Abstract
The nucleotide composition of genomes undergoes dramatic variations among all three kingdoms of life. GC content, an important characteristic for a genome, is related to many important functions, and therefore GC content and its distribution are routinely reported for sequenced genomes. Traditionally, GC content distribution is assessed by computing GC contents in windows that slide along the genome. Disadvantages of this routinely used window-based method include low resolution and low sensitivity. Additionally, different window sizes result in different GC content distribution patterns within the same genome. We proposed a windowless method, the GC profile, for displaying GC content variations across the genome. Compared to the window-based method, the GC profile has the following advantages: 1) higher sensitivity, because of variation-amplifying procedures; 2) higher resolution, because boundaries between domains can be determined at one single base pair; 3) uniqueness, because the GC profile is unique for a given genome and 4) the capacity to show both global and regional GC content distributions. These characteristics are useful in identifying horizontally-transferred genomic islands and homogenous GC-content domains. Here, we review the applications of the GC profile in identifying genomic islands and genome segmentation points, and in serving as a platform to integrate with other algorithms for genome analysis. A web server generating GC profiles and implementing relevant genome segmentation algorithms is available at: www.zcurve.net.
Keywords: GC profile, Genomic island, Genome segmentation.
1. INTRODUCTION
The nucleotide composition of genomes undergoes dramatic variations among all three kingdoms of life. GC content, an important characteristic for a genome, is related to many important genome functions [1]. Therefore GC content and its distribution along the genome are routinely reported for sequenced genomes.
In prokaryotic genomes, assessing GC content change is useful in identifying horizontally-transferred genes, which usually have distinct GC content from the host genome. A growing body of evidence suggests that horizontal gene transfer is a universal event throughout bacterial evolution [2-4]. Genomic islands comprise relatively large genomic regions, e.g., 10-200 Kb, which are acquired by core genomes via horizontal gene transfer. Depending on their functions, genomic islands can be classified as pathogenicity islands, metabolic islands, secretion islands, resistance islands and symbiosis islands [5-7]. For instance, pathogenicity islands carry genes encoding one or more virulence factors including adhesins, toxins, invasins, capsule synthesis, iron uptake and effectors secreted by the type III and IV secretion apparatuses [5, 8].
In eukaryotic genomes, GC content is related to isochore structures, a phenomenon that the genome is organized into mosaics of long domains of relatively homogenous GC content [9]. The isochore structure has been shown to correlate with many genomic features, such as gene density [10, 11], intron length [12], replication timing [13], recombination frequency [14], methylation pattern [15], repeat elements [16], and transposable elements [17]. Thus, determining the underlying mechanism driving the evolution of isochores is helpful in understanding the organization of genomes.
Traditionally, GC content distribution of a genome is usually assessed by computing GC content in sliding windows moving along the genome [11]. The disadvantage of this routinely-used window-based method is that the resolution is low, e.g., the method is not sufficiently sensitive to detect small changes in GC content. In addition, the distribution pattern of GC content obtained is largely dependent on the window size, and therefore different window sizes lead to different patterns.
We proposed a windowless method to calculate GC content, called the GC profile [18]. In contrast to the traditionally used sliding-window based method, the GC profile is a windowless method that has high resolution. Because no window is used, the GC profile is unique for a given genome. Importantly, this method shows a global GC content distribution. Therefore, the GC profile method has been successfully used for identification of genomic islands and genome segmentation points, and in comparative genomics.
2. THE GC PROFILE METHOD, A WINDOWLESS METHOD IN CALCULATING GC CONTENT
Based on the Z-curve, any DNA sequence can be uniquely described by three independent distributions, i.e., those of the bases of purine/pyrimidine (xn), amino/keto (yn), and weak/strong hydrogen bonds (zn). In particular, zn displays the distribution of bases of GC/AT types along the sequence, which is calculated as follows [19, 20].
| (1) |
where An,Cn, Gn and Tn are the cumulative numbers of the bases A, C, G and T, respectively, Based on zn, GC content can be calculated using a windowless technique [18]. The curve of zn ~ n is fitted by a straight line using the least square technique, z = kn, where (z, n) is the coordinate of a point on the fitted straight line and k is its slope. Then we have Let denote the average GC content within a region Δn in a sequence, we find
| (2) |
where is the average slope of the z’ curve within the region Δn, which is a DNA segment, e.g., a genomic island. The above method is called the windowless technique for GC content computation [18], and the z’ curve is also called the cumulative GC profile. In the z’ curve, a jump counter-intuitively denotes a drop in GC content, and therefore, we define GC profile as negative z’. Strait lines of the GC profile denote a region with relatively homogenous GC content. An upward line indicates an abrupt increase in GC content, whereas a drop indicates an abrupt decrease in GC content.
