Abstract
The precursor of Escherichia coli RTEM beta-lactamase was purified to homogeneity on a milligram scale by a procedure independent of the binding properties of the protein and refolded to an active, reduced form. For comparing the folding kinetics, the wild-type enzyme was reduced and a mutant was constructed, in which the two cysteines that form a very stable disulfide bond in the RTEM enzyme were both changed into alanines. The rate of folding was determined by directly measuring the increase in enzymatic activity. The reduced precursor folds at least 15 times more slowly than either the reduced mature enzyme or the mature Cys----Ala double mutant under identical conditions. The wild-type enzyme, the Cys----Ala double mutant and the precursor protein all had similar KM values, demonstrating a very similar native state. The slow folding of the precursor compared with the mature form may be an essential and general feature to secure a transport competent conformation necessary for the translocation through a membrane in protein export. This folding assay of a precursor by directly following its enzymatic activity may facilitate the characterization of putative folding modulators in bacterial membrane transport.
Full text
PDF
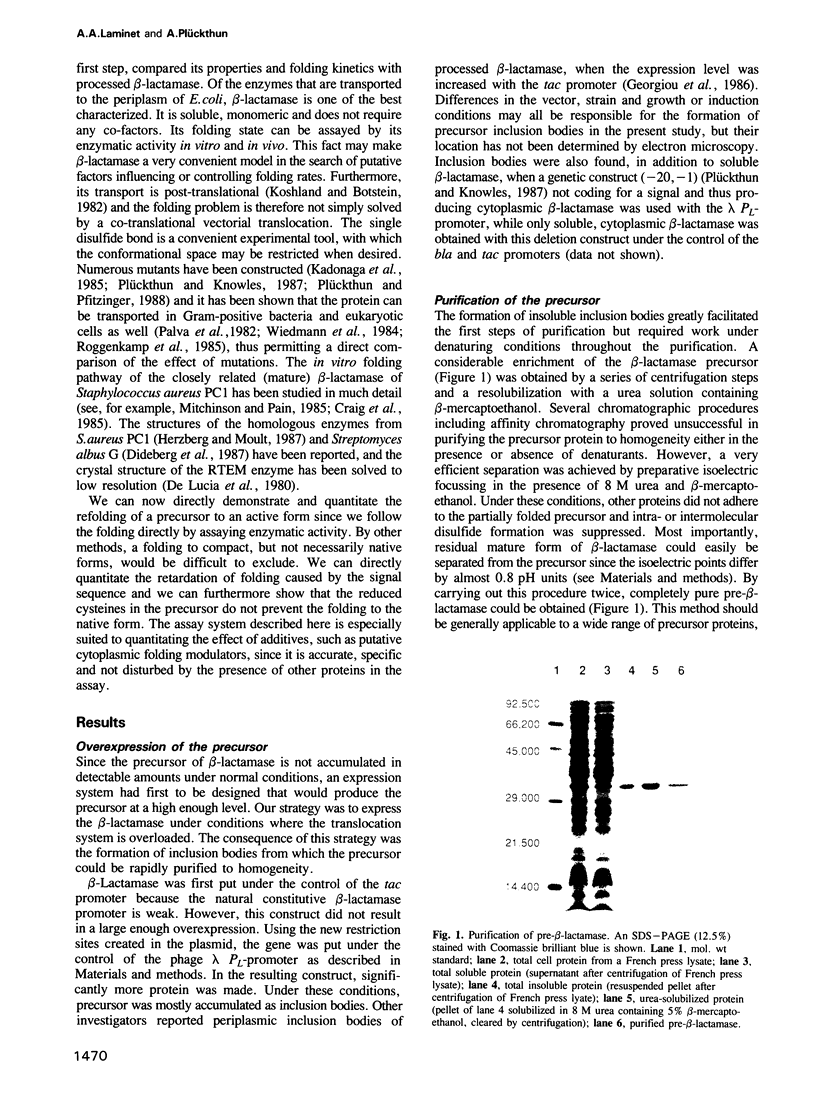


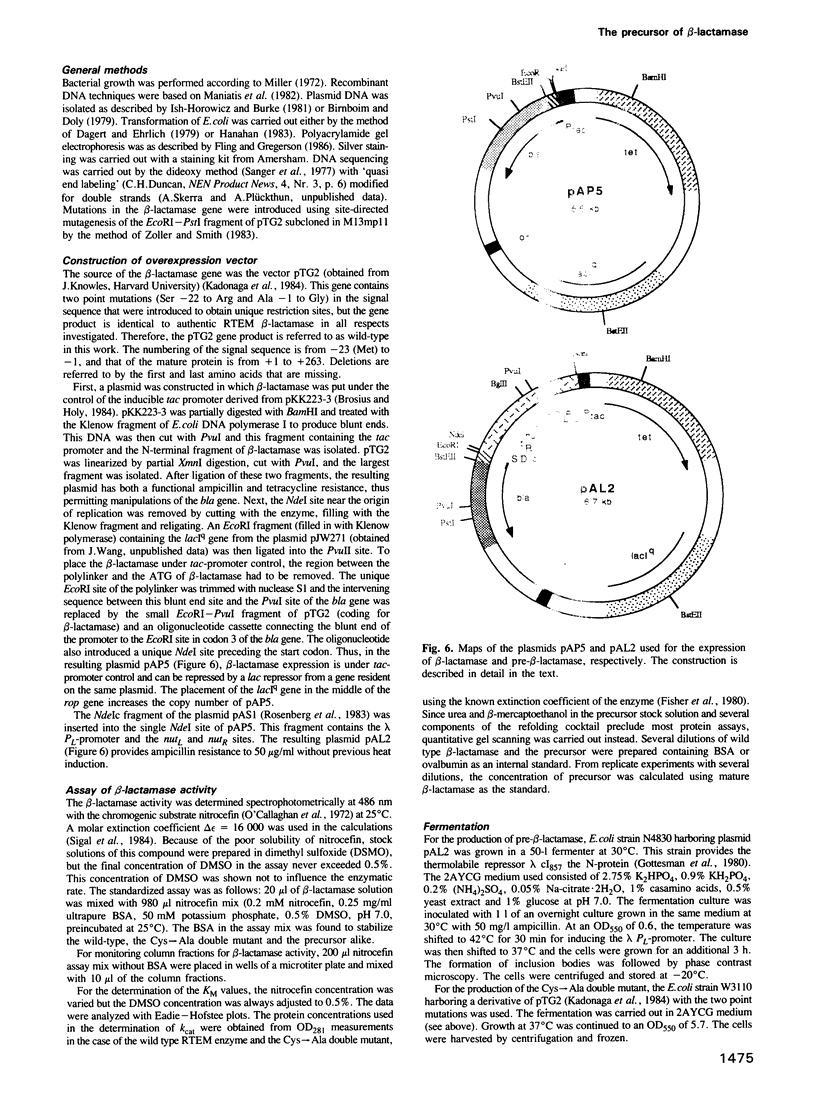


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ainger K. J., Meyer D. I. Translocation of nascent secretory proteins across membranes can occur late in translation. EMBO J. 1986 May;5(5):951–955. doi: 10.1002/j.1460-2075.1986.tb04308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Anfinsen C. B. Principles that govern the folding of protein chains. Science. 1973 Jul 20;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Benson S. A., Hall M. N., Silhavy T. J. Genetic analysis of protein export in Escherichia coli K12. Annu Rev Biochem. 1985;54:101–134. doi: 10.1146/annurev.bi.54.070185.000533. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs M. S., Gierasch L. M. Molecular mechanisms of protein secretion: the role of the signal sequence. Adv Protein Chem. 1986;38:109–180. doi: 10.1016/s0065-3233(08)60527-6. [DOI] [PubMed] [Google Scholar]
