Abstract
Polycomb-mediated chromatin repression modulates gene expression during development in metazoans. Binding of multiple sequence-specific factors at discrete Polycomb response elements (PREs) is thought to recruit repressive complexes that spread across an extended chromatin domain. To dissect the structure of PREs, we applied high-resolution mapping of nonhistone chromatin proteins in native chromatin of Drosophila cells. Analysis of occupied sites reveal interactions between transcription factors that stabilize Polycomb anchoring to DNA, and implicate the general transcription factor ADF1 as a novel PRE component. By comparing two Drosophila cell lines with differential chromatin states, we provide evidence that repression is accomplished by enhanced Polycomb recruitment both to PREs and to target promoters of repressed genes. These results suggest that the stability of multifactor complexes at promoters and regulatory elements is a crucial aspect of developmentally regulated gene expression.
The expression of many genes during development is controlled by the Polycomb system of chromatin-mediated repression (Orlando 2003; Schuettengruber and Cavalli 2009). This system is thought to respond to changes in gene expression programs and epigenetically maintain patterns as cells differentiate. Polycomb-regulated domains typically encompass multiple genes and are characterized by methylation of histone H3 at K27 (H3K27me3). The chromodomain of Polycomb binds this modification and may lead to the compaction of chromatin (Min et al. 2003; Francis et al. 2004; Simon and Kingston 2013). Genetic analysis in Drosophila and mammals has shown that Polycomb response elements (PREs) within domains are essential to establish and maintain repression (Busturia et al. 1997; Sengupta et al. 2004). PREs are short segments of DNA with a high density of binding sites for transcription factors, and an appealing model for control of Polycomb-regulated domains asserts that, in some cell types, multiple DNA-sequence-specific factors bind at a PRE and recruit the E(z) histone methyltransferase. This leads to H3K27 trimethylation and repression across the domain (Breiling et al. 2004; Muller and Kassis 2006). Mounting evidence suggests that PREs may also loop to interact with target promoters within repressed domains (Comet et al. 2011). However, it remains mysterious that both repressing (Polycomb-group [PcG]) and activating trithorax-group (trxG) factors are recruited to PREs in all cell types (Strutt et al. 1997; Grimaud et al. 2006; Papp and Muller 2006; Beisel et al. 2007; Schwartz et al. 2010). It is unclear how developmental signals are integrated to switch PREs between repressive and derepressed states.
Here, we apply high-resolution methods using native chromatin to characterize the factors bound at PREs in Drosophila cells. Contrary to how Polycomb is recruited across domains, we identify a specific transcription factor—ADF1—that coincides with Polycomb at many PREs and at Polycomb-marked promoters throughout the genome. By comparing differential Polycomb-repressed domains in two Drosophila cell lines, we assign a role for factor complexes in stabilizing interactions with Polycomb specifically at repressive PREs.
Results
Polycomb is recruited to PREs by DNA-binding factors
The chromodomain of Polycomb binds to the histone modification H3K27me3, but there have been suspicions that Polycomb interacts with nonhistone proteins specifically at PREs (Muller and Kassis 2006; Schwartz et al. 2006). To address this, we applied a high-resolution technique to map Polycomb in Drosophila S2R+ cells. Digestion of native chromatin with micrococcal nuclease (MNase) produces large fragments (120–160 bp) derived from nucleosomes, as well as shorter (20–75 bp) segments that correspond to DNA protected by bound transcription factors (Henikoff et al. 2011; Kent et al. 2011). Reasoning that factor–DNA particles may remain associated after MNase digestion, we implemented a simple native chromatin immunoprecipitation (N-ChIP) protocol (Kasinathan et al. 2014). In this method, MNase-digested chromatin is solubilized at low-salt concentrations to enhance electrostatic stability of protein–DNA complexes without covalent cross-linking (Lohman and Mascotti 1992). We then use affinity purification and paired-end sequencing without size selection to recover specific protein–DNA complexes. The method is stringent, in that immunoprecipitation of native chromatin particles requires tight binding of DNA. We expected that N-ChIP with an antibody to the Polycomb protein would identify its binding sites in the genome, and the size of the recovered DNA molecules would define whether Polycomb binds nucleosomes or smaller nonhistone factors at those sites. In all N-ChIP experiments here we performed two biological replicates, mapping ∼20–100 million reads in each replicate.
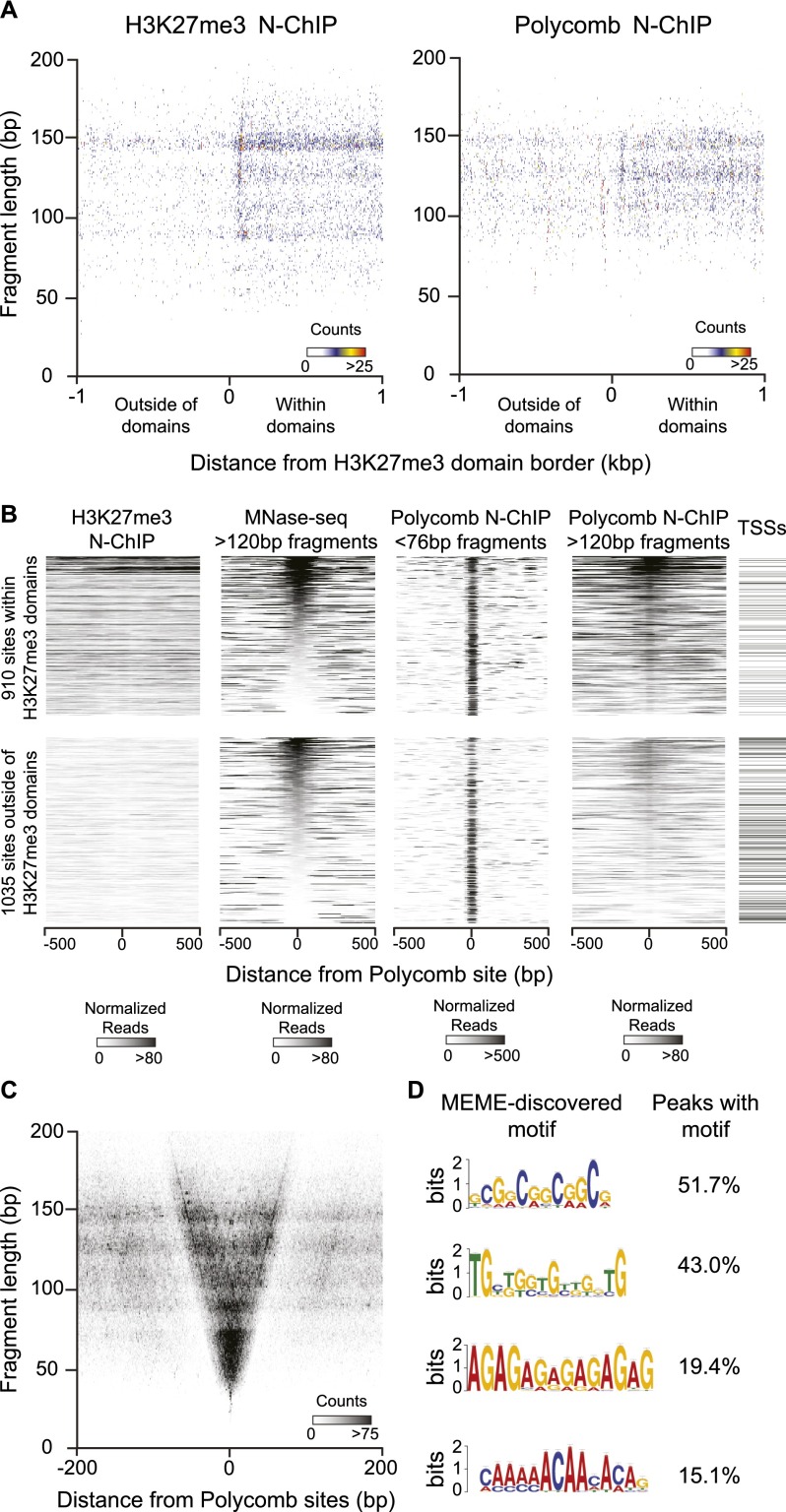
We first determined the localization of H3K27-trimethylated nucleosomes in S2R+ cells to define Polycomb-regulated domains. We selected domains as regions >5 kb with high contiguous H3K27me3 signals, and aligned these by their edges to compare chromatin features within and around domains (Li and Zhou 2013; see Methods). We displayed fragments recovered in N-ChIP experiments in a midpoint plot, where the central base pair of each fragment is positioned relative to domain edge on the x-axis, and by its size on the y-axis (Fig. 1A). Native-ChIP recovers a low level of background from nucleosomes and nucleosome-derived fragments that are abundant in the input material (Henikoff et al. 2011; Teves and Henikoff 2011), but nucleosomes within domains are enriched approximately twofold by N-ChIP for H3K27me3 (Fig. 1A). We then compared the enrichment of fragments by N-ChIP with a Polycomb antibody. Polycomb shows a more subtle enrichment (∼1.3×) of nucleosome-sized fragments within H3K27me3 domains (Fig. 1A). This is consistent with binding of the chromodomain protein to modified nucleosomes, but relatively weak enrichment suggests that the interaction may only partially survive under the conditions of native ChIP.
