Abstract
The Systemic Capillary Leak Syndrome (SCLS) is an extremely rare, orphan disease that resembles, and is frequently erroneously diagnosed as, systemic anaphylaxis. The disorder is characterized by repeated, transient, and seemingly unprovoked episodes of hypotensive shock and peripheral edema due to transient endothelial hyperpermeability. SCLS is often accompanied by a monoclonal gammopathy of unknown significance (MGUS). Using Affymetrix Single Nucleotide Polymorphism (SNP) microarrays, we performed the first genome-wide SNP analysis of SCLS in a cohort of 12 disease subjects and 18 controls. Exome capture sequencing was performed on genomic DNA from nine of these patients as validation for the SNP-chip discoveries and de novo data generation. We identified candidate susceptibility loci for SCLS, which included a region flanking CAV3 (3p25.3) as well as SNP clusters in PON1 (7q21.3), PSORS1C1 (6p21.3), and CHCHD3 (7q33). Among the most highly ranked discoveries were gene-associated SNPs in the uncharacterized LOC100130480 gene (rs6417039, rs2004296). Top case-associated SNPs were observed in BTRC (rs12355803, 3rs4436485), ARHGEF18 (rs11668246), CDH13 (rs4782779), and EDG2 (rs12552348), which encode proteins with known or suspected roles in B cell function and/or vascular integrity. 61 SNPs that were significantly associated with SCLS by microarray analysis were also detected and validated by exome deep sequencing. Functional annotation of highly ranked SNPs revealed enrichment of cell projections, cell junctions and adhesion, and molecules containing pleckstrin homology, Ras/Rho regulatory, and immunoglobulin Ig-like C2/fibronectin type III domains, all of which involve mechanistic functions that correlate with the SCLS phenotype. These results highlight SNPs with potential relevance to SCLS.
Keywords: systemic capillary leak syndrome; genetics, genome-wide SNP study; cell junction; cell adhesion; cytoskeleton; vascular permeability
Introduction
The systemic capillary leak syndrome (SCLS) is a very rare disorder of unknown etiology first described in 1960 by Clarkson.1 The disease occurs sporadically, primarily in Caucasian adults, who experience acute, reversible attacks of hypotension and anasarca due to the loss of plasma volume into peripheral tissues, particularly to the extremities.2 The diagnosis of SCLS is strictly clinical and is made on the basis of characteristic symptoms and evidence of decreased plasma volume (hemoconcentration) and protein loss from the intravascular space (hypoalbuminemia) during acute episodes.3 Although SCLS is probably underreported due to the difficulty of diagnosis and its resemblance to more prevalent diseases such as sepsis and systemic anaphylaxis, fewer than 250 cases have been reported since its original description in 1960. Trigger(s) for attacks are often not apparent, and most patients are asymptomatic between episodes. Treatment of acute disease is primarily supportive (IV fluids, vasopressors); more recently intravenous immunoglobulin (IVIG) administered on a monthly basis has shown promise in reducing the frequency and severity of SCLS episodes.4,5
The pathogenesis of SCLS is unknown. Although 85–95% of patients with SCLS also have MGUS, a plasma cell dyscrasia characterized by the presence of a serum monoclonal immunoglobulin (typically IgG), the reactivity of this “paraprotein” and its role in the disease are unknown. In a recent study that included 23 patients with confirmed SCLS, we found elevated levels of canonical mediators of endothelial hyperpermeability (VEGF and Ang2) in episodic sera only.6 Likewise, acute sera induced hyperpermeability of human microvascular endothelial cell (HMVEC) monolayers in functional assays.6 These results suggest that circulating blood factors induce the clinical symptoms of SCLS by inciting transient hyperpermeability in the microvasculature of peripheral extremities.
Because of the rarity of the disease and the paucity of reported cases, it is not possible to accumulate adequate numbers of SCLS patients for a genome-wide association study. However, new cost-effective technologies such as microarray SNP-chip typing and whole exome sequencing (WES) allowed us to identify candidate genes, mutations, and potential targets for further study and validation.10 We performed a genome-wide SNP analysis using SNP-chips on 12 SCLS patients and 18 controls. Exome capture sequencing was done using genomic DNA from nine of the patients for validation of these SNPs and for discovery of SNPs not present on the SNP-chips. Interestingly, we found haplotypes significantly associated with SCLS, whose annotated functions provide insight into our previous mechanistic studies of SCLS sera. These preliminary findings provide the basis for further experiments to characterize the genetic factors responsible for SCLS susceptibility and disease.
Results
SNPs associated with SCLS by SNP-chip analysis
Our study cohort included 12 patients with SCLS. The control group consisted of 15 age, sex and ethnicity-matched healthy donors, two relatives of SCLS patients, and a reference sample supplied by the SNP-chip manufacturer (Affymetrix), which was from a Caucasian male of unknown age. 33% of the patient group (n = 4) and 44% (n = 8) of the control group were female (Table 1). The mean ages of the case and control subsets at the time of sample collection were not significantly different (median age 52.5 y [range 40–67] and 49 y [range 19–67], respectively) (P = 0.07). Consistent with the demographics of SCLS, 100% of the disease group was Caucasian, as was 89% of the control group (16 of 18).
Table 1. Characteristics of study population.
| Disease (n = 12)* | Controls (n = 18) | |
|---|---|---|
| Males |
8 |
10 |
| Females |
4 |
8 |
| Mean age at enrollment (years ± S.D.)** |
53.8 ± 6.9 |
46.5 ± 13.8*** |
| Caucasian |
12 |
16& |
| Classic acute SCLS |
11 |
NA |
| Chronic SCLS |
1 |
NA |
| MGUS | 10 (83%) | NA |
Diagnosis of SCLS made according to established criteria: at least one episode of hypotension, hemoconcentration (elevated Hgb/Hct), and hypoalbuminemia or chronic edema and hypoalbuminemia; **P = 0. 07, Mann-Whitney test;***does not include reference sample; &includes 1 Hispanic, 1 African-American.