3. IDENTIFICATION OF GENOMIC ISLANDS BY THE GC PROFILE METHOD
3.1. Genomic Islands in Rhodopseudomonas Palustris
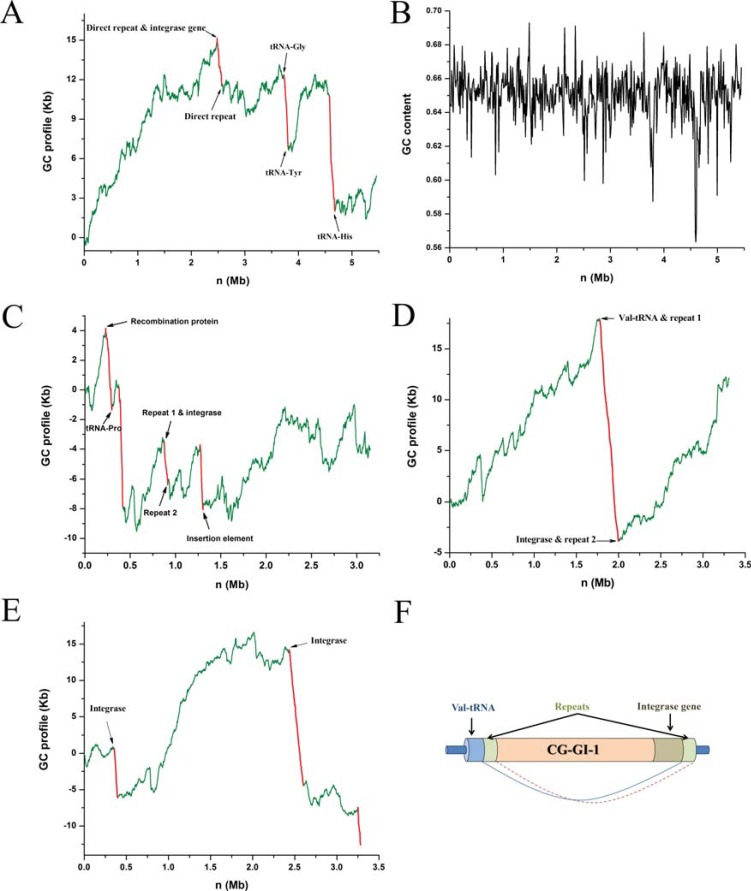
R. palustris, a purple photosynthetic bacterium, is extremely metabolically versatile, as it can use both light and chemical inorganic compounds for energy, and can grow with or without oxygen. Therefore it is a model to study how organisms integrate metabolic modules in response to environmental changes [21]. In the R. palustris genome, based on the GC profile, we have identified three genomic islands, which are associated with three abrupt drops in the GC profile (Fig. 1A) [22], indicating that they have an abrupt decrease of GC content, while this is not illustrative based on GC content distribution using 20 Kb windows (Fig. 1B). The identified regions have many conserved features of genomic islands. For instance, the averaged GC content of these genomics islands is 0.616, much lower than that of the genome, 0.652. RPAGI-1 is flanked by two directly repeated sequences. An integrase gene is located at a site following the repeat at the 5’ junction. RPAGI-2 is flanked by two t-RNA genes and RPAGI-3 has a t-RNA gene at its 3’ junction.
Fig. (1).
Identification of horizontally-transferred genomic islands by using GC profiles. A) GC profiles and B) the GC content distribution based on 20Kb sliding windows for the genome of Rhodopseudomonas palustris. GC profiles of C) Corynebacterium efficiens, D) Corynebacterium glutamicum and E) Vibrio vulnificus CMCP6. F) A typical structure of the genomic island. The schematic representation of CGGI-1, a genomic island in C. glutamicum. Figure not drawn to scale. Segments of GC profiles for genomic island are shown in red, and those for host gnomes are shown in green.
These three genomic islands appear to play a role in facilitating this bacterium to adapt to versatile environmental changes. For instance, the only 2 sets of type IV secretion genes for conjugal transfer of DNA are all located in the genomic islands. Because each of the two gene sets has all the 10 components required for a type IV secretion pathway, and because these two type IV secretion systems are the only such systems in the genome, they are very likely to be functional. RPAGI-3 has 5 multidrug efflux transport related genes and RPAGI-1 has two genes that encode an arsenate reductase pump modifier and an arsenical pump membrane protein. The presence of metal efflux transporters, multidrug efflux pumps and type IV secretion systems in horizontally-transferred genomic islands helps to explain why R. palustris survives in a great variety of environments by acquiring necessary nutrients while resisting toxic compounds [22].
3.2. Genomic Islands in Corynebacterium Efficiens
C. efficiens, a close relative of C. glutamicum, is widely used for fermentative production of amino acids on an industrial scale [23]. C. efficiens can still grow and produce glutamate at 40°C or above, whereas C. glutamicum cannot. The remarkable thermostability of C. efficiens is a useful trait from an industrial viewpoint as it reduces the considerable cost of cooling needed to dissipate the heat generated during glutamate fermentation [24]. The complete genome sequence of C. efficiens provides an opportunity to investigate the mechanisms for the high thermostability of C. efficiens at the sequence level.
Using the GC profile, we have identified 4 genomic islands (Fig. 1C), which have multiple conserved genomic-island specific features, such as low GC content, biased codon usage, the presence of mobile genes, direct repeats and tRNA loci at junctions [25]. Nishio et al. [24] compared the thermal stability of 13 orthologous enzymes of C. efficiens and C. glutamicum. Although most of the tested enzymes from C. efficiens were more thermostable than their orthologs of C. glutamicum, unexpectedly, however, an aspartate kinase in C. efficiens was found to be comparatively less stable than in C. glutamicum [24]. We found that the gene encoding this enzyme from C. efficiens is located in one of the identified genomic islands. Therefore, one explanation for its reduced stability is that due to the recent horizontal transfer of this gene, the adaptive mutations of the C. efficiens genome have not yet been performed sufficiently to increase thermal stability of this aspartate kinase, and indeed, it lacks the biased amino acid substitutions that increase protein stability [25].