Figure 1.
Polycomb N-ChIP recovers nucleosomes and factor binding sites. N-ChIP was performed on MNase-digested chromatin from S2R+ cells with antisera to the H3K27me3 histone modification or to the Polycomb protein. (A) Polycomb binds nucleosomes within H3K27me3 domains; 555 domains were defined by contiguous H3K27me3 enrichment (see Methods), and the midpoints of each fragment recovered in N-ChIP experiments are plotted around aligned borders of those domains. Coincident midpoints are pseudo-colored. Plotting fragments from H3K27me3 (left) and from Polycomb (right) N-ChIP show the relative enrichment of nucleosome-sized fragments within domains in each experiment. (B) Chromatin features of focal Polycomb binding sites. Heat maps of 1945 aligned focal Polycomb binding sites, displaying read counts normalized within nucleosome (>120 bp) or factor (<76 bp) fragment size classes; 910 sites fall within H3K27me3 domains, and 1035 sites fall outside of domains. Each group is sorted by nucleosome occupancy at the binding site based on MNase-seq fragments >120 bp in length. Heat maps from left to right display the distributions of H3K27me3-marked nucleosomes (H3K27me3 N-ChIP fragments >120 bp), of nucleosomes (MNase-seq fragments >120 bp), of Polycomb-bound small fragments (Polycomb N-ChIP fragments <76 bp), and of Polycomb-bound nucleosomes (Polycomb N-ChIP fragments >120 bp). Intervals that include a gene TSS are annotated on the right. (C) Polycomb binds small DNA fragments at focal sites; 1945 focal Polycomb binding sites were called from Polycomb N-ChIP data, and midpoints of each fragment overlapping these sites are plotted. Polycomb N-ChIP strongly enriches DNA fragments ranging from 30 to 75 bp in length, and more moderately fragments up to 150 bp in length. (D) MEME discovery of motifs enriched at focal Polycomb binding sites. The four most frequent motifs are shown, with the percentages of focal sites with a significant (P < 5 × 10−4) match to each motif on the right; 77.7% of focal Polycomb binding sites contain at least one of these four motifs.
N-ChIP with Polycomb antibody also recovers short DNA fragments, implying that the protein is recruited to chromatin through non-nucleosomal interactions. We mapped 2380 sites in the Drosophila genome where Polycomb is associated with fragments <76 bp in length (Supplemental Table 1). Of these focal sites, 912 fall within H3K27me3 domains (Fig. 1B). The majority of Polycomb binding sites fall outside of H3K27me3 domains, including 241 gene promoters. This is consistent with previous mapping of Polycomb to sites both within and outside of H3K27me3 domains and to numerous promoters in the Drosophila genome (Schwartz et al. 2006; Tolhuis et al. 2006; Enderle et al. 2011).
To characterize the chromatin context of focal Polycomb binding sites, we used MNase digestion coupled with paired-end deep sequencing (MNase-seq). MNase-seq is a general method for chromatin profiling, in that no factor of interest is specified in advance. Instead, all occupied sites are profiled based simply on protection of DNA (Henikoff et al. 2011; Teves and Henikoff 2011). While Polycomb binds small fragments at all focal sites, MNase-seq landscapes reveal that sites can be nucleosome-enriched or nucleosome-depleted (Fig. 1B). While non-nucleosomal factors may stably occupy nucleosome-depleted sites and recruit Polycomb, factors may only transiently bind at nucleosome-occupied sites. We noted that Polycomb-bound nucleosomal fragments are enriched around nucleosome-occupied focal sites (Fig. 1B), suggesting that nucleosomal binding of Polycomb stabilizes interactions at some sites. H3K27 methylation appears to enhance binding, as Polycomb-bound nucleosomal fragments are more abundant within H3K27me3 domains (Supplemental Fig. 1).
We next displayed all Polycomb-bound fragments around focal Polycomb binding sites on midpoint plots (Henikoff et al. 2011). Small fragments ranging from 35 to 75 bp are most abundant after Polycomb N-ChIP, although fragments as large as 150 bp are also enriched (Fig. 1C). Larger fragments are primarily derived from focal sites within H3K27me3 domains (Supplemental Fig. 2), consistent with enhanced nucleosomal binding of Polycomb around these sites. To infer how Polycomb is bound to non-nucleosomal fragments, we searched the immunoprecipitated short DNA for shared sequence motifs. Four common motifs are found at Polycomb sites (Fig. 1D). The most frequent motif is found at 51.7% of sites and is similar to the consensus motif for ADF1, a general MADF-domain transcription factor that was first identified as a regulator of the Adh gene (England et al. 1992; DeZazzo et al. 2000). The other motifs include one similar to that for the TRL transcription factor, which has been previously implicated in PRE function (Hagstrom et al. 1997; Strutt et al. 1997), and motifs resembling those for the KLU and AEF1 transcription factors. These predictions suggest that these four factors may contribute to Polycomb targeting, although we note that the predictions are limited by the quality and completeness of motif libraries.
PREs are clusters of highly occupied binding sites
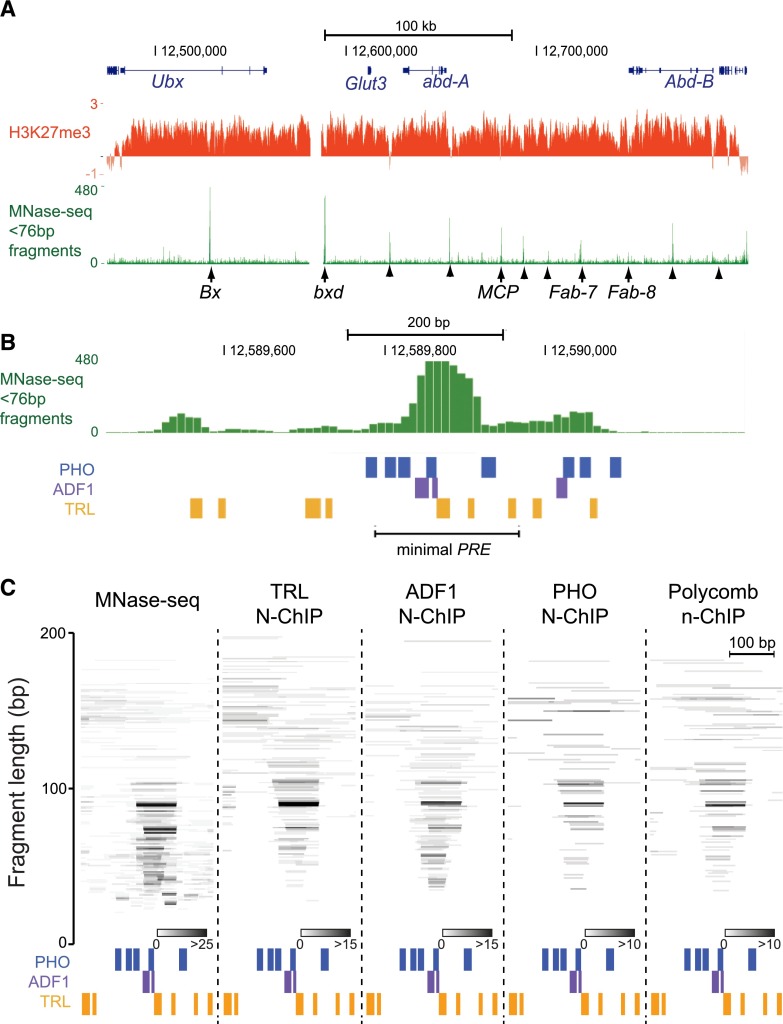
To construct a more comprehensive picture of the transcription factors bound at PREs, we first examined MNase-seq landscapes spanning the well-studied Bithorax-complex (BX-C) region, a 330-kb Polycomb-regulated domain that is repressed in S2 cells. We displayed landscapes as normalized read counts, where each small fragment is represented as the fraction of total mapped fragments ranging from 0 to 75 bp in length. Landscapes reveal that a cluster of highly occupied sites corresponds to each of the Bx, bxd, MCP, Fab-7, and Fab-8 PREs in the BX-C (Fig. 2A; Supplemental Fig. 3). Additional clusters of occupied sites are observed in the domain; many of these coincide with inferred PREs and boundaries between regulatory regions (Schwartz et al. 2006; Holohan et al. 2007), suggesting the clusters mark additional regulatory elements within the BX-C.
Figure 2.