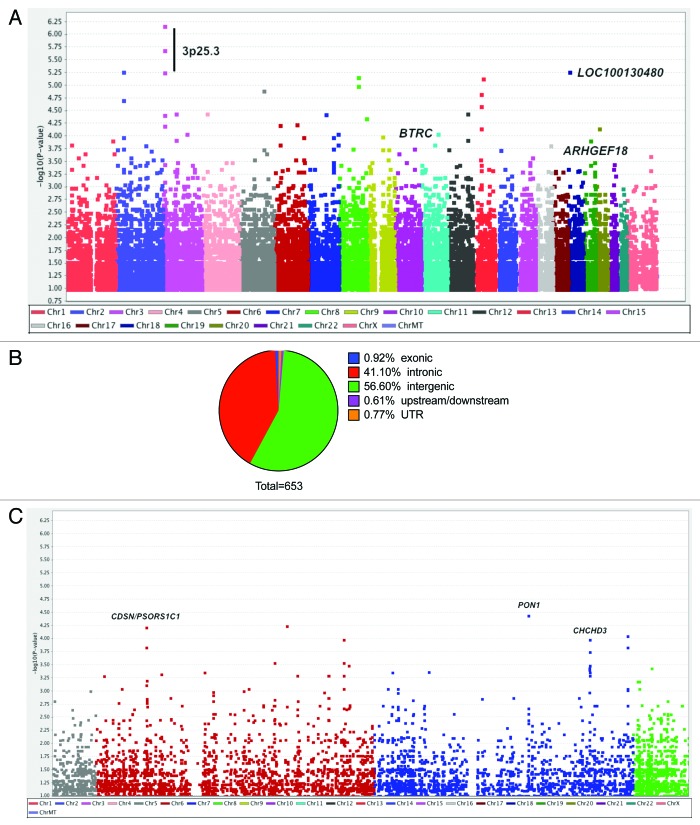
After quality control, 873,246 SNPs across 24 chromosomes in 12 SCLS and 18 control subjects were discovered. 23,652 SNPs had association P values less than 0.05 by Fisher’s exact test (Fig. 1A). Because our studies were limited by the small number of patients with SCLS, we set an initial threshold for genome-wide significance of less than 10−3 by Chi-square test and analyzed variants meeting this criterion in further detail. A total of 653 SNPs in 134 genes had association P values of less than 10−3, and 74 SNPs had P values of 10−4 or less by PLINK analysis. Six of the 653 SNPs were exonic, three of which encoded non-synonymous mutations. Nine SNPs were located upstream or downstream of coding sequences potentially within regulatory regions while the majority of these SNPs were intronic or intergenic (Fig. 1B).
Figure 1. Genome-wide analysis of 875,967 SNPs in 12 cases of SCLS and 18 control subjects. (A) Manhattan plot showing negative log-transformed P values of the case-control allele frequency significance on the y-axis. The color scale of the x-axis denotes chromosome numbers. Gene names associated with individual dots indicate SNPs of greatest significance or with potential disease relevance. (B) Distribution of 653 SNPs meeting the established criterion for genome-wide significance (P ≤ 1 × 10−3, Chi square test). (C) Expanded view of chromosomes 6–7, which contain regions of neighboring SNPs highly associated with SCLS.
Several of the most highly ranked SNPs associated with SCLS are presented in Table 2. SNPs located in genes with unknown functions or without obvious importance to disease pathogenesis are not discussed here. Nonetheless, SNPs were detected in genes with potential relevance to SCLS by virtue of their known or suspected functions in endothelial cell signaling, adhesion, or barrier function. For example, rs12355803 (P = 8.38 × 10−5, OR 13) and rs4436485 (P = 2.88 × 10−4, OR 9.46) were two highly ranked SNPs in BTRC (10q24.32). This gene encodes β transducin repeat-containing E3 ubiquitin protein ligase, βTRCP, a proteasomal factor implicated in inflammation and in growth factor receptor recycling, as discussed further below.11,12 Two SCLS-associated SNPs were discovered in AFAP1 (rs6446627 and rs7682611; both with P = 4.1 × 10−4, OR 21). AFAP1 encodes actin filament binding protein one hundred and ten (AFAP110), a Src-phosphorylated adaptor that regulates growth factor-induced actin stress fiber formation and adhesion.13 We also identified SNPs in introns of membrane-associated proteins related to endothelial cell growth and/or permeability such as rs919751 in PDGFRB (P = 2.7 × 10−4, OR 8.33) and rs4782779 in CDH13 (P = 7.67 × 10−4, OR 9.31) (Table 2). A trio of SNPs in EDG2, which encodes lysophosphatidic acid (LPA) receptor one (LPAR1), was detected only in SCLS cases with a case allelic frequency of 0.29 (rs12552348, rs1326890, rs2025766; P = 5.65 × 10−4).