- Brosius J., Holy A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6929–6933. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright S. J., Waley S. G. Purification of beta-lactamases by affinity chromatography on phenylboronic acid-agarose. Biochem J. 1984 Jul 15;221(2):505–512. doi: 10.1042/bj2210505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirico W. J., Waters M. G., Blobel G. 70K heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988 Apr 28;332(6167):805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- Collier D. N., Bankaitis V. A., Weiss J. B., Bassford P. J., Jr The antifolding activity of SecB promotes the export of the E. coli maltose-binding protein. Cell. 1988 Apr 22;53(2):273–283. doi: 10.1016/0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- Cover W. H., Ryan J. P., Bassford P. J., Jr, Walsh K. A., Bollinger J., Randall L. L. Suppression of a signal sequence mutation by an amino acid substitution in the mature portion of the maltose-binding protein. J Bacteriol. 1987 May;169(5):1794–1800. doi: 10.1128/jb.169.5.1794-1800.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig S., Hollecker M., Creighton T. E., Pain R. H. Single amino acid mutations block a late step in the folding of beta-lactamase from Staphylococcus aureus. J Mol Biol. 1985 Oct 20;185(4):681–687. doi: 10.1016/0022-2836(85)90053-1. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Experimental studies of protein folding and unfolding. Prog Biophys Mol Biol. 1978;33(3):231–297. doi: 10.1016/0079-6107(79)90030-0. [DOI] [PubMed] [Google Scholar]
- Crooke E., Brundage L., Rice M., Wickner W. ProOmpA spontaneously folds in a membrane assembly competent state which trigger factor stabilizes. EMBO J. 1988 Jun;7(6):1831–1835. doi: 10.1002/j.1460-2075.1988.tb03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
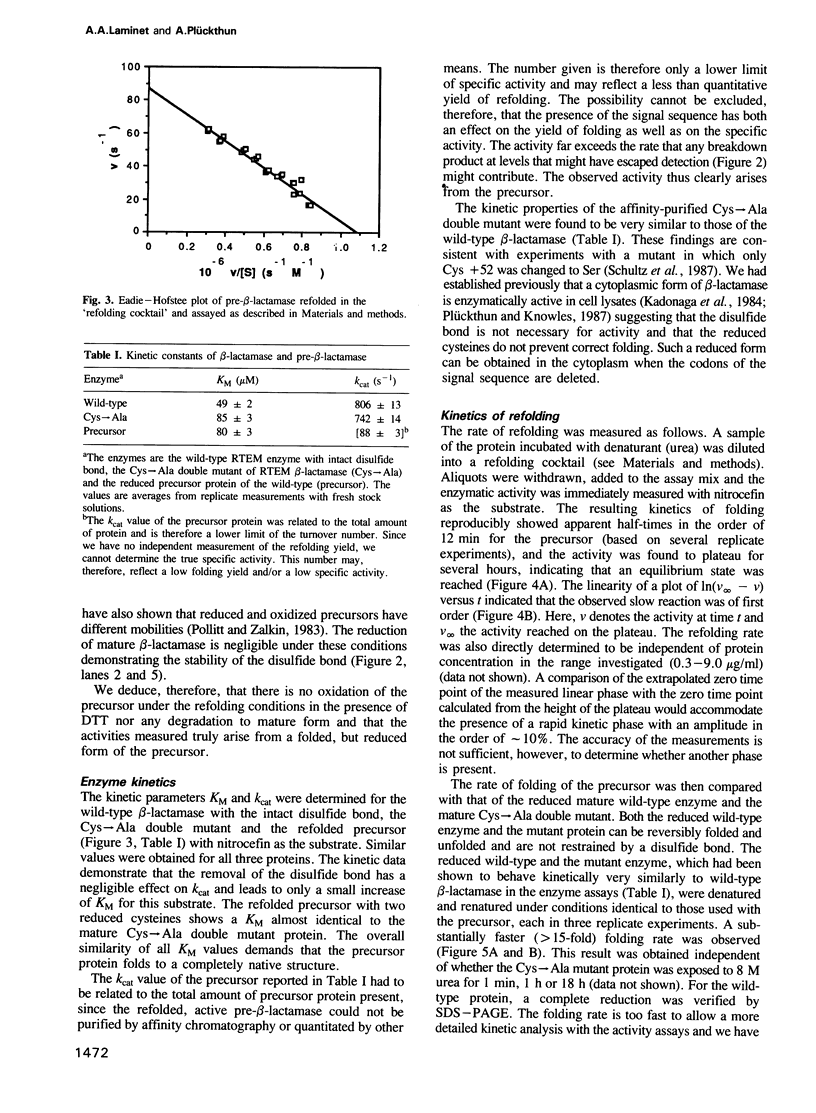
- Crooke E., Guthrie B., Lecker S., Lill R., Wickner W. ProOmpA is stabilized for membrane translocation by either purified E. coli trigger factor or canine signal recognition particle. Cell. 1988 Sep 23;54(7):1003–1011. doi: 10.1016/0092-8674(88)90115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke E., Wickner W. Trigger factor: a soluble protein that folds pro-OmpA into a membrane-assembly-competent form. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5216–5220. doi: 10.1073/pnas.84.15.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- DeLucia M. L., Kelly J. A., Mangion M. M., Moews P. C., Knox J. R. Tertiary and secondary structure analysis of penicillin-binding proteins. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):374–376. [PubMed] [Google Scholar]