Regulatory elements within the BX-C are clusters of factor-occupied sites. (A) UCSC Genome Browser snapshot of the 330-kb H3K27me3 domain containing the BX-C on chromosome 3R in S2 cells. The distributions of H3K27me3 (modENCODE data) and non-nucleosomal MNase-seq fragments (<76-bp fragments) are shown. Genetically defined PREs (arrows) and regulatory elements inferred from X-ChIP mapping (Schwartz et al. 2006) (arrowheads) are indicated. (B) Browser snapshot of non-nucleosomal MNase-seq fragments at the bxd PRE (chromosome 3R:12,589,450–12,590,250). The positions of high-scoring (P < 5 × 10−4) TRL, PHO, and ADF1 motifs. The segment containing the minimal functional PRE was previously defined (Sipos et al. 2007). (C) Individual fragments in MNase-seq or recovered in the indicated N-ChIP experiments that map to the protected structure (chromosome 3R:12,589,700–12,590,000) at the bxd PRE. Positions of high-scoring (P < 5 × 10−4) TRL, PHO, and ADF1 motifs are indicated. Fragments are positioned on the y-axis by their lengths, and coincident fragments are indicated in shades of gray.
MNase-seq features for each of the known PREs within the BX-C are diverse. Each PRE displays clusters of highly occupied sites with quantitatively less protection nearby, suggesting that there are highly occupied sites flanked by sites with more transient factor binding. The MCP PRE is the smallest region, where four distinct high-occupancy sites span ∼400 bp (Supplemental Fig. 3C). In contrast, the Fab-7 and Fab-8 elements are larger, with protected sites spread over 1000 bp (Supplemental Fig. 3D,E). These two elements have been genetically subdivided into PREs and boundary insulators (Hagstrom et al. 1997; Mihaly et al. 1997; Ciavatta et al. 2007), and indeed Fab-8 appears to contain two clusters of highly occupied sites. These clusters imply that multiple factors are closely juxtaposed and may interact in complexes at PREs.
The high resolution of MNase-seq enables motif analysis to predict what factors occupy individual sites. We could generate high-confidence predictions for ∼1/2 of the highly occupied peaks in BX-C PREs. These include motifs for the TRL and PHO factors, both of which have been previously localized to PREs (Supplemental Fig. 3). We also frequently observed occupancy over ADF1 motifs at PREs within the BX-C, and at other Polycomb binding sites throughout the genome. Enrichment of a similar but unassigned motif has been previously noted (Ringrose et al. 2003; Schuettengruber et al. 2009). A variety of transcription factors including ZESTE, DSP1, GRH, and SP1 have been previously linked to PRE function (Déjardin and Cavalli 2004; Brown et al. 2005; Déjardin et al. 2005; Blastyák et al. 2006). While occupancy over motifs for these factors is occasionally seen, it is less common genome-wide. Together with the recovery of factor motifs by Polycomb N-ChIP, these results imply that Polycomb may be anchored to PREs through interactions with a specific set of transcription factors, including ADF1.
We noted that highly occupied sites within PREs often have closely juxtaposed motifs for multiple factors. Individual DNA fragments detected by MNase-seq data suggest that these sites are simultaneously occupied. We focused on the bxd PRE within the BX-C as an example. The functional core of the bxd PRE has been genetically defined (Sipos et al. 2007; Kozma et al. 2008) and coincides with a highly protected 70-bp segment in MNase-seq data (Fig. 2B). The segment contains two motifs for ADF1, three motifs for TRL, and one motif for PHO. Fragments covering individual motifs and combinations of adjacent motifs are produced after MNase digestion (Fig. 2C). The larger fragments suggest that a complex of simultaneously bound factors protects the segment. Smaller fragments may result from cleavage within the complex, or alternatively may be due to transient factor binding at individual sites.
The ADF1, TRL, and PHO factors localize to Polycomb binding sites
To determine if the factors predicted from motif analysis are present at PREs, we performed high-resolution N-ChIP mapping. We used antibodies specific to the TRL, ADF1, and PHO factors, and identified ∼1300–2300 binding sites (from fragments 20–75 bp in length) for each factor in the genome (Supplemental Table 2). Consensus motifs derived for each factor from these sets of binding sites resemble previously defined motifs, but as tandem repeats of a simpler consensus (Supplemental Fig. 4A). For TRL, this may provide higher affinity sites for binding (van Steensel et al. 2003). For each factor we counted the number of binding sites within and outside of Polycomb-regulated domains (Supplemental Table 1; Supplemental Fig. 4B). ADF1 binding sites are enriched approximately threefold at promoters (±500 bp of annotated transcriptional start sites [TSSs]) within domains, although it also marks many promoters throughout the genome. The other ADF1 binding sites are more distant from promoters and found both within and outside of Polycomb-regulated domains. Similar enrichments are seen for TRL and PHO binding, consistent with general functions for all three factors in transcriptional regulation.
The bxd PRE contains a highly protected structure in MNase-seq data, and this structure coincides with bound Polycomb, ADF1, TRL, and PHO from N-ChIP experiments. However, there are differences in the sizes of DNA fragments associated with each factor (Fig. 2C). Displaying individual fragments in the bxd interval reveals that DNA fragments ranging from 30 to 100 bp are associated with ADF1, mirroring the pattern of fragments in MNase-seq data. In contrast, TRL and PHO are associated predominantly with overlapping fragments 60–100 bp in length, suggesting that multiple factors bind together on these longer fragments. Although small fragments bound only with ADF1 are detected, this does not necessarily mean that ADF1 is sometimes present alone at the PRE, as smaller fragments may also result from MNase cleavage within a multi-subunit complex. We also note that small fragments detected on the right side of the occupied structure in MNase-seq data are not accounted for in any N-ChIP experiment, implying that an additional factor may bind this site. Finally, as N-ChIP for Polycomb predominantly recovers the larger DNA fragments from the bxd PRE, it appears that Polycomb only stably binds the multifactor complex at the PRE.
ADF1 interacts with Polycomb
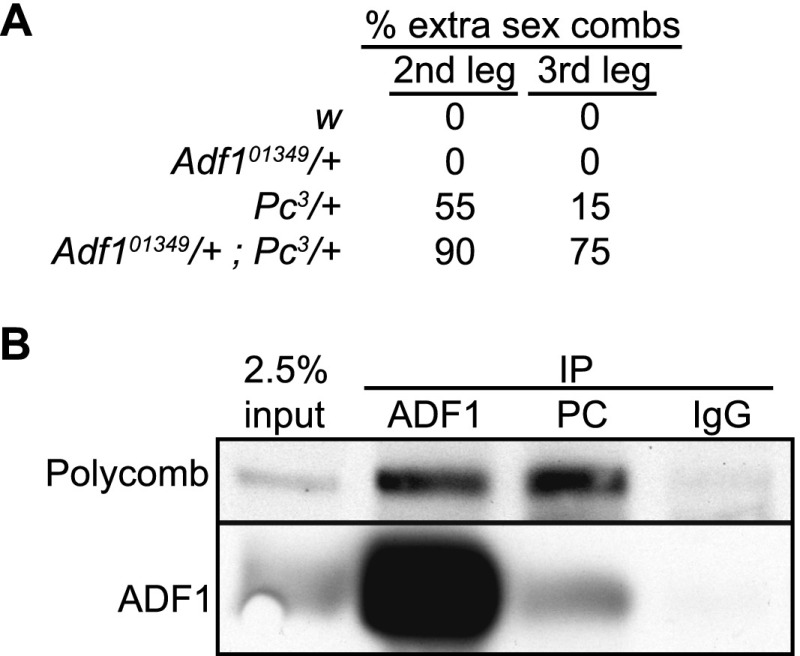
To test if ADF1 is important for Polycomb-mediated repression in vivo, we used the Adf101349 hypomorphic allele (Parrish et al. 2006) to test for genetic interactions with Polycomb (Pc) mutations in flies. Adult males heterozygous for a Pc3 allele show infrequent sex combs on second and third legs, reflecting transformation into first legs and sporadic derepression of homeotic genes (Duncan 1982). While animals with the Adf101349 allele alone show no homeotic transformation, males trans-heterozygous for Pc3 and Adf101349 have significantly increased transformation of both the second and third legs to anterior identities (Fig. 3A). Similar synergistic interactions with Polycomb alleles have been previously described for the Trl gene (Strutt et al. 1997) and pho (Kwon et al. 2003).
Figure 3.
ADF1 interacts with Polycomb. (A) Adult males were scored for the presence of extra sex combs on second and third leg pairs, an indicator of homeotic transformation to first leg identity. The percentages of legs with extra sex combs in each genotype are shown. (B) Coimmunoprecipitation of Polycomb and ADF1 from S2R+ cells. Proteins were immunoprecipitated with anti-Polycomb, anti-ADF1, or nonspecific IgG antisera, separated on SDS gels, and transferred to nitrocellulose for immunoblotting with the antisera indicated to the left; 2.5% of input was compared with material recovered after immunoprecipitation.