Table 2. Highly ranked SNPs associated with SCLS.
| SNP | Gene | Chromosome | bp* | location | Minor allele (case frequency) | OR (95% CI) | P value** |
|---|---|---|---|---|---|---|---|
| rs6417039 |
LOC100130480 |
18 |
6568416 |
intron |
A (0.54) |
41.4 (4.8–352.9) |
4.2 × 10−6 |
| rs3917490 |
PON1 |
7 |
94948841 |
intron |
T (0.75) |
12.4 (3.6–42.9) |
1.9 × 10−5 |
| rs3130981 |
CDSN |
6 |
31083813 |
exon (N527D) |
T (0.58) |
15.4 (3.7–64.6) |
2.6 × 10−5 |
| rs12489170 |
PARP15 |
3 |
122354792 |
exon (G394R) |
A (0.38) |
NA*** |
6.8 × 10−5 |
| rs12355803 |
BTRC |
10 |
103176449 |
intron |
A (0.54) |
13 (3.1–54.3) |
8.4 × 10−5 |
| rs10239315 |
CHCHD3 |
7 |
132672067 |
intron |
C (0.71) |
8.5 (2.6–27.7) |
1.8 × 10−4 |
| rs919751 |
PDGFRB |
5 |
149505489 |
intron |
G (0.63) |
8.3 (2.5–27.8) |
2.7 × 10−4 |
| rs11668246 |
ARHGEF18 |
19 |
7492991 |
intron |
A (0.63) |
8.7 (2.5–30.2) |
2.9 × 10−4 |
| rs2364281 |
KBTBD8 |
3 |
67050203 |
intron |
T (0.71) |
7.3 (2.3–23.2) |
4.5 × 10−4 |
| rs12552348 |
EDG2 |
9 |
113705204 |
intron |
T (0.29) |
NA |
5.7 × 10−4 |
| rs4782779 |
CDH13 |
16 |
83402943 |
intron |
A (0.46) |
9.3 (2.2–38.9) |
7.7 × 10−4 |
| rs12911414 | RASGRF1 | 15 | 79343534 | intron | G (0.58) | 7 (2.1–23.1) | 7.9 × 10−4 |
GRCh37.p5 assembly;**Chi-square test; ***Not applicable, detected only in cases.
Whole exome sequencing
We performed exome capture sequencing to validate the SNP-chip data and to discover SNPs not present on the array. Cost considerations limited this analysis to the first nine patients enrolled in the study. Based on WES data alone, we discovered 10, 955 SNPs that were detected in all nine patients tested. We compared data across the two technologies and found a total of 1,416 SNPs in common. 61 of these SNPs were significantly associated with SCLS based on PLINK analysis of the SNP-chip data (P < 0.05, Chi square test). Ten of the SCLS-associated SNPs encoded exonic, nonsynonymous mutations whereas 23 variants encoded exonic, synonymous mutations. The remaining 28 SNPs were located in intronic, untranslated, or intergenic regions. Several of the top ranked SNPs detected by both methods are presented in Table 3. This subset included SNPs in genes involved in protein degradation and/or the proteasome (SERPINB1, CPN1, CTSL1, KLK7, PSMB3). In particular, deficiency of carboxypeptidase 1 (encoded by CPN1), which is a proteolytic inactivator of serum kinins and anaphylatoxins, has been associated with increased susceptibility to C5a-mediated anaphylactic shock in mice, which is a clinical phenotype that closely resembles SCLS.14 Functional annotation of SNPs present in both SNP-chip and WES also revealed genes involved prominently in cell-cell junctions and adhesion (MYOF, DSG4, SIGLEC12, SLC24A3), suggesting potential impact on the endothelial barrier dysfunction that characterizes SCLS.
Table 3. SCLS associated SNPs detected by both exome sequencing and SNP array.
| SNP | Gene | Chromosome | bp* | location | Minor allele (case frequency) | OR (95% CI) | P value** |
|---|---|---|---|---|---|---|---|
| rs386915 |
SERPINB1 |
6 |
2834265 |
intron |
C (0.125) |
0.16 (0.04–0.63) |
5.2 × 10−3 |
| rs228274 |
PSMB3 |
17 |
36909061 |
5UTR# |
A (0.083) |
0.161 (0.03–0.79) |
0.015 |
| rs1569767 |
SLC24A3 |
20 |
19261623 |
exon (V55I) |
A (0.583) |
3.64 (1.22- 10.8) |
0.018 |
| rs10883439 |
CPN1 |
10 |
101841153 |
intron |
A (0.208) |
9.21 (1–84.7) |
0.022 |
| rs1614065 |
MYOF |
10 |
95097537 |
intron |
T (0.167) |
0.25 (0.07–0.88) |
0.025 |
| rs3752135 |
SIGLEC12 |
19 |
52000624 |
exon (Y376S) |
T (0.25) |
5.67 (1.03–31) |
0.03 |
| rs4799570 |
DSG4 |
18 |
28986333 |
exon (I644L) |
A (0.375) |
3.72 (1.06–13.05) |
0.034 |
| rs2274611 |
CTSL1 |
9 |
90342675 |
intron |
T (0.667) |
3.14 (1.07–9.27) |
0.035 |
| rs12185460 | ARHGAP28 | 18 | 6898415 | intron | C (0.292) | 0.329 (0.11–0.99) | 0.044 |
GRCh37.p5 assemby; **Chi-square test; #5′ untranslated region
We determined the possible ramifications of amino acid changes conferred by SNPs discovered in the SNP-chip and WES data using the PolyPhen software algorithm. Of the three SNPs in the SNP-chip data set associated with non-synonymous exonic mutations, those in PARP15 and OR5A2 resulted in potentially deleterious amino acid changes, as shown in Table 4. Nine of ten non-synonymous exonic mutations detected by both WES and SNP-chip were classified as benign whereas rs8181512 in OR52N2 is predicted to lead to a damaging amino acid change (H791R) in an olfactory receptor.