3.3. Genomic Islands in Corynebacterium Glutamicum
C. glutamicum is a Gram-positive soil bacterium that has the ability to excrete large amounts of certain amino acids, and therefore this bacterium is most widely used for fermentative production of amino acids on an industrial scale [26]. We identified 2 genomic islands in the genome of C. glutamicum, and its GC profile shows 2 drops that indicate an abrupt decrease in GC content [27] (Fig. 1D). The CGGI-1 has a GC content of 0.49, whereas that of the genome is 0.55, and it is flanked by repeat elements of about 500 bp. It is noteworthy that the 500 bp repeat elements surrounding a genomic island are considered to be long. The tRNA gene has been repeatedly found to be the integration site of genomic islands, and indeed, a cluster of 7 tRNA genes at the 5’ junction immediately follows the repeat element, and the other repeat is preceded by an integrase gene [27]. Therefore CGGI-1 has a typical structure of the genomic island (Fig. 1F).
3.4. Genomic Islands in Vibrio vulnificus CMCP6
V. vulnificus is a Gram-negative, curved rod-shaped bacterium that has been known to cause both foodborne and wound infections throughout the world [28]. In the United States, the diseases caused by this bacterium make it the leading cause of seafood-associated fatalities [29]. These diseases include invasive septicemia, with a mortality rate of approximately 60% [29]. We have identified 3 genomic islands VVGI-1, VVGI-2 and VVGI-3, which are associated with an abrupt decrease in GC content (drops) in the GC profile for V. vulnificus (Fig. 1E). VVGI-1 harbors two genes encoding invasion-associated proteins (cell wall-associated hydrolases), which can be involved in cellular processes, such as cell-wall turnover, motility, protein secretion and pathogenicity [30]. VVGI-2 harbors a gene encoding hemolysin, which denotes a class of toxins that have lytic activity on red blood cells (RBC). Hemolysins, which can form pores in the membranes of many types of cells, play a role in colonization and multiplication in the shellfish by breaking down the host tissues. Therefore, VVGI-2 is a potential pathogenicity island. VVGI-3 has a gene coding for a multidrug resistance efflux pump, which is a system that enables the bacteria to expel a wide variety of antimicrobial compounds, suggesting the VVGI-3 to be a resistance island [27].
4. USE OF THE GC PROFILE ON ISOCHORE IDENTIFICATION
Based on density gradient ultracentrifugation experiments, more than 3 decades ago, Bernardi and coworkers discovered that mammalian genomes are organized into mosaics with fairly constant average GC content over scales of hundreds of kilobases and by relatively abrupt changes to another fairly constant GC content region, referred to as the isochore structure [31], and many important biological properties, such as replication timing and recombination rates, are linked to the isochore structure of the genome [9].
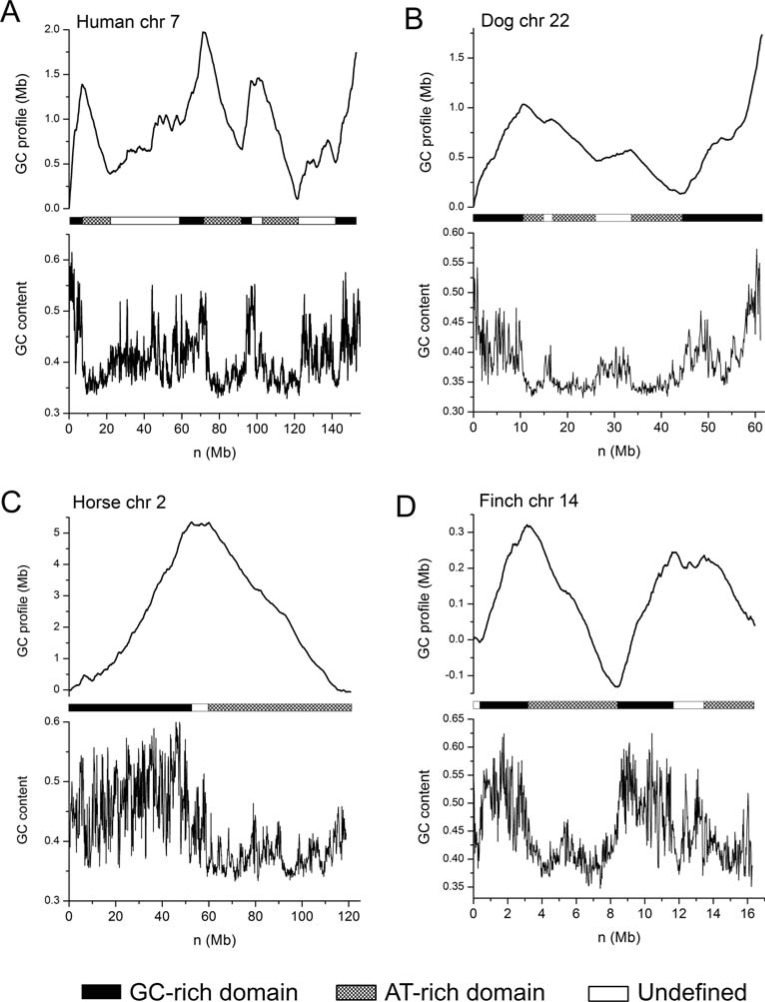
The GC profile method provides an effective way to reveal GC content organization, and is therefore useful in identifying isochore structures of the genome. (Fig. 2) shows GC profiles for some typical isochores in human, horse, dog and Finch genomes. By visual inspection, it can be seen that in human chromosome 7, there are clearly low-GC content and high-GC content regions that alternatively appear along the genome (Fig. 2A). For example, the first 20 Mb region can be divided into 2 regions with a 7 Mb GC-rich domain and a 13 Mb GC-poor domain, with GC contents being 0.50 and 0.36, respectively (Fig. 2A). Similar organization is also present in dog genome (Fig. 2B), horse genome (Fig. 2C) and Finch genome (Fig. 2D). For instance, the horse chromosome 2 can be roughly divided into 2 domains. The first GC rich domain has an average GC content of 0.47, and the GC-poor domain has an average GC content of 0.38, resulting in a GC content difference of 0.9. Both domains are about 50 Mb in size. Likewise, the finch chromosome 14 can be divided into 4 domains with 2 being GC-rich and the other 2 being GC-poor, and GC-rich and GC-poor domains appear alternatively throughout the genome. Based on GC profile, statistical methods can be used to examine homogeneity and to identify genome segmentation points. It should be noted that the GC profile is unique for a given genome, but isochores are not, due to different definitions.