Since ADF1 frequently coincides with Polycomb binding, we tested if Polycomb and ADF1 physically interact. Indeed, we found that an immunoprecipitate of Polycomb from MNase-digested chromatin is enriched for ADF1 (Fig. 3B). Enrichment of Polycomb in an immunoprecipitate with ADF1 antibody is also observed. This physical interaction suggests that ADF1 may directly recruit Polycomb to its DNA binding sites. TRL and Polycomb also physically interact (Mishra et al. 2003), as do PHO complexes and Polycomb (Mohd-Sarip et al. 2002). We detected a set of Polycomb binding sites where only one of these factors is also detected, suggesting that all three transcription factors may independently recruit Polycomb to focal binding sites. Additional transcription factors may also recruit Polycomb, as 61% of focal sites do not bind ADF1, TRL, or PHO in N-ChIP experiments (Supplemental Table 2). However, we note that while 1002 Polycomb sites contain ADF1 motifs, N-ChIP for ADF1 recovers only 42% of these sites. It is possible that incomplete recall by N-ChIP underestimates the number of factor binding sites, or that motif analysis overpredicts factor binding.
Transcription factors cooperatively bind PREs
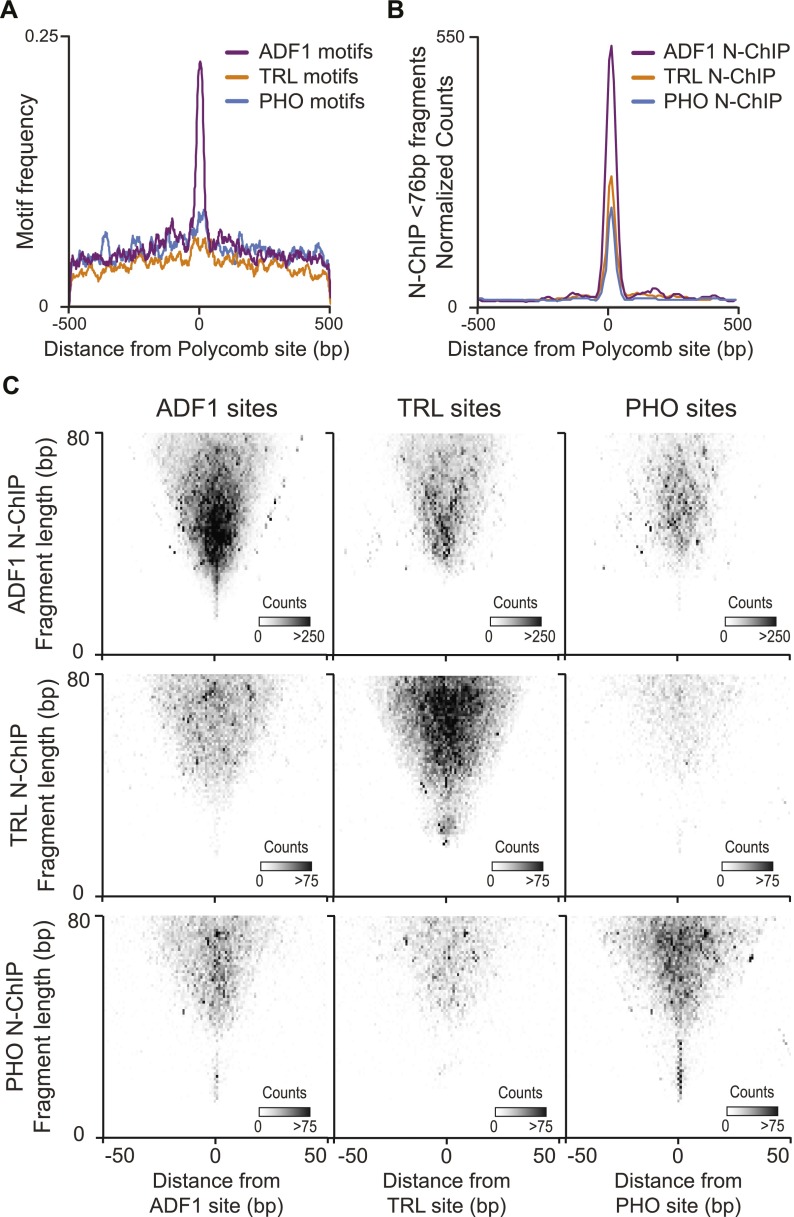
The Polycomb binding sites we identified by N-ChIP often contain an ADF1 motif, but TRL and PHO motifs are only slightly enriched (Fig. 4A). In contrast to expectations from motif distributions, N-ChIP signals for TRL and PHO show dramatic enrichment at Polycomb binding sites (Fig. 4B). This is also apparent in individual sites. For example, the bxd PRE contains a number of PHO and TRL binding motifs that are not protected from MNase digestion (Fig. 2C). These must be sites that are not stably engaged with their cognate factors. Notably, the TRL and PHO sites that are occupied are adjacent to the bound ADF1 motifs. This implies that ADF1 is a key determinant for occupancy of TRL and PHO motifs.
Figure 4.
Coincidence of recovered fragments from N-ChIP. (A) Distribution of high-scoring (P < 5 × 10−4) ADF1, TRL, and PHO motifs centered on focal Polycomb binding sites. (B) Distribution of normalized read counts from N-ChIP experiments for ADF1, TRL, and PHO centered on focal Polycomb binding sites. (C) Midpoint plots showing pairwise combinations of ADF1, TRL, or PHO N-ChIP-recovered fragments with their called binding sites. ADF1 sites are predominantly enriched for fragments recovered by ADF1 N-ChIP. TRL sites are enriched for both TRL and ADF1-bound fragments. PHO sites are enriched for both PHO- and ADF1-bound fragments.
If factors simultaneously bind at PREs, we expect that the fragments recovered from N-ChIP experiments would span multiple factor binding sites. We therefore analyzed the recovery of factor-bound fragments in one N-ChIP around called sites of other factors (Fig. 4C). Midpoint plots around ADF1 sites reveal frequent binding of TRL and PHO. Interestingly, fragments recovered by TRL and PHO N-ChIP tend to be larger (40–80 bp) than the bulk of ADF1-bound sites. Longer fragments are expected from sites where simultaneous binding of multiple factors protects DNA from MNase digest. Similar co-occupancy of longer fragments is apparent at TRL sites and PHO sites, and predominantly with ADF1 in both cases (Fig. 4C). Thus, at some sites ADF1 promotes the binding of TRL and PHO.
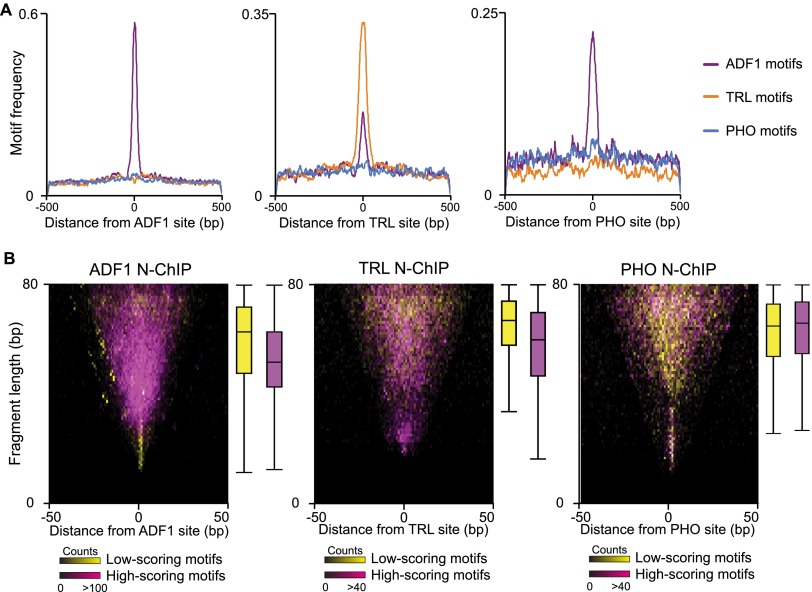
Promotion of factor binding by ADF1 is supported by motif analysis of each group of sites. As expected, ADF1 sites called from N-ChIP data display strong enrichment of high-scoring ADF1 motifs (Fig. 5A). Both TRL and ADF1 motifs are enriched at called TRL sites from N-ChIP; this implies that ADF1 contributes to TRL binding at its target sites. This is dramatic for PHO sites recovered by N-ChIP. High-scoring PHO motifs are only marginally enriched at PHO binding sites, but ADF1 sites are common. This argues that PHO binding relies on the assistance of co-bound factors.
Figure 5.
Motif qualities are related to fragment sizes recovered in N-ChIP. (A) The distribution of high-scoring motifs (P < 5 × 10−4) around factor binding sites from N-ChIP experiments. (B) Midpoint plots of small DNA (<80 bp) fragments recovered from N-ChIP experiments around called binding sites. Each site was ranked by motif scores for the respective factors. Fragments from sites with motifs in the top quintile (magenta) and bottom quintile (yellow) of motif scores are displayed. Box plots show quartile distribution of fragment sizes for each group.