Table 4. Impact of non-synonymous mutations detected by SNP-chip and/or WES.
| SNP | Protein | Mutation | Frequency by WES (# positive/total) |
PolyPhen annotation (score) |
P value |
|---|---|---|---|---|---|
| rs3130981 |
CDSN |
N527D |
0/9 |
benign (0.004) |
2.6 × 10−5 |
| rs12489170 |
PARP15 |
G394R / G628R* |
4/9 |
probably damaging (1.0) |
6.7 × 10−5 |
| rs1453547 |
OR5A2 |
P172L |
7/9 |
probably damaging (0.997) |
8.5 × 10−4 |
| rs8181512 | OR52N2 | H791R | 9/9 | probably damaging (0.991) | 4 × 10−3 |
alternative splice variants; **Chi-square test
Functional annotation of SCLS-associated variants
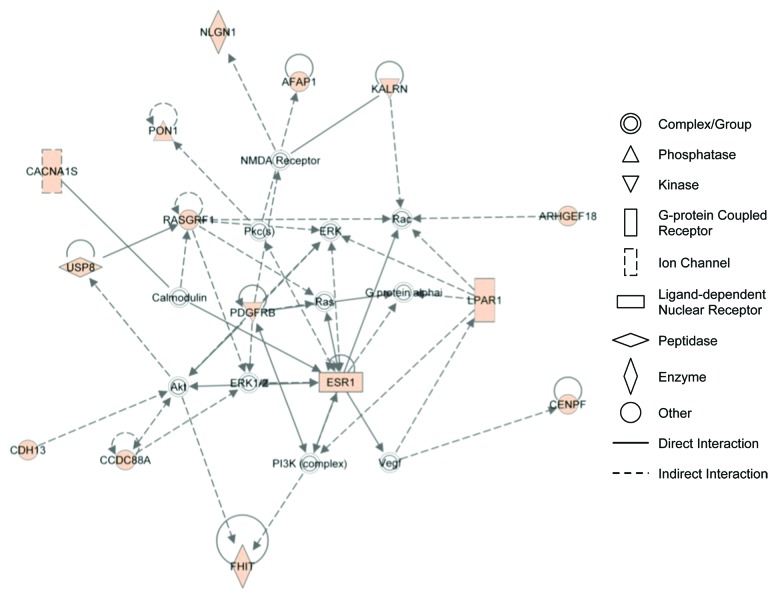
We analyzed the 134 genes containing 653 SNPs significantly associated with SCLS by SNP-chip analysis using the DAVID functional clustering algorithm (Table 5). This study revealed enrichment of genes related to endothelial barrier function including those involving cell projections, cell adhesion, cell junctions, and synapses (Fig. 2A and B). Functional categories enriched at least 2-fold in this subset included cell adhesion (3.3-fold, P = 5.5 × 10−3) and cell junctions (3.9-fold, P = 1 × 10−3). Several gene ontology terms were enriched including regulation of Rho signal transduction (6.6-fold, P = 6.9 × 10−3), biological and cell adhesion (2.4-fold, P = 6.8 × 10−3), cadherin binding (23.6-fold, P = 6.8 × 10−3), and cell adhesion molecule binding (13.4-fold, P = 0.02). Additional functional and network evaluation using IPA revealed enrichment of functional categories including cell-to-cell signaling and interaction and cellular assembly and maintenance (Fig. 3). These enriched categories were closely related to the mechanisms implicated by our previous functional studies of SCLS sera, as discussed below.
Table 5. Functional enrichment analysis of top ranked SNPs.
| Annotation terms | Enrichment score | #Genes* | Fold enrichment* | Select clustered genes | P value** |
|---|---|---|---|---|---|
| cell projection organization, neuron projection, cell projection |
3.26 |
13 |
4.6 |
ATP2B2, NCK2, RASGRF1, DSCAM, CDH13, CTNNA2, CHRM3, KALRN, LPAR1 |
2.3 × 10−5 |
| synapse, cell junction, postsynaptic cell membrane |
2.64 |
11 |
4 |
AFAP1, CTNNA2, CHRM3, CDSN, GRID1, GRIK1 |
3.9 × 10−4 |
| plasma membrane (part, integral, intrinsic) |
2.6 |
34 |
2 |
ATP2B2, DSCAM, AFAP1, ADCY3, CDH13, CTNNA2, CHRM3, CDSN, EMCN, ITGA9, GNA14, LPAR1, PDGFRB, TRPA1 |
4.6 × 10−5 |
| pleckstrin homology domain, Rho and Ras regulation, RhoGEF |
2.43 |
5 |
21 |
ARAP1, ASAP2, RASGRF1, ARHGEF18, AFAP1, ADCY3, BTRC, DGKB, KALRN, LPAR1, PON1 |
8.9 × 10−5 |
| fibronectin type III domain, immunoglobulin domain Ig-like C2 (types 1–2), |
2.35 |
8 |
3.88 |
ADAMTSL1, CNTN3, NEO1, PDGFRB, PTPRD, ITGA9, CDH13, PCDH15, PCDH19 |
1.4 × 10−4 |
| cell adhesion, biological adhesion, cell-cell adhesion | 2.19 | 9 | 3.3 | DSCAM, CDH13, CTNNA2, CNTN3, CDSN, EMCN, ITGA9 | 5.5 × 10−3 |
For top cluster term; **EASE score (modified Fisher's exact test) for top cluster term
Figure 2. Functional enrichment of SCLS-associated SNPs. (A, B) 2-D heat maps representing genes containing SNPs significantly associated with SCLS and their associated functional annotation terms, as determined by the DAVID software algorithm. Related genes are portrayed on the x-axis and their corresponding annotations on the y-axis. Green areas represent positively reported gene-term association whereas black areas denote no gene-term association yet reported. Functionally enriched categories related to endothelial barrier dysregulation in SCLS included “cell projections” (A) and “cell adhesion” (B).