Fig. (2).
Isochore structures revealed by GC profiles. GC profiles (upper panel) and GC content distribution based on 100 Kb sliding windows (lower panel) for A) human chromosome 7, B) dog chromosome 22, C) horse chromosome 2 and D) finch chromosome 14.
When the draft human genome sequence became available, it was hoped that human isochores could be determined at the sequence level, unexpectedly, however, no isochore was identified in the human genome [11]. Specifically, the hypothesis of homogeneity was rejected in each 300-Kb window in the human genome, resulting in a debate about whether or not isochores exist [32]. We think the debate was due to a misunderstanding of the isochore concept. The homogeneity of GC content should be considered to be relative, rather than absolute. That is, homogeneity of the isochore GC content should be considered by taking into account of GC content variations of the entire chromosome, rather than independently performing homogeneity tests within sliding windows of a fixed size, e.g., 300 Kb. The GC profile method shows a global GC content distribution, in contrast to window-based methods, and therefore it is useful in identifying isochore structure, and in identifying global GC content variations in the genome.
5. INTEGRATION WITH OTHER ALGORITHMS ON THE PLATFORM OF THE GC PROFILE
5.1. Integration of the GC Profile into MobilomeFINDER
MobilomeFINDER is a useful bioinformatics server that facilitates high-throughput in-silico and experimental discovery of bacterial mobile regions [33]. The analysis tools for tRNA/tmRNA gene contents and contexts (tRNAcc) exploit the Mauve aligner [34] to survey whether tRNA/tmRNA gene sites across multiple genomes are occupied by anomalous strain-specific DNA segments. The GC profile method has been integrated into MobilomeFINDER to locate putative genomic islands [33]. For instance, most alien DNA fragments appear to map to a limited number of species-specific insertion ‘hotspots’ in bacterial genomes, with the most common hotspots being tRNA/tmRNA gene sites [35]. Most insertion hotspots occur in drops (sudden decrease in GC content) of GC profiles.
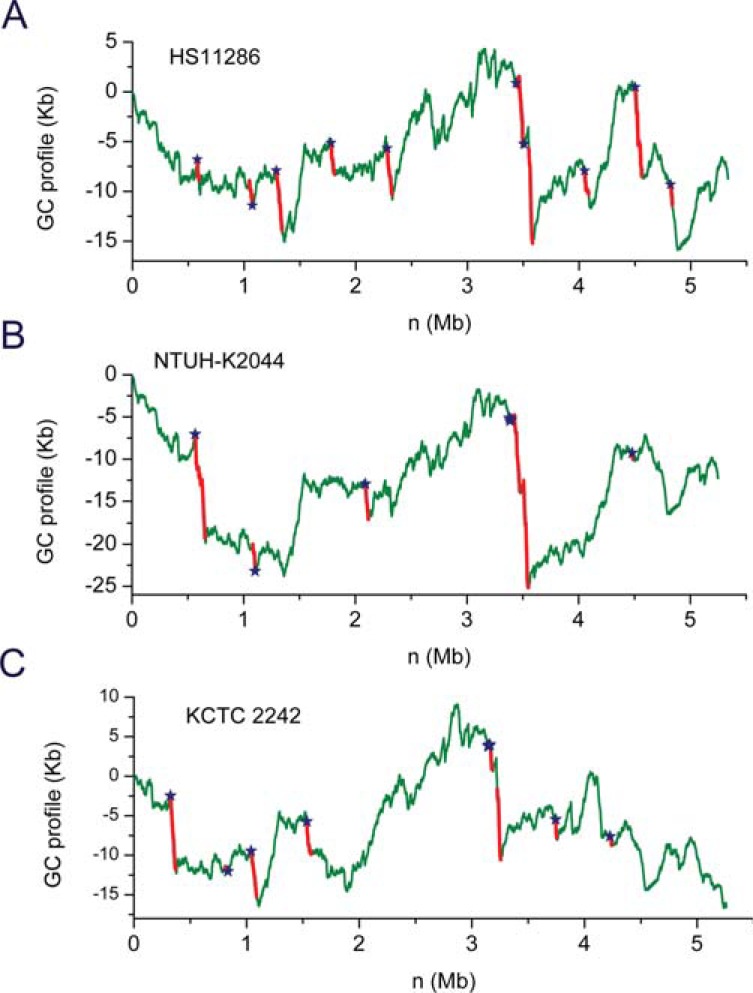
We used MobilomeFINDER to examine genome sequences of 6 Klebsiella pneumoniae strains (MGH 78578, Kp342, NTUH-K2044, HS11286, KCTC2242 and Kp1084), and identified 35 island-like regions that are inserted into 10 tRNA/tmRNA gene hotspots, which are located on drops of GC profiles (Fig. 3). K. pneumoniae strain HS11286 is a carbapenemase-producing clinical isolate obtained from the sputum of a patient [36]. NTUH-K2044 was isolated from a patient with liver abscess and meningitis, and KCTC 2242 is the 2,3-Butanediol-producing strain. In the GC profile of HS11286, two putative integrative conjugative elements (ICEs) were identified [37]: an asn34 tRNA gene-associated, ICEKp1-like element ICEKpnHS11286-1 (62 Kb in size, 52.5% GC content) and a phe55 tRNA gene-associated element ICEKpn HS11286-2 (56 Kb, 50.2% GC content). For the NTUH-K2044 strain, the experimentally reported 76-Kb ICEKp1 [38] was inserted into the asn36 tRNA gene sites while a putative element ICEKpnK2044-2 (79 Kb, 51.4% GC content) was found in the leu82 tRNA gene locus. Therefore, GC profile is useful as a tool that is used in conjunction with other methods to identify horizontally-transferred genes.