If factor co-binding contributes to occupancy of sites, we expect that this will be more important at sites with poor-scoring motifs for a factor. We therefore divided sites by the scores associated with underlying motifs. We displayed the sizes of fragments recovered in N-ChIP experiments for these two groups in midpoint plots (Fig. 5B). We reasoned that high-scoring motifs may allow solitary binding of a factor, thereby protecting short segments at binding sites. If low-scoring sites require co-bound factors, longer protected fragments are expected. These predictions are true for both ADF1 and TRL, implying that these factors can bind on their own at high-quality motifs or together with other factors at poor motifs. In contrast, PHO-bound fragments with high-scoring or low-scoring motifs are relatively large, averaging ∼70 bp. This suggests that PHO often co-binds with additional DNA-binding proteins. While the consensus motif we derived for PHO from N-ChIP experiments resembles previously published motifs (Supplemental Fig. 4), it contains mismatches to the in vitro-derived ideal PHO binding sequence (Brown et al. 1998; Fritsch et al. 1999; Mishra et al. 2001). Previous studies have also observed that PHO-bound sites often contain one or two mismatches to the ideal motif, and at least at some of these sites PHO relies on cooperative interactions with transcription factors for stable binding (Mahmoudi et al. 2003; Mohd-Sarip et al. 2005; Blastyák et al. 2006).
Changes in factor occupancy are associated with switching of PRE states
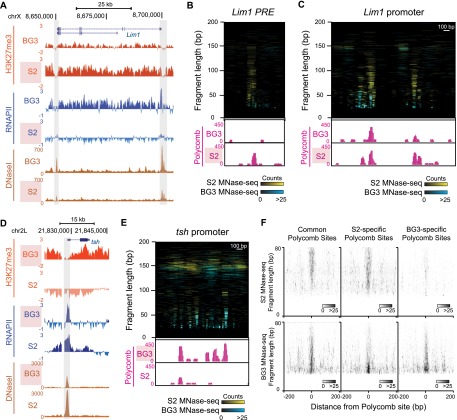
Previous studies have concluded that PcG and trxG factors are bound at PREs in both repressed and derepressed cell types, even though the activity of the regulatory element must change (Strutt et al. 1997; Papp and Muller 2006; Beisel et al. 2007; Schwartz et al. 2010; Langlais et al. 2012). As our methods allow us to examine factors and complexes at high resolution, we compared MNase-seq landscapes and Polycomb binding at PREs in alternate states. We surveyed H3K27me3 and expression patterns in S2 and DmBG3-c2 (BG3) cell lines to identify domains that are repressed in one cell line and derepressed in the other line. We identified two extended H3K27me3 domains that are present in S2 cells but absent in BG3 cells (encompassing the Lim1 and hth genes), and one domain present in BG3 cells but absent in S2 cells (encompassing the tsh gene). Like other regulatory elements, PREs are DNase I nuclease hypersensitive sites (DHS) (Galloni et al. 1993; Karch et al. 1994; Hagstrom et al. 1997); therefore, we located putative PREs for these domains by examining genomic maps of DHS in S2 and BG3 cell lines (Kharchenko et al. 2011). The complete domains and chromatin features for each of these regions are shown in Supplemental Figure 5.
The repressed Lim1 gene is the simplest example. Lim1 is included within a 140-kb H3K27me3 domain in S2 cells, but histone methylation is reduced and the gene is transcribed in BG3 cells (Fig. 6A). The repressed domain contains five DHS: a doublet DHS at the Lim1 promoter, another DHS at the CG12075 promoter within the domain, and two DHS in the 3′ end of the Lim1 gene. The Lim1 promoter and the leftmost DHS are significant binding sites for Polycomb (Supplemental Fig. 5A). We infer that the leftmost DHS is a PRE that may regulate the Lim1 promoter. Both the promoter and the putative PRE have extensive factor binding as detected by MNase-seq, displaying both small fragments and larger overlapping fragments that implies multifactor complexes stably occupy these sites. Strikingly, fragment sizes and read counts are greatly reduced at the PRE in BG3 cells where Lim1 is transcribed (Fig. 6B). Some small fragments are common between the repressed and derepressed states, suggesting that many of the same factors bind. However, the decrease in read counts suggest that these factors bind with reduced occupancy or stability. Finally, Polycomb binding at the PRE is abolished in BG3 cells, indicating that Polycomb binding is conditional, and that the derepressed state corresponds to absence of this protein. These results imply that the stability of factor complexes is an important determinant of Polycomb recruitment at PREs.
Figure 6.
Architectural changes at Polycomb binding sites in alternate repression states. (A) Genome Browser snapshot of a differential H3K27me3 domain that includes the Lim1 gene. The distributions of H3K27me3 (modENCODE X-ChIP), RNAPII (modENCODE X-ChIP), and DNase-seq count density (modENCODE) are shown for S2 cells and BG3 cells. The domain has abundant H3K27me3 methylation and Lim1 is silent in S2 cells, but methylation is reduced and the gene is active in BG3 cells. The two major DHS near Lim1 that correspond to the promoter (right) and a putative PRE (left) are shaded. (B) Comparative plots of MNase-seq fragments (<200 bp) at the Lim1 PRE in S2 (cyan) and BG3 (yellow) cells. Genome Browser snapshots below display normalized read counts from Polycomb N-ChIP (pink) for each cell line. The PRE is extensively occupied in S2 cells, but protection is reduced in BG3 cells. (C) Comparative plots of MNase-seq fragments (<200 bp) at the Lim1 promoter in S2 (cyan) and BG3 (yellow) cells. Browser snapshots below display normalized read counts from Polycomb N-ChIP (pink) for each cell line. The promoter has extensive protection in S2 cells where the gene is repressed. Four Polycomb binding sites are present in S2 cells, only one of which is constitutive and present in BG3 cells. (D) Browser snapshot of a differential H3K27me3 domain that includes the tsh gene, showing the distributions of H3K27me3, RNAPII, and DNase-seq count density for S2 cells and BG3 cells. The domain has H3K27me3 methylation and tsh is silent in BG3 cells, but abolished, and the gene is active in S2 cells. The putative PRE (shaded) defined by DHS and Polycomb binding coincides with the promoter. (E) Comparative plots of MNase-seq fragments (<200 bp) at the tsh PRE in S2 (cyan) and BG3 (yellow) cells. Browser snapshots below display normalized read counts from Polycomb N-ChIP (pink) for each cell line. The PRE is extensively occupied in BG3 cells, but the region is predominantly occupied by nucleosomes in S2 cells. (F) Midpoint plots of MNase-seq fragments at 70 focal Polycomb binding sites common to both S2 and BG3 cells (left), 133 sites found only in S2 cells (middle), and 91 sites found only in BG3 cells (right). Larger fragments are characteristic of Polycomb-bound sites.
The chromatin structure of the Lim1 promoter is more complex. The promoter displays high factor occupancy when the gene is repressed, and four Polycomb binding sites within a 1000-bp interval (Fig. 6C). When Lim1 is derepressed, only three sites of Polycomb binding disappear. Thus, the promoter contains both conditional Polycomb binding sites like the PRE and constitutive binding sites that are occupied in both chromatin states. Notably, the constitutive Polycomb sites coincide with protected high-scoring ADF1 motifs, suggesting that solitary binding of ADF1 at this site is sufficient for Polycomb recruitment. Interestingly, the overall occupancy of the promoter is reduced when the gene is derepressed, implying that repression involves more elaborate structures at promoters than active transcription.
The H3K27me3 domain containing the tsh gene is complicated by potentially redundant PREs. Regulation of tsh expression uses two distant alternative PREs during development (Zirin and Mann 2004; Oktaba et al. 2008), but the only major Polycomb binding site in BG3 cells lies at the tsh promoter, suggesting that it contains both PRE and promoter functions (Fig. 6D; Supplemental Fig. 5B). In spite of this, this single putative PRE-promoter hybrid in BG3 cells has high factor occupancy underlying a cluster of four Polycomb binding sites, and both are reduced in S2 cells where tsh is active. Finally, the hth domain also appears complex, with multiple potential PREs. These show high factor occupancy and Polycomb binding in both cell lines (Supplemental Fig. 5C). We note, however, that hth is not dependent on Polycomb repression in embryos (Zirin and Mann 2004), and thus the functional effect of Polycomb binding in this region is unclear.
We analyzed the occupancy at Polycomb binding sites more comprehensively by identifying individual focal sites with a conservative threshold. Seventy focal Polycomb binding sites are common between S2 and BG3 cells, and these show similar MNase-seq fragment size distributions in both cell lines (Fig. 6F). In contrast, cell line-specific Polycomb binding sites display fragments ranging from 30 to 80 bp, but these same sites in the other cell line have reduced read counts and fragment sizes, implying that factor complexes only occupy the site when Polycomb is bound.