Figure 3. IPA network analysis reveals association of genes related to cytoplasmic/cytoskeletal organization. A list of 134 genes containing 653 SNPs associated with SCLS were uploaded for core analysis using Ingenuity Pathways Knowledge Base as reference. The network has been simplified for clear illustration of genes of interest. Colored symbol are genes contained on the list. Symbol shapes correspond to molecular functions as indicated. Direct or indirect interactions are shown by complete or dashed lines, respectively
Discussion
SCLS is a rare disorder of unknown etiology that causes substantial morbidity and mortality to those affected. During an acute SCLS attack, patients may present in hypotensive shock due to the loss of up to 70% of their plasma volume into the tissues. Life-threatening complications result from hypovolemia and/or hypoperfusion (renal failure, stroke), increased serum viscosity (thrombosis), increased pressures in the edematous upper and lower extremities (compartment syndromes), and heart failure due to the recruitment of fluids into the intravascular compartment during the post-leak phase associated with the acute SCLS episode. Our previous research demonstrated that application of acute SCLS sera to normal HMVECs induced barrier breakdown, suggesting that exogenous factors present in acute SCLS sera induce the transient clinical symptoms of vascular leak. Accordingly, we found elevated levels of endothelial permeability mediators, VEGF and Ang2, in acute, but not baseline, SCLS sera. However, key questions remain. What are the cellular sources of these mediators, and what is the trigger for their secretion? Do primary genetic abnormalities in endothelial cells of SCLS subjects induce aberrant responses to VEGF, Ang2, or other factors? Does the monoclonal IgG present in SCLS sera elicit the endothelial hyperpermeability associated with acute SCLS episodes, either directly or indirectly?
A significant obstacle in addressing these questions is the extreme rarity of the disease. Currently, worldwide prevalence is estimated to be ~50–100 cases with a confirmed diagnosis. In spite of these challenges, we performed the first genome-wide SNP and exome-capture sequencing analysis of an SCLS cohort to uncover potential genetic abnormalities associated with the disease. Our pilot study identified haplotypes significantly associated with the SCLS trait and several candidate susceptibility loci. Most strikingly, functional annotation of SCLS-associated SNPs revealed significant enrichment of cell junctions, cell-to-cell interactions, Rho-Rac signaling, and adhesion, among others, which are cellular constituents or processes that affect formation and maintenance of the vascular barrier.
We found previously that SCLS sera induced endothelial barrier disruption through internalization of the membrane junctional protein VE-cadherin.6 Our functional annotation of SNP-chip data suggests that gene polymorphisms present in SCLS patients could lead to intrinsically abnormal endothelial responses to known permeability mediators. As but one example, rs4782779 in CDH13 was strongly associated with SCLS. CDH13 encodes T-cadherin, a glycophosphatidyloinositol (GPI)-linked junctional adhesion protein whose physiological functions include protecting endothelial cells from reactive oxygen species (ROS)-induced damage. In published study, SCLS sera elicited endothelial production of ROS, which are known to induce vascular hyperpermeability,15,16 suggesting that mutations in CDH13 could affect endothelial responses to oxidative stress in this disorder.
We also discovered SNPs in genes that could lead to impaired responses to VEGF, a permeability mediator present in increased amounts in acute SCLS sera. rs12355803 and several other SNPs in BTRC were detected in the SCLS group. BTRC encodes βTRCP, the F box protein component of SCF (SKP1-cullin-F-box) ubiquitin protein ligase complex.17,18,19 A published study demonstrated that βTRCP promoted ubiquitylation and degradation of the endothelial VEGF receptor (VEGFR2), and βTRCP knockdown enhanced VEGFR2 signaling.12 We also detected a disease-associated SNP (rs1614065) in MYOF,, which encodes dysferlin, an Ig C2 domain containing protein (also a functionally enriched category) that promotes endothelial cell adhesion by inhibiting VEGFR2 degradation.20,21 Thus, polymorphisms in BRTC and/or MYOF could affect responses to VEGF in SCLS patients through modulation of VEGFR2 expression.
Our prior mechanistic studies revealed that acute SCLS sera induced endothelial retraction through actin-stress fiber formation and cell morphological changes. Canonical permeability mediators such as VEGF and Ang2 induce actin polymerization and cytoskeletal rearrangement by activating Rho- and Rac-dependent signaling pathways.22 We detected SCLS-associated SNPs in ARAP1, which encodes a PH domain-containing RhoGAP,23 in KALRN, encoding a RhoGEF with kinase activity,24 and in EDG2, which encodes LPAR1. All three gene products may affect endothelial barrier function through Rho-Rac activation.25
Finally, we detected SCLS associated SNPs in genes potentially related to the clonal plasma cell expansion that typifies MGUS.26-32 A trio of SNPs in PARP15 (3q21) had an allelic frequency of 0.33 in SCLS patients, but was not detected in the control group. PARP15 is a transcriptional repressor associated with B cell lymphoma.33 One such SCLS-associated SNP in PARP discovered by the SNP-chip analysis (rs12489170) was also detected in 4 of 9 patients by WES, at a frequency of 42–100%. Because this SNP encodes a damaging missense mutation in the PARP15 protein (G394R or G628R depending on the splice variant), further examination of PARP15 sequence in the SCLS cohort may be warranted.