Fig. (3).
GC profiles for genomes of the Klebsiella pneumoniae strains A) HS11286, B) NTUH-K2044 and C) KCTC 2242. The GC profile is integrated into MobilomeFINDER, which was used to identify horizontally-transferred genes (red). The tRNA /tmRNA genes are identified as insertion sites (blue stars).
5.2. Integration of GC Profile with Genome Segmentation Algorithms
The genome order index S is defined as S = a2 + c2 + g2 + t2, where a, c, g and t denote the occurrence frequencies of A, C, G and T, respectively, in a genome or a DNA sequence. The genome order index S, derived from the Z-curve theory, is a useful statistical quantity to reflect the compositional characteristics of a genome [39], which can serve as an appropriate divergence measure to quantify the compositional difference between two DNA sequences. Based on GC profile and the genome order index S, a new segmentation algorithm has been developed to partition a given genome or DNA sequence into compositionally distinct domains [40]. Therefore, GC profile can be a platform on which other algorisms are integrated for genome analysis, such as segmentation.
6. USE OF THE GC PROFILE METHOD IN COMPARATIVE GENOMICS
Because the cumulative GC profile has high resolution in displaying GC content changes, it is a useful tool for comparative genomics. For instance, by comparing cumulative GC profiles of the genomes of Bacillus cereus and B. anthracis, 3 genomic islands were identified in the former [41]. Likewise, by comparing cumulative GC profiles of genomes of 2 strains of B. cereus, some genomic islands were identified [42]. By comparing GC profiles of 3 pneumonia pathogens, Chlamydophila pneumoniae, Mycoplasma pneumoniae and Streptococcus pneumoniae, Guo and Wei identified 8 GIs, and 3 of which contain clusters of genes coding for virulence factors [43].
The high sensitivity of GC profiles in detecting GC content change is exemplified by comparative analysis of the GC profiles for genomes of B. cereus ATCC 10987 and B. cereus ATCC 14579. Although GC profiles for both genomes indicate that their overall GC content distributions are similar, each GC profile contains drops that are missing in the other. Indeed, the 2 drops (sharp decreases in GC content) in the GC profile of the B. cereus ATCC 10987 genome are identified as genomic islands, BCEGI-1 and BCEGI-2, which have GC content of 0.31 and 0.32, respectively, much lower than that of the genome, 0.36. Due to low resolution and low sensitivity, these genomic islands could not be identified using the window based method. By comparing the genes and genomic sequences around the 2 genomic islands, their integration sites were determined precisely at the single nucleotide level [42].