Extensive factor occupancy of Polycomb-repressed promoters
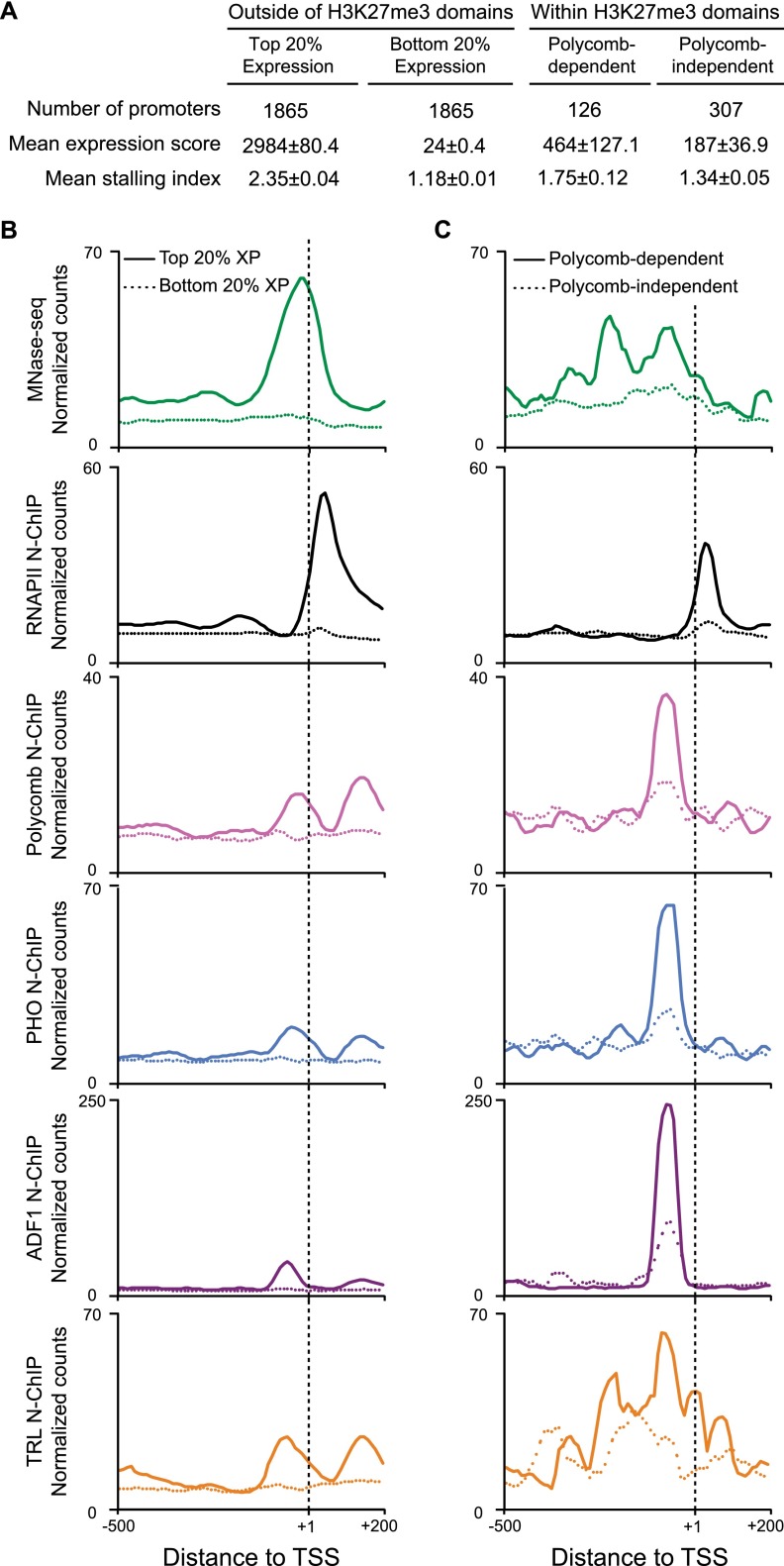
Polycomb repression has been attributed to chromatin compaction (Francis et al. 2004), inhibition of RNAPII initiation (Dellino et al. 2004), or inhibition of RNAPII elongation (Chopra et al. 2009). In many of these models, Polycomb binding at repressed target promoters is expected, and in fact this is observed (Fig. 6; Breiling et al. 2001). However, it is surprising that MNase-seq profiling of the repressed Lim1 and tsh promoters also show more extensive factor occupancy than when these genes are transcribed. This implies that Polycomb repression does not render chromatin inaccessible to factor binding. We examined Polycomb-repressed genes more generally to determine if they shared common features. To distinguish genes in H3K27me3 domains that are repressed by Polycomb from genes lacking activating factors in S2 cells, we selected genes within domains that are derepressed upon Polycomb knockdown (Lagarou et al. 2008). We then aligned genes by their annotated TSSs and plotted the fragments detected by MNase-seq and N-ChIP experiments (Fig. 7). We compared these to features of active and inactive promoters outside of H3K27me3 domains.
Figure 7.
Polycomb-regulated promoters have extensive factor occupancy and stalled RNAPII. (A) Expression and stalling indexes of genes in S2 cells outside of H3K27me3 domains, genes within H3K27me3 domains and derepressed by Polycomb knockdown (Lagarou et al. 2008), and genes within domains but unaffected by Polycomb knockdown. (B,C) Aggregation plots showing factor distributions around promoters in S2 cells. Normalized read counts for MNase-seq fragments (<76 bp, green) and fragments recovered in N-ChIP experiments for the large subunit of RNAPII (Teves and Henikoff 2011), Polycomb, ADF1, TRL, and PHO are shown. (B) Factor distributions at promoters outside of H3K27me3 domains. Promoters in the top quintile of expression (solid lines) are compared with promoters in the bottom quintile (dashed lines) genes. Highly expressed genes display high occupancy in MNase-seq localized around the TSS, and a peak of RNAPII centered ∼50 bp downstream from the TSS. (C) Factor distributions at promoters within H3K27me3 domains. Promoters that are derepressed after RNAi knockdown of Polycomb (solid lines) are compared with promoters where expression is unchanged after Polycomb knockdown (dashed lines).
As expected from earlier results (Chopra et al. 2009), plots of N-ChIP data for RNAPII using the 8WG16 monoclonal antibody to the largest RNAPII subunit (Teves and Henikoff 2011) show that Polycomb-repressed promoters are enriched for RNAPII ∼30–40 bp downstream from the TSS (Fig. 7), even though they are only expressed at low levels. This is consistent with the idea that Polycomb repression is accomplished at least in part by pausing of RNAPII (Enderle et al. 2011). Plotting MNase-seq data reveals that these promoters are dramatically more occupied than inactive promoters (Fig. 7). Protection extends broadly even compared with active promoters, spanning ∼400 bp. These promoters are enriched for Polycomb, ADF1, PHO, and TRL binding, with distinctive characteristics. Polycomb, ADF1, and PHO coincide ∼70 bp upstream of the TSS. Thus, these factors are positioned to potentially interact with general transcription factors at the core promoter. In contrast, TRL is enriched broadly across ∼400 bp throughout the promoter region in Polycomb-regulated genes, consistent with previous correlations of motif distributions and RNAPII pausing (Hendrix et al. 2008). Thus, it appears that Polycomb-repressed promoters have a complex architecture that must be determined by factor motif composition.
Discussion
Using high-resolution native-ChIP, we show here that the Polycomb protein binds chromatin in two modes: first, through nucleosomal interactions within H3K27me3 chromatin domains, and second at focal sites through interactions with DNA-binding factors. Focal binding sites include many PREs and promoters of silenced genes within H3K27me3 domains. However, Polycomb is also recruited to promoters and other sites throughout the genome, where it is not associated with H3K27 methylation or transcriptional repression. Thus, it appears that focal binding of Polycomb marks many genomic loci, and only a fraction of these are converted into repressed domains. At least some focal binding sites depend on the chromatin state of a domain, as derepression is accompanied by the loss of Polycomb. This suggests that conditional Polycomb binding may depend on multiple interactions within domains, perhaps nucleosomal binding of H3K27me3-modified nucleosomes.
The DNA-binding components that characterize Polycomb binding sites offer some clues as to how conditional and constitutive binding sites are determined. We identified the ADF1 and TRL transcription factors as common DNA-binding proteins at most focal Polycomb binding sites. ADF1 and TRL are ubiquitous factors that promote chromatin accessibility, both through the recruitment of ATP-dependent chromatin remodelers (Xiao et al. 2001) and through trapping of transiently exposed DNA within nucleosomes (Gao and Benyajati 1998). Increased accessibility may expose motifs for additional factors, facilitating binding. Indeed, TRL is required for PHO and additional factor binding to chromatinized templates in vitro (Mahmoudi et al. 2003; Mulholland et al. 2003). Our evidence further suggests bound ADF1 promotes the co-binding of both PHO and TRL to juxtaposed motifs. Proximity may allow physical interactions between the three factors, but ADF1 can also noncooperatively enhance binding of TRL through site exposure of nucleosomal DNA (Gao and Benyajati 1998). Such dependencies imply that the arrangement of individual low-affinity sites is critical for regulated factor binding at conditional Polycomb binding sites, including PREs. Conditional sites may be important to allow switching of repressive states during development.