Although our SNP analysis points to disease associations with a number of gene variants, the contribution of hereditary factors to the SCLS phenotype is currently unknown.34,35 Most SCLS patients are adults with no family history of disease, who manifest symptoms in mid-life. A Jewish family with familial SCLS was reported in 2010.36 The index case was an eight-year old boy with repeated attacks of hypotensive shock accompanied by hemoconcentration and hypoalbuminemia consistent with the diagnosis of SCLS. However, the sparse family history provided makes it unclear whether this patient and his family had vertically transmitted SCLS or a variant of the disorder that was originally described by Clarkson. Further approaches such as whole genomic sequencing or RNA-Seq of plasma cell- or endothelial cell-enriched sources (e.g., bone marrow or blood-outgrowth endothelial cells) could reveal the presence of acquired somatic mutations in adult SCLS patients.
In summary, we performed a genome-wide SNP analysis of a poorly studied disorder, SCLS. We discovered polymorphisms in genes whose functional annotations suggest involvement in cell-cell junctions and signaling, cell adhesion, and cytoskeletal organization, which correlate with our previous mechanistic studies of SCLS sera. Such annotations provide a framework for future allelic discrimination strategies to validate top ranked SNPs discovered here as well as novel SNPs unique to the SCLS cohort detected by exome capture sequencing. We stress that although this is a preliminary survey, our study will facilitate the acquisition of research funds that will allow for an in-depth genetic survey of more patients, using more expensive technology such as whole genomic sequencing. This paper, which focuses on genetic characterization of a rare and unusual disease, is the first effort of its kind for SCLS. Although our findings must be corroborated in a larger cohort, they provide a springboard for discovery of underlying pathophysiological mechanisms, biomarkers, and avenues for therapy.
Patients, Methods, and Materials
Study subjects
SCLS patients were included in the study based upon meeting the following established clinical criteria: at least one episode of reversible hypotension, hemoconcentration, and hypoalbuminemia, or chronic edema and hypoalbuminemia in the absence of secondary causes.2,4 All patients were seen at the Clinical Center of the National Institutes of Health. Written informed consent was obtained from each patient, and the study protocol (09-I-0184) conformed to the ethical guidelines of the 2008 Declaration of Helsinki as reflected in a priori approval from the Institutional Review Board of the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH). Age-, sex-, and ethnicity-matched blood samples were obtained from the NIH Blood Bank.
DNA isolation and SNP array
Mononuclear cells were isolated from patient and healthy control human peripheral blood samples using Lymphocyte Separation Medium (MP Biomedicals). Because a high proportion of patients with SCLS have MGUS, which indicates that chromosomal mutations or translocations may have occurred during early B lineage development, we used purified B lymphocytes for genome-wide SNP analysis. B cells were isolated from peripheral blood during flow sorting using anti-CD19 antibody (BD Biosciences). DNA targets from patients and healthy controls were prepared and hybridized to Affymetrix Genome-Wide Human SNP array 6.0 chips following the manufacturer’s recommendations. Quality analysis was performed by importing the CEL files into Genotyping Console (GTC 4.0) according to the “Quality Control Assessment in Genotyping Console” (Affymetrix revision 1). An association analysis was performed for the control vs. case using PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/anal.shtml#cc) software utilizing the Fisher’s exact test, Chi-square and the estimated odds ratio. Pearson Chi square values and P values (Chi square and Fisher’s exact test) were calculated using Haploview 4.2. PLINK data were deposited into GEO (Accession number GSE40495). The data can be viewed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=lvsdbiagqmgewhi&acc=GSE49045
Exome capture sequencing
Genomic DNA was isolated from peripheral blood mononuclear cells of nine SCLS subjects and randomly digested into fragments with a length of approximately 150 to 200 base pairs (bp). Adapters were ligated to each end of the fragments, and adaptor-ligated templates were purified using Agencourt AMPure SPRI beads (Beckman Coulter). Purified DNA fragments were amplified by ligation-mediated polymerase chain reaction (LM-PCR), purified, and hybridized to the SureSelect Biotinylated RNA Library (BAITS) for enrichment (Agilent Technologies). The magnitude of enrichment of captured LM-PCR products was estimated using the Agilent 2100 Bioanalyzer. Each captured library was then loaded onto the Illumina Hiseq2000 sequencer. Whole exome sequence was obtained from all nine SCLS patients, with a mean target read depth of 54.19X, such that 88% of targeted reads had at least 10X coverage. Raw image files were processed by Illumina base calling Software 1.7 for base calling with default parameters, and the sequences of each individual were generated as 90 bp paired-end reads. Burrows-Wheeler Aligner was used to align sequences to the reference genome (GRCh37/hg19 assembly). SNPs were analyzed using SOAPsnp, and Small Insertion-Deletions (InDels) were detected by SAMtools. ANNOVAR was used to annotate variants. PolyPhen-2 (Polymorphism Phenotyping v2; http://genetics.bwh.harvard.edu/pph2/) software was used to predict possible impact of amino acid substitutions on the structure and function of individual proteins.
Functional annotation
DAVID7,8 and Enrichr9 (http://amp.pharm.mssm.edu/Enrichr/enrich) algorithms, and Ingenuity Pathways Analysis (Ingenuity Systems) software tools were utilized for functional enrichment analysis and hierarchical clustering of SNPs detected by SNP Chip and/or whole exome sequencing. In DAVID, P values were calculated at medium classification stringency using a modified Fisher’s exact test (EASE score), and terms with P values < 0.05 were considered significantly enriched. ChIP Enrichment Analysis (ChEA, http://amp.pharm.mssm.edu/lib/chea.jsp) was used to analyze genome-wide transcription factor binding to trait-associated loci9.
Disclosure of Potential Conflicts of Interest
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health (Project AI001830). Support by M.Y. for this project was funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The authors declare no competing financial interests.