- Deshaies R. J., Koch B. D., Werner-Washburne M., Craig E. A., Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988 Apr 28;332(6167):800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- Dideberg O., Charlier P., Wéry J. P., Dehottay P., Dusart J., Erpicum T., Frère J. M., Ghuysen J. M. The crystal structure of the beta-lactamase of Streptomyces albus G at 0.3 nm resolution. Biochem J. 1987 Aug 1;245(3):911–913. doi: 10.1042/bj2450911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M., Hwang S., Schatz G. Unfolding and refolding of a purified precursor protein during import into isolated mitochondria. EMBO J. 1988 Apr;7(4):1139–1145. doi: 10.1002/j.1460-2075.1988.tb02923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M., Schatz G. Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature. 1986 Jul 17;322(6076):228–232. doi: 10.1038/322228a0. [DOI] [PubMed] [Google Scholar]
- Eilers M., Schatz G. Protein unfolding and the energetics of protein translocation across biological membranes. Cell. 1988 Feb 26;52(4):481–483. doi: 10.1016/0092-8674(88)90458-8. [DOI] [PubMed] [Google Scholar]
- Endo T., Schatz G. Latent membrane perturbation activity of a mitochondrial precursor protein is exposed by unfolding. EMBO J. 1988 Apr;7(4):1153–1158. doi: 10.1002/j.1460-2075.1988.tb02925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
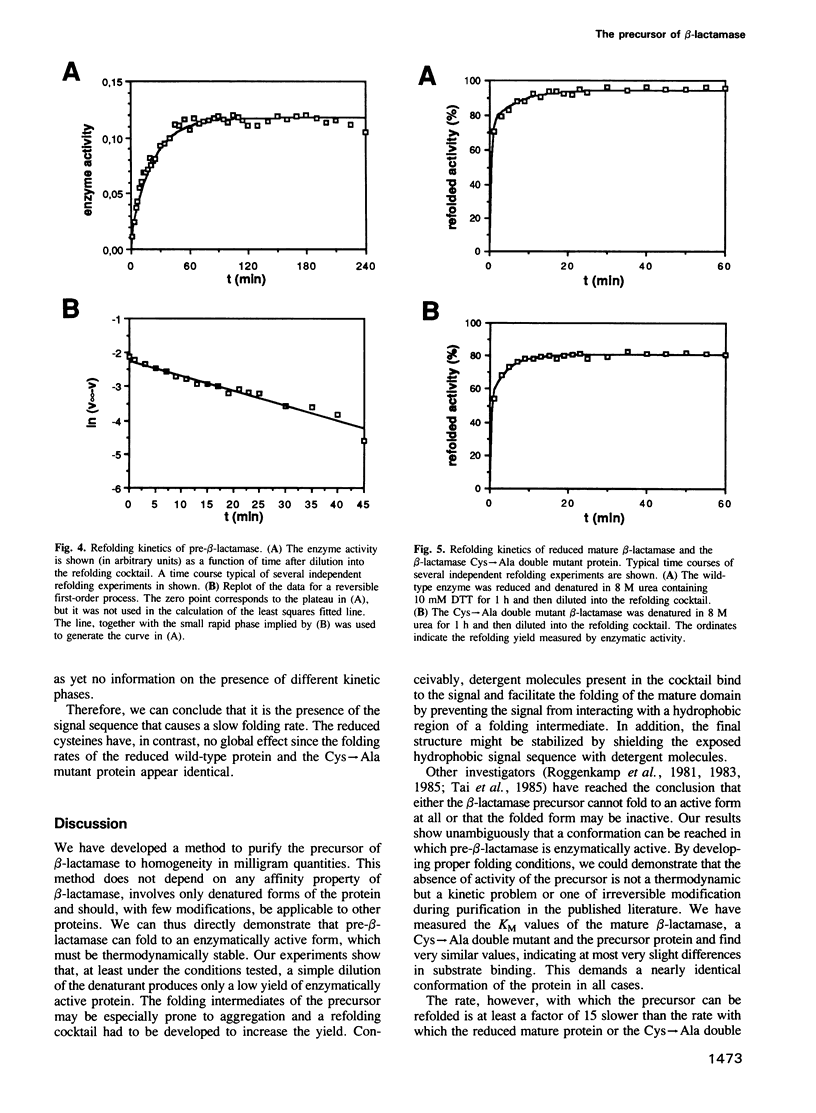
- Fisher J., Belasco J. G., Khosla S., Knowles J. R. beta-Lactamase proceeds via an acyl-enzyme intermediate. Interaction of the Escherichia coli RTEM enzyme with cefoxitin. Biochemistry. 1980 Jun 24;19(13):2895–2901. doi: 10.1021/bi00554a012. [DOI] [PubMed] [Google Scholar]
- Fitts R., Reuveny Z., van Amsterdam J., Mulholland J., Botstein D. Substitution of tyrosine for either cysteine in beta-lactamase prevents release from the membrane during secretion. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8540–8543. doi: 10.1073/pnas.84.23.8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fling S. P., Gregerson D. S. Peptide and protein molecular weight determination by electrophoresis using a high-molarity tris buffer system without urea. Anal Biochem. 1986 May 15;155(1):83–88. doi: 10.1016/0003-2697(86)90228-9. [DOI] [PubMed] [Google Scholar]