We emphasize that focal Polycomb recruitment does not appear to require a specific combination of recruiting factors. While conditional sites coincide with binding of multiple factors, many constitutive sites display only ADF1, TRL, or PHO binding. While other unknown factors may also bind at these sites, Polycomb must interact with multiple common transcription factors. However, these factors are not sufficient for Polycomb recruitment, because not all ADF1 and TRL binding sites recruit Polycomb. Our results suggest that factor turnover at binding sites may limit stable Polycomb recruitment. At constitutive sites high occupancy by a single factor recruits Polycomb. Many of these sites are gene promoters, and may mark potential targets for Polycomb repression. At conditional sites formation of a stable factor complex is required to recruit Polycomb, and in derepressed domains individual factors only transiently bind. Repression at constitutive Polycomb-bound promoters may only occur if nearby conditional sites are also stably bound.
A requirement for stable factor binding to recruit Polycomb suggests a mechanism for how PREs are linked to expression states within regulated domains. In H3K27me3 domains PREs loop to target promoters, leading to repression (Cléard et al. 2006; Lanzuolo et al. 2007). If bringing chromatin complexes together through a loop enhances factor stability, switching of a PRE could be accomplished by disrupting a loop, for example if enhancers compete with PREs for promoters. In this way, regulatory elements would integrate developmental transcriptional programs to modulate Polycomb binding at conditional sites.
Previous studies have demonstrated that general transcription factors and RNAPII are bound at Polycomb-repressed promoters (Breiling et al. 2001; Dellino et al. 2004; Chopra et al. 2009). However, short transcripts from these genes imply that RNAPII is often paused (Enderle et al. 2011). It is intriguing that Polycomb-regulated promoters appear more extensively occupied by factors than those of active genes. The characteristic arrangement of Polycomb, ADF1, and PHO binding close to core promoter sequences suggests that these factors will contact transcriptional machinery. Recent work has demonstrated that precise spacing within promoters is critical for RNAPII pausing, suggesting that factors bound near the core promoter capture promoter-proximal RNAPII and prevent elongation (Kwak et al. 2013). Perhaps looping of a PRE to a target promoter stabilizes a factor complex that includes RNAPII, thereby inhibiting transcription.
Methods
Cell culture and native chromatin immunoprecipitation
Drosophila S2 and S2R+ cells were grown to log phase in Schneider’s media (Gibco) supplemented with 10% FBS (HyClone). Drosophila DmBG3-c2 (BG3) cells were grown in M3 media (Sigma) supplemented with BPYE, 10% FBS, and 10 µg/mL insulin. Nuclei were harvested and digested with MNase as previously described (Henikoff et al. 2009). MNase-digested chromatin was then extracted and solubilized in 80 mM NaCl buffer by pushing through a 26-gauge needle as described (Kasinathan et al. 2014). Antibodies against Polycomb (Schuettengruber et al. 2009), TRL (Melnikova et al. 2004), ADF1 (Lang and Juan 2010), PHO (Klymenko et al. 2006), and H3K27me3 (Abcam, ab6002) were used for immunoprecipitation. Illumina sequencing libraries, paired-end sequencing, and base calling were performed as described using TruSeq primers (Henikoff et al. 2011).
Sequence analysis
Binding sites were called from N-ChIP data using normalized read counts for 20- to 75-bp long fragments and setting a threshold of the genomic mean plus an arbitrary number of standard deviations as described (Kasinathan et al. 2014). Larger fragments dominate calls in the 20- to 75-bp grouping, so we called peaks a second time using normalized read counts for 20- to 35-bp fragments and combined the lists of sites. Sites for different factors within 100 bp of each other were considered coincident, while sites ±500 bp of an annotated TSS in the r5.23 genome build (FlyBase.org) were considered coincident with promoters. Midpoint-plots of fragment sizes around called binding sites were drawn as previously described (Henikoff et al. 2011). For browser tracks of MNase-seq and N-ChIP data, we normalized read counts in the 0- to 75-bp and >120-bp size classes to display factor and nucleosome distributions, respectively, and binned data with 10-bp averaging of normalized read counts for visualization using the UCSC Genome Browser (http://genome.ucsc.edu). Previously published MNase-seq data for S2 cells (Teves and Henikoff 2011) are available from GEO under accession GSE30755. MNase-seq data for BG3 cells have been deposited with modENCODE (modMine.org), under accession modENCODE_3964.
H3K27me3 domains were manually defined in S2 and BG3 cells using modENCODE X-ChIP data for H3K27me3 (S2, modENCODE_298; BG3, modENCODE_297; Riddle et al. 2011) as regions at least 5 kb in length >2× H3K27me3 enrichment. We identified 98 domains in S2 cells and 125 domains in BG3 cells. H3K27me3 domain and domain borders in S2R+ cells were identified from N-ChIP paired-end sequenced data with a calling procedure based on previously described methods (Li and Zhou 2013). Regions with nucleosome-sized reads (120–160 bp) >2× the genomic average of H3K27me3 N-ChIP signals with gaps <500 bp were designated as H3K27me3 domains, if the regions were at least 5 kb in length. Domains separated by <5 kb were grouped together.
We used gene expression calls in S2 and BG3 cells derived from cDNA microarray profiling (Cherbas et al. 2010). The stalling index of RNAPII was calculated as described (Zeitlinger et al. 2007) using RNAPII ChIP data sets modENCODE_329 (S2 cells) and modENCODE_950 (DmBG3-c2 cells). Gene expression changes before and after Polycomb knockdown in S2 cells were from Lagarou et al. (2008).
Motif analysis
A window of ±50 bp around each N-ChIP called site coordinate was used for motif discovery using MEME (Bailey et al. 2009). For Polycomb sites, only nonredundant motifs were considered. For ADF1, TRL, and PHO sites, MEME-discovered position weight matrices were compared to existing transcription factor binding motifs in the FlyFactorSurvey database (Zhu et al. 2011) using TOMTOM: Only the best-scoring match (i.e., the lowest E-value) was considered. Motif instances at individual loci were annotated using FIMO with a threshold of P < 5 × 10−4, using the FlyFactorSurvey motif entries together with N-ChIP-discovered motifs.
Fly crosses
Fly crosses were performed using standard methods at 25°C; 10–20 flies were scored for each genotype.
Coimmunoprecipitation
For Polycomb–ADF1 coimmunoprecipitation, N-ChIP targeting Polycomb in S2R+ cells was repeated, except proteins were recovered from beads in SDS buffer. Western blot detection was performed on a Nitrocellulose membrane using antibodies against Polycomb (1:1000, from Papp and Muller 2006) and ADF1 (1:2000), and HRP-recombinant Protein A (1:5000, Invitrogen) as secondary detection reagent.
Data access
Illumina HiSeq data for N-ChIPs from S2R+, S2, and BG3 cell lines have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE47829.
Acknowledgments
We thank W. Bender and J. Kassis for insightful discussions. We acknowledge Christine Codomo for technical assistance. G.A.O. was supported by a Marie Curie Actions International Outgoing Fellowship (IOF-300710) from the European Union 7th Framework Program, Research Executive Agency. This work was supported by the NIH-NHGRI modENCODE project (U01 HG004274).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.163642.113.