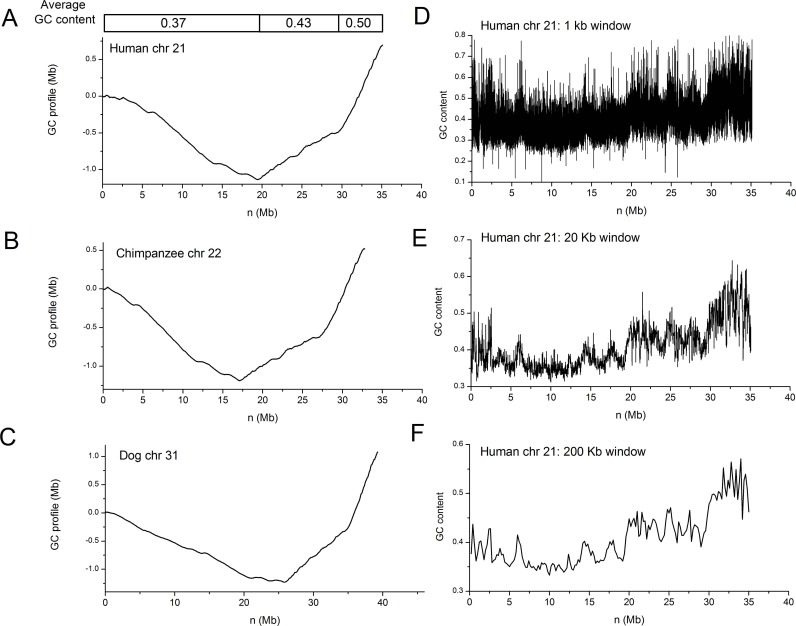
Global as well as regional changes in GC content can be easily grasped by comparing GC profiles. For instance, the GC profile for human chromosome 21 clearly shows that the chromosome is organized into 3 domains, with average GC contents being 0.37, 0.43 and 0.50 (Fig. 4A). It is also apparent that human chromosome 21, chimpanzee chromosome 22 and dog chromosome 31 are homologous to each other (Fig. 4A-C). The GC profile is unique for a chromosome, but the window-based method, when different window sizes are used, results in different GC content distributions. For human chromosome 21, for instance, a small window size (1 Kb) leads to large statistical fluctuation, while large window sizes (200 Kb) average out detailed changes in GC content (Fig. 4D-F).
Fig. (4).
Comparative analysis of GC profiles for chromosomes 21, 22, and 31 for human, chimpanzee and dog, respectively. GC profiles for A) human chromosome 21, B) chimpanzee chromosome 22, C) dog chromosome 31. GC content distributions for human chromosome 21 based on sliding windows of D) 1 Kb, E) 20 Kb and F) 200 Kb. GC content changes are masked by high degree of variations when the window size is small, and GC content changes are averaged when the window size is large.
7. WEB SERVER FOR GENERATING THE GC PROFILE
To facilitate the use of the GC profile and the new genome segmentation algorithm, a web server has been constructed, which is implemented on Apache server and the web interface using Common Gateway Interface (CGI) Perl scripts [44]. The segmentation algorithms are written in C++. The output graphs are generated by gnuplot graphics routine. GC-Profile has a user-friendly and intuitive input interface. Users can choose to paste a sequence into the box or upload the sequence (FASTA format) in a file. By default, GC-Profile generates four files for each job: two tables and two figures. The output web page shows the process of GC-Profile, and provides links to the results of sequence segmentation: (i) coordinates, sizes and G + C contents of the segmented domains as an HTML table; (ii) number, coordinates, segmentation strength, segmentation times and segmented contig of the segmentation points as an HTML table; (iii) the cumulative GC profile and (iv) GC content of the input sequence in PNG format. If upload options are chosen, the density distribution or coordinate points labeled to the GC profile can also be obtained. This web server can be accessed at http://tubic.tju.edu.cn/GC-Profile/, or http://www.zcurve.net then click ‘Genome segmentation’.
8. CONCLUDING REMARKS
Below is a comparison between window-based and window-less methods for displaying GC content distribution.
The GC profile has higher sensitivity, because of the special subtraction procedure, i.e. Equation (3), which amplifies the variation of GC content. Therefore, even an extremely small change in GC content can be detected. This characteristic is especially useful if the difference between GC content of the horizontally-transferred elements and that of the host genome is small.
The GC profile has high resolution. GC content can be calculated at single-base pair resolution. Because of its high sensitivity, regions with GC content changes can be detected, and the boundary can be identified accurately.
The GC profile is unique for a genome, while the window-based method leads to different GC content distribution if the window size is changed.
The GC profile shows a global, as well as regional, GC content change, while the window-based method shows only a regional GC content. This characteristic is especially important for identifying isochores, which, we suggest should be examined in the context of the entire chromosome.
The GC profile is also intuitive in identifying homogenous regions in GC content variation, and this is useful in identifying isochores (Table 1). It is noteworthy that the GC profile is not equal to the distribution curve of GC content itself, but its derivative is proportional to GC content.
Table 1.
Comparison between the window-based method and the windowless method (GC profile) for displaying GC content dis-tributions.
| Window-based Method | Window-less Method (GC Profile) | Note | |
|---|---|---|---|
| Method | Overlapping or non-overlapping sliding windows | Cumulative number of G and C along the genome, the z’ component of Z-curve. | The 2 methods are complementary |
| Sensitivity | Low, because GC content variation is averaged within a window | High, because of the special subtraction procedure that amplifies GC content variation. | The high sensitivity is useful especially for detecting horizontally-transferred genomic islands. |
| Resolution | Low | High, because no window is used, the boundary between domains can be determined at one single nucleotide. | The high resolution proves valuable in identifying genome segmentation points. |
| Uniqueness | Different window size leads to different results | Unique for a given genome | The uniqueness is a useful feature for comparative genomics. |
| Regional or global | Regional, GC content within a window | Global as well as regional, because cumulative GC content is calculated. | The global GC content distribution is a useful characteristic in identifying isochores. |
| Examining homogeneity | Not intuitive | Intuitive, because a straight line denotes a homogenous region, as opposed to GC content variations of the entire genome. Homogeneity can be examined statistically as well. | Examining homogeneity based on the GC profile is helpful in identifying isochores. |
Although the advantages of cumulative GC profiles are highlighted above, we do not mean to replace the window-based method with the GC profile. In contrast, in many cases, the two methods are complementary and should be used together to visualize GC content distribution along genomic sequences.