Acknowledgments
We thank Stacy Ricklefs for her help with SNP chip assays and Drs. Todd Wilson and Helene Rosenberg (NIAID/NIH) for helpful discussions and critical review of the manuscript.
Author Contributions
Recruited and cared for patients and analyzed data: Wisch L, Young M, and Nelson CM. Conceived and designed experiments: Xie Z, Sturdevant D, Porcella SF, and Druey KM. Performed experiments: Xie Z and Sturdevant D. Analyzed data: Xie Z, Nagarajan V, Sturdevant D, Chan E, Porcella SF, and Druey KM. Wrote the paper: Xie Z, Porcella SF, and Druey KM.
Glossary
Abbreviations:
- SCLS
systemic capillary leak syndrome
- MGUS
monoclonal gammopathy of unknown significance
- OR
odds ratio
- SNP
single nucleotide polymorphism
- VEGF
vascular endothelial growth factor
- Ang2
angiopoietin 2
- MAF
minor allele frequency
- VE-cadherin
vascular endothelial cadherin
- GEF
guanine nucleotide exchange factor
- GAP
GTPase activating protein
- HDL
high density lipoprotein
- LDL
low density lipoprotein
- GWAS
genome-wide association studies
- WES
whole exome capture sequencing
References
- 1.Clarkson B, Thompson D, Horwith M, Luckey EH. Cyclical edema and shock due to increased capillary permeability. Am J Med. 1960;29:193–216. doi: 10.1016/0002-9343(60)90018-8. [DOI] [PubMed] [Google Scholar]
- 2.Druey KM, Greipp PR. Narrative review: the systemic capillary leak syndrome. Ann Intern Med. 2010;153:90–8. doi: 10.7326/0003-4819-153-2-201007200-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapoor P, Greipp PT, Schaefer EW, Mandrekar SJ, Kamal AH, Gonzalez-Paz NC, Kumar S, Greipp PR. Idiopathic systemic capillary leak syndrome (Clarkson’s disease): the Mayo clinic experience. Mayo Clin Proc. 2010;85:905–12. doi: 10.4065/mcp.2010.0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gousseff M, Arnaud L, Lambert M, Hot A, Hamidou M, Duhaut P, Papo T, Soubrier M, Ruivard M, Malizia G, et al. Capillary Leak Syndrome Registry The systemic capillary leak syndrome: a case series of 28 patients from a European registry. Ann Intern Med. 2011;154:464–71. doi: 10.7326/0003-4819-154-7-201104050-00004. [DOI] [PubMed] [Google Scholar]
- 5.Pecker M, Adams M, Graham W. The systemic capillary leak syndrome. Ann Intern Med. 2011;155:335–, author reply 335-6. doi: 10.7326/0003-4819-155-5-201109060-00016. [DOI] [PubMed] [Google Scholar]
- 6.Xie Z, Ghosh CC, Patel R, Iwaki S, Gaskins D, Nelson C, Jones N, Greipp PR, Parikh SM, Druey KM. Vascular endothelial hyperpermeability induces the clinical symptoms of Clarkson disease (the systemic capillary leak syndrome) Blood. 2012;119:4321–32. doi: 10.1182/blood-2011-08-375816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 8.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–44. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darshi M, Mendiola VL, Mackey MR, Murphy AN, Koller A, Perkins GA, Ellisman MH, Taylor SS. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem. 2011;286:2918–32. doi: 10.1074/jbc.M110.171975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–4. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 12.Shaik S, Nucera C, Inuzuka H, Gao D, Garnaas M, Frechette G, Harris L, Wan L, Fukushima H, Husain A, et al. SCF(β-TRCP) suppresses angiogenesis and thyroid cancer cell migration by promoting ubiquitination and destruction of VEGF receptor 2. J Exp Med. 2012;209:1289–307. doi: 10.1084/jem.20112446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorfleutner A, Stehlik C, Zhang J, Gallick GE, Flynn DC. AFAP-110 is required for actin stress fiber formation and cell adhesion in MDA-MB-231 breast cancer cells. J Cell Physiol. 2007;213:740–9. doi: 10.1002/jcp.21143. [DOI] [PubMed] [Google Scholar]
- 14.Mueller-Ortiz SL, Wang D, Morales JE, Li L, Chang JY, Wetsel RA. Targeted disruption of the gene encoding the murine small subunit of carboxypeptidase N (CPN1) causes susceptibility to C5a anaphylatoxin-mediated shock. J Immunol. 2009;182:6533–9. doi: 10.4049/jimmunol.0804207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philippova M, Joshi MB, Pfaff D, Kyriakakis E, Maslova K, Erne P, Resink TJ. T-cadherin attenuates insulin-dependent signalling, eNOS activation, and angiogenesis in vascular endothelial cells. Cardiovasc Res. 2012;93:498–507. doi: 10.1093/cvr/cvs004. [DOI] [PubMed] [Google Scholar]
- 16.Assaly R, Olson D, Hammersley J, Fan PS, Liu J, Shapiro JI, Kahaleh MB. Initial evidence of endothelial cell apoptosis as a mechanism of systemic capillary leak syndrome. Chest. 2001;120:1301–8. doi: 10.1378/chest.120.4.1301. [DOI] [PubMed] [Google Scholar]
- 17.Humbert R, Adler DA, Disteche CM, Hassett C, Omiecinski CJ, Furlong CE. The molecular basis of the human serum paraoxonase activity polymorphism. Nat Genet. 1993;3:73–6. doi: 10.1038/ng0193-73. [DOI] [PubMed] [Google Scholar]