References
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C, Buness A, Roustan-Espinosa IM, Koch B, Schmitt S, Haas SA, Hild M Katsuyama T, Paro R 2007. Comparing active and repressed expression states of genes controlled by the Polycomb/Trithorax group proteins. Proc Natl Acad Sci 104: 16615–16620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyák A, Mishra RK, Karch F, Gyurkovics H 2006. Efficient and specific targeting of Polycomb group proteins requires cooperative interaction between Grainyhead and Pleiohomeotic. Mol Cell Biol 26: 1434–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiling A, Turner BM, Bianchi ME, Orlando V 2001. General transcription factors bind promoters repressed by Polycomb group proteins. Nature 412: 651–655 [DOI] [PubMed] [Google Scholar]
- Breiling A, O’Neill LP, D’Eliseo D, Turner BM, Orlando V 2004. Epigenome changes in active and inactive Polycomb-group-controlled regions. EMBO Rep 5: 976–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell 1: 1057–1064 [DOI] [PubMed] [Google Scholar]
- Brown JL, Grau DJ, DeVido SK, Kassis JA 2005. An Sp1/KLF binding site is important for the activity of a Polycomb group response element from the Drosophila engrailed gene. Nucleic Acids Res 33: 5181–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busturia A, Wightman CD, Sakonju S 1997. A silencer is required for maintenance of transcriptional repression throughout Drosophila development. Development 124: 4343–4350 [DOI] [PubMed] [Google Scholar]
- Cherbas L, Willingham A, Zhang D, Yang L, Zou Y, Eads BD, Carlson JW, Landolin JM, Kapranov P, Dumais J, et al. 2010. The transcriptional diversity of 25 Drosophila cell lines. Genome Res 21: 301–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Hong JW, Levine M 2009. Regulation of Hox gene activity by transcriptional elongation in Drosophila. Curr Biol 19: 688–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciavatta D, Rogers S, Magnuson T 2007. Drosophila CTCF is required for Fab-8 enhancer blocking activity in S2 cells. J Mol Biol 373: 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cléard F, Moshkin Y, Karch F, Maeda RK 2006. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet 38: 931–935 [DOI] [PubMed] [Google Scholar]
- Comet I, Schuettengruber B, Sexton T, Cavalli G 2011. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc Natl Acad Sci 108: 2294–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déjardin J, Cavalli G 2004. Chromatin inheritance upon Zeste-mediated Brahma recruitment at a minimal cellular memory module. EMBO J 23: 857–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déjardin J, Rappailles A, Cuvier O, Grimaud C, Decoville M, Locker D, Cavalli G 2005. Recruitment of Drosophila Polycomb group proteins to chromatin by DSP1. Nature 434: 533–538 [DOI] [PubMed] [Google Scholar]
- Dellino GI, Schwartz YB, Farkas G, McCabe D, Elgin SC, Pirrotta V 2004. Polycomb silencing blocks transcription initiation. Mol Cell 13: 887–893 [DOI] [PubMed] [Google Scholar]
- DeZazzo J, Sandstrom D, de Belle S, Velinzon K, Smith P, Grady L, DelVecchio M, Ramaswami M, Tully T 2000. nalyot, a mutation of the Drosophila myb-related Adf1 transcription factor, disrupts synapse formation and olfactory memory. Neuron 27: 145–158 [DOI] [PubMed] [Google Scholar]
- Duncan IM 1982. Polycomblike: a gene that appears to be required for the normal expression of the bithorax and antennapedia gene complexes of Drosophila melanogaster. Genetics 102: 49–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderle D, Beisel C, Stadler MB, Gerstung M, Athri P, Paro R 2011. Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Res 21: 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- England BP, Admon A, Tjian R 1992. Cloning of Drosophila transcription factor Adf-1 reveals homology to Myb oncoproteins. Proc Natl Acad Sci 89: 683–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL 2004. Chromatin compaction by a Polycomb group protein complex. Science 306: 1574–1577 [DOI] [PubMed] [Google Scholar]
- Fritsch C, Brown JL, Kassis JA, Muller J 1999. The DNA-binding Polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development 126: 3905–3913 [DOI] [PubMed] [Google Scholar]
- Galloni M, Gyurkovics H, Schedl P, Karch F 1993. The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J 12: 1087–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Benyajati C 1998. Specific local histone-DNA sequence contacts facilitate high-affinity, non-cooperative nucleosome binding of both adf-1 and GAGA factor. Nucleic Acids Res 26: 5394–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Negre N, Cavalli G 2006. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res 14: 363–375 [DOI] [PubMed] [Google Scholar]
- Hagstrom K, Muller M, Schedl P 1997. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics 146: 1365–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS 2008. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc Natl Acad Sci 105: 7762–7767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K 2009. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res 19: 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S 2011. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci 108: 18318–18323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R 2007. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet 3: e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch F, Galloni M, Sipos L, Gausz J, Gyurkovics H, Schedl P 1994. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res 22: 3138–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinathan S, Orsi GA, Zentner GE, Ahmad K, Henikoff S 2014. High-resolution mapping of transcription factor binding sites on native chromatin. Nat Methods 11: 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent NA, Adams S, Moorhouse A, Paszkiewicz K 2011. Chromatin particle spectrum analysis: a method for comparative chromatin structure analysis using paired-end mode next-generation DNA sequencing. Nucleic Acids Res 39: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, Wild B, Wilm M, Muller J 2006. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev 20: 1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma G, Bender W, Sipos L 2008. Replacement of a Drosophila Polycomb response element core, and in situ analysis of its DNA motifs. Mol Genet Genomics 279: 595–603 [DOI] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT 2013. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339: 950–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SH, Kim SH, Chung HM, Girton JR, Jeon SH 2003. The Drosophila pleiohomeotic mutation enhances the Polycomblike and Polycomb mutant phenotypes during embryogenesis and in the adult. Int J Dev Biol 47: 389–395 [PubMed] [Google Scholar]
- Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Verrijzer CP 2008. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev 22: 2799–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M, Juan E 2010. Binding site number variation and high-affinity binding consensus of Myb-SANT-like transcription factor Adf-1 in Drosophilidae. Nucleic Acids Res 38: 6404–6417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlais KK, Brown JL, Kassis JA 2012. Polycomb group proteins bind an engrailed PRE in both the “ON” and “OFF” transcriptional states of engrailed. PLoS ONE 7: e48765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V 2007. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol 9: 1167–1174 [DOI] [PubMed] [Google Scholar]
- Li G, Zhou L 2013. Genome-wide identification of chromatin transitional regions reveals diverse mechanisms defining the boundary of facultative heterochromatin. PLoS ONE 8: e67156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman TM, Mascotti DP 1992. Thermodynamics of ligand-nucleic acid interactions. Methods Enzymol 212: 400–424 [DOI] [PubMed] [Google Scholar]
- Mahmoudi T, Zuijderduijn LM, Mohd-Sarip A, Verrijzer CP 2003. GAGA facilitates binding of Pleiohomeotic to a chromatinized Polycomb response element. Nucleic Acids Res 31: 4147–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova L, Juge F, Gruzdeva N, Mazur A, Cavalli G, Georgiev P 2004. Interaction between the GAGA factor and Mod(mdg4) proteins promotes insulator bypass in Drosophila. Proc Natl Acad Sci 101: 14806–14811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch F 1997. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 124: 1809–1820 [DOI] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17: 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RK, Mihaly J, Barges S, Spierer A, Karch F, Hagstrom K, Schweinsberg SE, Schedl P 2001. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol Cell Biol 21: 1311–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra K, Chopra VS, Srinivasan A, Mishra RK 2003. Trl-GAGA directly interacts with lola like and both are part of the repressive complex of Polycomb group of genes. Mech Dev 120: 681–689 [DOI] [PubMed] [Google Scholar]
- Mohd-Sarip A, Venturini F, Chalkley GE, Verrijzer CP 2002. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol Cell Biol 22: 7473–7483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A, Cleard F, Mishra RK, Karch F, Verrijzer CP 2005. Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes Dev 19: 1755–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland NM, King IF, Kingston RE 2003. Regulation of Polycomb group complexes by the sequence-specific DNA binding proteins Zeste and GAGA. Genes Dev 17: 2741–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Kassis JA 2006. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr Opin Genet Dev 16: 476–484 [DOI] [PubMed] [Google Scholar]
- Oktaba K, Gutiérrez L, Gagneur J, Girardot C, Sengupta AK, Furlong EE, Müller J 2008. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev Cell 15: 877–889 [DOI] [PubMed] [Google Scholar]
- Orlando V 2003. Polycomb, epigenomes, and control of cell identity. Cell 112: 599–606 [DOI] [PubMed] [Google Scholar]
- Papp B, Muller J 2006. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev 20: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Kim MD, Jan LY, Jan YN 2006. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev 20: 820–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Minoda A, Kharchenko PV, Alekseyenko AA, Schwartz YB, Tolstorukov MY, Gorchakov AA, Jaffe JD, Kennedy C, Linder-Basso D, et al. 2011. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res 21: 147–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Rehmsmeier M, Dura JM, Paro R 2003. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev Cell 5: 759–771 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G 2009. Recruitment of Polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136: 3531–3542 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G 2009. Functional anatomy of Polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V 2010. Alternative epigenetic chromatin states of Polycomb target genes. PLoS Genet 6: e1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta AK, Kuhrs A, Muller J 2004. General transcriptional silencing by a Polycomb response element in Drosophila. Development 131: 1959–1965 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE 2013. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 49: 808–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos L, Kozma G, Molnár E, Bender W 2007. In situ dissection of a Polycomb response element in Drosophila melanogaster. Proc Natl Acad Sci 104: 12416–12421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H, Cavalli G, Paro R 1997. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J 16: 3621–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, Henikoff S 2011. Heat shock reduces stalled RNA polymerase II and nucleosome turnover genome-wide. Genes Dev 25: 2387–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M 2006. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet 38: 694–699 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Bussemaker HJ 2003. Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc Natl Acad Sci 100: 2580–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C 2001. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell 8: 531–543 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Christensen RG, Kazemian M, Hull CJ, Enuameh MS, Basciotta MD, Brasefield JA, Zhu C, Asriyan Y, Lapointe DS, et al. 2011. FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res 39: D111–D117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin JD, Mann RS 2004. Differing strategies for the establishment and maintenance of teashirt and homothorax repression in the Drosophila wing. Development 131: 5683–5693 [DOI] [PubMed] [Google Scholar]