, Huang T, Lin S, Huang K. 2009. ChIP-Seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J 28: 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao H, Chin L, Garraway LA, Fisher DE 2012. Melanoma: from mutations to medicine. Genes Dev 26: 1131–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Björklund M, Wei G, Yan J, Niittymäki I, et al. 2009. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet 41: 885–890 [DOI] [PubMed] [Google Scholar]
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35: W71–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman NF, Aneas I, Nobrega MA 2010. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res 20: 1191–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellbrock C, Rana S, Paterson H, Pickersgill H, Brummelkamp T, Marais R 2008. Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PLoS ONE 3: e2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J, et al. 2012. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 487: 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JB, Brown SJ, Cole MD 2010. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol Cell Biol 30: 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Hemesath TJ, Takemoto CM, Horstmann MA, Wells AG, Price ER, Fisher DZ, Fisher DE 2000. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev 14: 301–312 [PMC free article] [PubMed] [Google Scholar]