ACKNOWLEDGEMENTS
RZ was supported in part by a startup fund from Wayne State University. HYO was supported in part by funding from the National Natural Science Foundation of China (31170082). FG was supported in part by funding from the National Natural Science Foundation of China (31171238, 30800642) and the Program for New Century Excellent Talents in University (No. NCET-12-0396).
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Sueoka N. On the genetic basis of variation and heterogeneity of DNA base composition. Proc. Natl. Acad. Sci. U S A. 1962;48:582–592. doi: 10.1073/pnas.48.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, McDonald L, Utterback TR, Malek JA, Linher KD, Garrett MM, Stewart AM, Cotton MD, Pratt MS, Phillips CA, Richardson D, Heidelberg J, Sutton GG, Fleischmann RD, Eisen JA, White O, Salzberg SL, Smith HO, Venter JC, Fraser CM. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399(6734):323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 3.Ochman H. Lateral and oblique gene transfer. Curr. Opin. Genet. Dev. 2001;11(6):616–619. doi: 10.1016/s0959-437x(00)00243-4. [DOI] [PubMed] [Google Scholar]
- 4.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 5.Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 6.Hentschel U, Hacker J. Pathogenicity islands the tip of the iceberg. Microbes Infect. 2001;3(7):545–548. doi: 10.1016/s1286-4579(01)01410-1. [DOI] [PubMed] [Google Scholar]
- 7.Hentschel U, Steinert M, Hacker J. Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol. 2000;8(5):226–231. doi: 10.1016/s0966-842x(00)01758-3. [DOI] [PubMed] [Google Scholar]
- 8.Bi D, Liu L, Tai C, Deng Z, Rajakumar K, Ou HY. SecReT4: a web-based bacterial type IV secretion system resource. Nucleic Acids Res. 2013;41(Database issue):D660–665. doi: 10.1093/nar/gks1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernardi G. The isochore organization of the human genome and its evolutionary history--a review. Gene. 1993;135(1-2):57–66. doi: 10.1016/0378-1119(93)90049-9. [DOI] [PubMed] [Google Scholar]
- 10.Zoubak S, Clay O, Bernardi G. The gene distribution of the human genome. Gene. 1996;174(1):95–102. doi: 10.1016/0378-1119(96)00393-9. [DOI] [PubMed] [Google Scholar]
- 11.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 12.Duret L, Mouchiroud D, Gautier C. Statistical analysis of vertebrate sequences reveals that long genes are scarce in GC-rich isochores. J. Mol. Evol. 1995;40(3):308–317. doi: 10.1007/BF00163235. [DOI] [PubMed] [Google Scholar]
- 13.Tenzen T, Yamagata T, Fukagawa T, Sugaya K, Ando A, Inoko H, Gojobori T, Fujiyama A, Okumura K, Ikemura T. Precise switching of DNA replication timing in the GC content transition area in the human major histocompatibility complex. Mol Cell Biol. 1997;17(7):4043–4050. doi: 10.1128/mcb.17.7.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenbarth I, Vogel G, Krone W, Vogel W, Assum G. An isochore transition in the NF1 gene region coincides with a switch in the extent of linkage disequilibrium. Am. J. Hum. Genet. 2000;67(4):873–880. doi: 10.1086/303085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caccio S, Jabbari K, Matassi G, Guermonprez F, Desgres J, Bernardi G. Methylation patterns in the isochores of vertebrate genomes. Gene. 1997;205(1-2):119–124. doi: 10.1016/s0378-1119(97)00560-x. [DOI] [PubMed] [Google Scholar]
- 16.Soriano P, Meunier-Rotival M, Bernardi G. The distribution of interspersed repeats is nonuniform and conserved in the mouse and human genomes. Proc. Natl. Acad. Sci. U S A. 1983;80(7):1816–1820. doi: 10.1073/pnas.80.7.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9(6):657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 18.Zhang CT, Wang J, Zhang R. A novel method to calculate the G+C content of genomic DNA sequences. J. Biomol. Struct. Dyn. 2001;19(2):333–341. doi: 10.1080/07391102.2001.10506743. [DOI] [PubMed] [Google Scholar]
- 19.Zhang CT, Zhang R. Analysis of distribution of bases in the coding sequences by a diagrammatic technique. Nucleic Acids Res. 1991;19(22):6313–6317. doi: 10.1093/nar/19.22.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang R, Zhang CT. Z-curves, an intuitive tool for visualizing and analyzing the DNA sequences. J. Biomol. Struct. Dyn. 1994;11(4):767–782. doi: 10.1080/07391102.1994.10508031. [DOI] [PubMed] [Google Scholar]
- 21.Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L, Land ML, Pelletier DA, Beatty JT, Lang AS, Tabita FR, Gibson JL, Hanson TE, Bobst C, Torres JL, Peres C, Harrison FH, Gibson J, Harwood CS. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 2004;22(1):55–61. doi: 10.1038/nbt923. [DOI] [PubMed] [Google Scholar]
- 22.Zhang CT, Zhang R. Genomic islands in Rhodopseudomonas palustris. Nat. Biotechnol. 2004;22(9):1078–1079. doi: 10.1038/nbt0904-1078b. [DOI] [PubMed] [Google Scholar]