- Georgiou G., Telford J. N., Shuler M. L., Wilson D. B. Localization of inclusion bodies in Escherichia coli overproducing beta-lactamase or alkaline phosphatase. Appl Environ Microbiol. 1986 Nov;52(5):1157–1161. doi: 10.1128/aem.52.5.1157-1161.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M. E., Adhya S., Das A. Transcription antitermination by bacteriophage lambda N gene product. J Mol Biol. 1980 Jun 15;140(1):57–75. doi: 10.1016/0022-2836(80)90356-3. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Herzberg O., Moult J. Bacterial resistance to beta-lactam antibiotics: crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.5 A resolution. Science. 1987 May 8;236(4802):694–701. doi: 10.1126/science.3107125. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke R. Folding and association of proteins. Prog Biophys Mol Biol. 1987;49(2-3):117–237. doi: 10.1016/0079-6107(87)90011-3. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Gautier A. E., Straus D. R., Charles A. D., Edge M. D., Knowles J. R. The role of the beta-lactamase signal sequence in the secretion of proteins by Escherichia coli. J Biol Chem. 1984 Feb 25;259(4):2149–2154. [PubMed] [Google Scholar]
- Kadonaga J. T., Plückthun A., Knowles J. R. Signal sequence mutants of beta-lactamase. J Biol Chem. 1985 Dec 25;260(30):16192–16199. [PubMed] [Google Scholar]
- Koshland D., Botstein D. Evidence for posttranslational translocation of beta-lactamase across the bacterial inner membrane. Cell. 1982 Oct;30(3):893–902. doi: 10.1016/0092-8674(82)90294-x. [DOI] [PubMed] [Google Scholar]
- Lill R., Crooke E., Guthrie B., Wickner W. The "trigger factor cycle" includes ribosomes, presecretory proteins, and the plasma membrane. Cell. 1988 Sep 23;54(7):1013–1018. doi: 10.1016/0092-8674(88)90116-x. [DOI] [PubMed] [Google Scholar]
- Melling J., Scott G. K. Preparation of gram quantities of a purified R-factor-mediated penicillinase from Escherichia coli strain W3310. Biochem J. 1972 Nov;130(1):55–62. doi: 10.1042/bj1300055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D. I. Signal recognition particle (SRP) does not mediate a translational arrest of nascent secretory proteins in mammalian cell-free systems. EMBO J. 1985 Aug;4(8):2031–2033. doi: 10.1002/j.1460-2075.1985.tb03888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsky A., Summers R. G., Knowles J. R. Secretion of beta-lactamase into the periplasm of Escherichia coli: evidence for a distinct release step associated with a conformational change. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4180–4184. doi: 10.1073/pnas.83.12.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchinson C., Pain R. H. Effects of sulphate and urea on the stability and reversible unfolding of beta-lactamase from Staphylococcus aureus. Implications for the folding pathway of beta-lactamase. J Mol Biol. 1985 Jul 20;184(2):331–342. doi: 10.1016/0022-2836(85)90384-5. [DOI] [PubMed] [Google Scholar]
- Müller M., Blobel G. In vitro translocation of bacterial proteins across the plasma membrane of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7421–7425. doi: 10.1073/pnas.81.23.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan C. H., Morris A., Kirby S. M., Shingler A. H. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972 Apr;1(4):283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. Protein secretion in Escherichia coli. Annu Rev Microbiol. 1985;39:615–648. doi: 10.1146/annurev.mi.39.100185.003151. [DOI] [PubMed] [Google Scholar]
- Palva I., Sarvas M., Lehtovaara P., Sibakov M., Käriäinen L. Secretion of Escherichia coli beta-lactamase from Bacillus subtilis by the aid of alpha-amylase signal sequence. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5582–5586. doi: 10.1073/pnas.79.18.5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Liu G., Topping T. B., Cover W. H., Randall L. L. Modulation of folding pathways of exported proteins by the leader sequence. Science. 1988 Feb 26;239(4843):1033–1035. doi: 10.1126/science.3278378. [DOI] [PubMed] [Google Scholar]
- Plückthun A., Knowles J. R. The consequences of stepwise deletions from the signal-processing site of beta-lactamase. J Biol Chem. 1987 Mar 25;262(9):3951–3957. [PubMed] [Google Scholar]
- Plückthun A., Pfitzinger I. Membrane-bound beta-lactamase forms in Escherichia coli. J Biol Chem. 1988 Oct 5;263(28):14315–14322. [PubMed] [Google Scholar]