- 18.Serrato M, Marian AJ. A variant of human paraoxonase/arylesterase (HUMPONA) gene is a risk factor for coronary artery disease. J Clin Invest. 1995;96:3005–8. doi: 10.1172/JCI118373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotani K, Tsuzaki K, Sakane N. Paraoxonase-1 gene Q192R polymorphism and reactive oxygen metabolites. J Int Med Res. 2012;40:1513–8. doi: 10.1177/147323001204000431. [DOI] [PubMed] [Google Scholar]
- 20.Bernatchez PN, Acevedo L, Fernandez-Hernando C, Murata T, Chalouni C, Kim J, Erdjument-Bromage H, Shah V, Gratton JP, McNally EM, et al. Myoferlin regulates vascular endothelial growth factor receptor-2 stability and function. J Biol Chem. 2007;282:30745–53. doi: 10.1074/jbc.M704798200. [DOI] [PubMed] [Google Scholar]
- 21.Sharma A, Yu C, Leung C, Trane A, Lau M, Utokaparch S, Shaheen F, Sheibani N, Bernatchez P. A new role for the muscle repair protein dysferlin in endothelial cell adhesion and angiogenesis. Arterioscler Thromb Vasc Biol. 2010;30:2196–204. doi: 10.1161/ATVBAHA.110.208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chavez A, Smith M, Mehta D. New insights into the regulation of vascular permeability. Int Rev Cell Mol Biol. 2011;290:205–48. doi: 10.1016/B978-0-12-386037-8.00001-6. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa J, Tsujita K, Takenawa T, Itoh T. ARAP1 regulates the ring size of circular dorsal ruffles through Arf1 and Arf5. Mol Biol Cell. 2012;23:2481–9. doi: 10.1091/mbc.E12-01-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu JH, Fanaroff AC, Sharma KC, Smith LS, Brian L, Eipper BA, Mains RE, Freedman NJ, Zhang L. Kalirin promotes neointimal hyperplasia by activating Rac in smooth muscle cells. Arterioscler Thromb Vasc Biol. 2013;33:702–8. doi: 10.1161/ATVBAHA.112.300234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panetti TS. Differential effects of sphingosine 1-phosphate and lysophosphatidic acid on endothelial cells. Biochim Biophys Acta. 2002;1582:190–6. doi: 10.1016/S1388-1981(02)00155-5. [DOI] [PubMed] [Google Scholar]
- 26.Cugno M, Castelli R, Cicardi M. Angioedema due to acquired C1-inhibitor deficiency: a bridging condition between autoimmunity and lymphoproliferation. Autoimmun Rev. 2008;8:156–9. doi: 10.1016/j.autrev.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Lettre G, Rioux JD. Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet. 2008;17(R2):R116–21. doi: 10.1093/hmg/ddn246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eriksson N, Tung JY, Kiefer AK, Hinds DA, Francke U, Mountain JL, Do CB. Novel associations for hypothyroidism include known autoimmune risk loci. PLoS One. 2012;7:e34442. doi: 10.1371/journal.pone.0034442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y, Wang B, Liu JL, Gao XH, Chen HD, Li YH. Association of -619C/T polymorphism in CDSN gene and psoriasis risk: a meta-analysis. Genet Mol Res. 2011;10:3632–40. doi: 10.4238/2011.October.21.6. [DOI] [PubMed] [Google Scholar]
- 30.Nair RP, Stuart PE, Nistor I, Hiremagalore R, Chia NV, Jenisch S, Weichenthal M, Abecasis GR, Lim HW, Christophers E, et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet. 2006;78:827–51. doi: 10.1086/503821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allanore Y, Saad M, Dieudé P, Avouac J, Distler JH, Amouyel P, Matucci-Cerinic M, Riemekasten G, Airo P, Melchers I, et al. Genome-wide scan identifies TNIP1, PSORS1C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet. 2011;7:e1002091. doi: 10.1371/journal.pgen.1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frech TM, Revelo MP, Drakos SG, Murtaugh MA, Markewitz BA, Sawitzke AD, Li DY. Vascular leak is a central feature in the pathogenesis of systemic sclerosis. J Rheumatol. 2012;39:1385–91. doi: 10.3899/jrheum.111380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aguiar RC, Takeyama K, He C, Kreinbrink K, Shipp MA. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J Biol Chem. 2005;280:33756–65. doi: 10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- 34.Calvo F, Sanz-Moreno V, Agudo-Ibáñez L, Wallberg F, Sahai E, Marshall CJ, Crespo P. RasGRF suppresses Cdc42-mediated tumour cell movement, cytoskeletal dynamics and transformation. Nat Cell Biol. 2011;13:819–26. doi: 10.1038/ncb2271. [DOI] [PubMed] [Google Scholar]
- 35.Niu J, Profirovic J, Pan H, Vaiskunaite R, Voyno-Yasenetskaya T. G Protein betagamma subunits stimulate p114RhoGEF, a guanine nucleotide exchange factor for RhoA and Rac1: regulation of cell shape and reactive oxygen species production. Circ Res. 2003;93:848–56. doi: 10.1161/01.RES.0000097607.14733.0C. [DOI] [PubMed] [Google Scholar]
- 36.Sion-Sarid R, Lerman-Sagie T, Blumkin L, Ben-Ami D, Cohen I, Houri S. Neurologic involvement in a child with systemic capillary leak syndrome. Pediatrics. 2010;125:e687–92. doi: 10.1542/peds.2009-1691. [DOI] [PubMed] [Google Scholar]
- 37.Abgueguen P, Chennebault JM, Pichard E. Immunoglobulins for treatment of systemic capillary leak syndrome. Am J Med. 2010;123:e3–4. doi: 10.1016/j.amjmed.2009.09.034. [DOI] [PubMed] [Google Scholar]