- Pollitt S., Zalkin H. Role of primary structure and disulfide bond formation in beta-lactamase secretion. J Bacteriol. 1983 Jan;153(1):27–32. doi: 10.1128/jb.153.1.27-32.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell. 1986 Sep 12;46(6):921–928. doi: 10.1016/0092-8674(86)90074-7. [DOI] [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J. Export of protein in bacteria. Microbiol Rev. 1984 Dec;48(4):290–298. doi: 10.1128/mr.48.4.290-298.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall L. L. Translocation of domains of nascent periplasmic proteins across the cytoplasmic membrane is independent of elongation. Cell. 1983 May;33(1):231–240. doi: 10.1016/0092-8674(83)90352-5. [DOI] [PubMed] [Google Scholar]
- Roggenkamp R., Dargatz H., Hollenberg C. P. Precursor of beta-lactamase is enzymatically inactive. Accumulation of the preprotein in Saccharomyces cerevisiae. J Biol Chem. 1985 Feb 10;260(3):1508–1512. [PubMed] [Google Scholar]
- Roggenkamp R., Hoppe J., Hollenberg C. P. Specific processing of the bacterial beta-lactamase precursor in Saccharomyces cerevisiae. J Cell Biochem. 1983;22(3):141–149. doi: 10.1002/jcb.240220303. [DOI] [PubMed] [Google Scholar]
- Roggenkamp R., Kustermann-Kuhn B., Hollenberg C. P. Expression and processing of bacterial beta-lactamase in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4466–4470. doi: 10.1073/pnas.78.7.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Ho Y. S., Shatzman A. The use of pKc30 and its derivatives for controlled expression of genes. Methods Enzymol. 1983;101:123–138. doi: 10.1016/0076-6879(83)01009-5. [DOI] [PubMed] [Google Scholar]
- Rothblatt J. A., Meyer D. I. Secretion in yeast: translocation and glycosylation of prepro-alpha-factor in vitro can occur via an ATP-dependent post-translational mechanism. EMBO J. 1986 May;5(5):1031–1036. doi: 10.1002/j.1460-2075.1986.tb04318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele G., Jacoby R. Conformational changes associated with proteolytic processing of presecretory proteins allow glutathione-catalyzed formation of native disulfide bonds. J Biol Chem. 1982 Oct 25;257(20):12277–12282. [PubMed] [Google Scholar]
- Scheele G., Jacoby R. Proteolytic processing of presecretory proteins is required for development of biological activities in pancreatic exocrine proteins. J Biol Chem. 1983 Feb 10;258(3):2005–2009. [PubMed] [Google Scholar]
- Schultz S. C., Dalbadie-McFarland G., Neitzel J. J., Richards J. H. Stability of wild-type and mutant RTEM-1 beta-lactamases: effect of the disulfide bond. Proteins. 1987;2(4):290–297. doi: 10.1002/prot.340020405. [DOI] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Tsunasawa S. Methionine or not methionine at the beginning of a protein. Bioessays. 1985 Jul;3(1):27–31. doi: 10.1002/bies.950030108. [DOI] [PubMed] [Google Scholar]
- Siegel V., Walter P. Elongation arrest is not a prerequisite for secretory protein translocation across the microsomal membrane. J Cell Biol. 1985 Jun;100(6):1913–1921. doi: 10.1083/jcb.100.6.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal I. S., DeGrado W. F., Thomas B. J., Petteway S. R., Jr Purification and properties of thiol beta-lactamase. A mutant of pBR322 beta-lactamase in which the active site serine has been replaced with cysteine. J Biol Chem. 1984 Apr 25;259(8):5327–5332. [PubMed] [Google Scholar]
- Sjöström M., Wold S., Wieslander A., Rilfors L. Signal peptide amino acid sequences in Escherichia coli contain information related to final protein localization. A multivariate data analysis. EMBO J. 1987 Mar;6(3):823–831. doi: 10.1002/j.1460-2075.1987.tb04825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai P. C., Zyk N., Citri N. In situ detection of beta-lactamase activity in sodium dodecyl sulfate-polyacrylamide gels. Anal Biochem. 1985 Jan;144(1):199–203. doi: 10.1016/0003-2697(85)90105-8. [DOI] [PubMed] [Google Scholar]
- Vestweber D., Schatz G. Point mutations destabilizing a precursor protein enhance its post-translational import into mitochondria. EMBO J. 1988 Apr;7(4):1147–1151. doi: 10.1002/j.1460-2075.1988.tb02924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Gilmore R., Blobel G. Protein translocation across the endoplasmic reticulum. Cell. 1984 Aug;38(1):5–8. doi: 10.1016/0092-8674(84)90520-8. [DOI] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W. T., Lodish H. F. Multiple mechanisms of protein insertion into and across membranes. Science. 1985 Oct 25;230(4724):400–407. doi: 10.1126/science.4048938. [DOI] [PubMed] [Google Scholar]
- Wickner W. Mechanisms of membrane assembly: general lessons from the study of M13 coat protein and Escherichia coli leader peptidase. Biochemistry. 1988 Feb 23;27(4):1081–1086. doi: 10.1021/bi00404a001. [DOI] [PubMed] [Google Scholar]
- Wiedmann M., Huth A., Rapoport T. A. Xenopus oocytes can secrete bacterial beta-lactamase. Nature. 1984 Jun 14;309(5969):637–639. doi: 10.1038/309637a0. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]