- 23.Fudou R, Jojima Y, Seto A, Yamada K, Kimura E, Nakamatsu T, Hiraishi A, Yamanaka S. Corynebacterium efficiens sp.nov. a glutamic-acid-producing species from soil and vegetables. Int. J. Syst. Evol. Microbiol. 2002;52(Pt 4):1127–1131. doi: 10.1099/00207713-52-4-1127. [DOI] [PubMed] [Google Scholar]
- 24.Nishio Y, Nakamura Y, Kawarabayasi Y, Usuda Y, Kimura E, Sugimoto S, Matsui K, Yamagishi A, Kikuchi H, Ikeo K, Gojobori T. Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Res. 2003;13(7):1572–1579. doi: 10.1101/gr.1285603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang R, Zhang CT. Genomic islands in the Corynebacterium efficiens genome. Appl. Environ. Microbiol. 2005;71(6):3126–3130. doi: 10.1128/AEM.71.6.3126-3130.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinoshita S, Udaka S, Shimono M. Studies on the amino acid fermentation.Part 1. Production of L-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 2004;50(6):331–343. [PubMed] [Google Scholar]
- 27.Zhang R, Zhang CT. A systematic method to identify genomic islands and its applications in analyzing the genomes of Corynebacterium glutamicum and Vibrio vulnificus CMCP6 chromosome I. Bioinform. 2004;20(5):612–622. doi: 10.1093/bioinformatics/btg453. [DOI] [PubMed] [Google Scholar]
- 28.Tacket CO, Brenner F, Blake PA. Clinical features and an epidemiological study of Vibrio vulnificus infections. J. Infect. Dis. 1984;149(4):558–561. doi: 10.1093/infdis/149.4.558. [DOI] [PubMed] [Google Scholar]
- 29.Strom MS, Paranjpye RN. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2000;2(2):177–188. doi: 10.1016/s1286-4579(00)00270-7. [DOI] [PubMed] [Google Scholar]
- 30.Loeffler JM, Nelson D, Fischetti VA. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science. 2001;294(5549):2170–2172. doi: 10.1126/science.1066869. [DOI] [PubMed] [Google Scholar]
- 31.Macaya G, Thiery JP, Bernardi G. An approach to the organization of eukaryotic genomes at a macromolecular level. J. Mol. Biol. 1976;108(1):237–254. doi: 10.1016/s0022-2836(76)80105-2. [DOI] [PubMed] [Google Scholar]
- 32.Bernardi G. Misunderstandings about isochores. Part 1. Gene. 2001;276(1-2):3–13. doi: 10.1016/s0378-1119(01)00644-8. [DOI] [PubMed] [Google Scholar]
- 33.Ou HY, He X, Harrison EM, Kulasekara BR, Thani AB, Kadioglu A, Lory S, Hinton JC, Barer MR, Deng Z, Rajakumar K. MobilomeFINDER web-based tools for in silico and experimental discovery of bacterial genomic islands. Nucleic Acids Res. 2007;35(Web Server issue):W97–W104. doi: 10.1093/nar/gkm380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5(6):e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ou HY, Chen LL, Lonnen J, Chaudhuri RR, Thani AB, Smith R, Garton NJ, Hinton J, Pallen M, Barer MR, Rajakumar K. A novel strategy for the identification of genomic islands by comparative analysis of the contents and contexts of tRNA sites in closely related bacteria. Nucleic Acids Res. 2006;34(1):e3. doi: 10.1093/nar/gnj005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu P, Li P, Jiang X, Bi D, Xie Y, Tai C, Deng Z, Rajakumar K, Ou HY. Complete genome sequence of Klebsiella pneumoniae subsp.pneumoniae HS a multidrug-resistant strain isolated from human sputum. J. Bacteriol. 2012; 194(7):1841–1842. doi: 10.1128/JB.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bi D, Xu Z, Harrison EM, Tai C, Wei Y, He X, Jia S, Deng Z, Rajakumar K, Ou HY. ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res. 2012;40(Database issue):D621–626. doi: 10.1093/nar/gkr846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin TL, Lee CZ, Hsieh PF, Tsai SF, Wang JT. Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J. Bacteriol. 2008;190(2):515–526. doi: 10.1128/JB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang CT, Zhang R. A nucleotide composition constraint of genome sequences. Comput. Biol. Chem. 2004;28(2):149–153. doi: 10.1016/j.compbiolchem.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Zhang CT, Gao F, Zhang R. Segmentation algorithm for DNA sequences. Phys. Rev. E. Stat. Nonlin. Soft Matter Phys. 2005;72(4 Pt 1):041917. doi: 10.1103/PhysRevE.72.041917. [DOI] [PubMed] [Google Scholar]
- 41.Zhang R, Zhang CT. Identification of genomic islands in the genome of Bacillus cereus by comparative analysis with Bacillus anthracis. Physiol. Genom. 2003;16(1):19–23. doi: 10.1152/physiolgenomics.00170.2003. [DOI] [PubMed] [Google Scholar]
- 42.Zhang R, Zhang CT. Accurate localization of the integration sites of two genomic islands at single-nucleotide resolution in the genome of Bacillus cereus ATCC 10987. Comp. Funct. Genom. 2008:451930. doi: 10.1155/2008/451930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo FB, Wei W. Prediction of genomic islands in three bacterial pathogens of pneumonia. Int. J. Mol. Sci. 2012;13(3):3134–3144. doi: 10.3390/ijms13033134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao F, Zhang CT. GC-Profile a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 2006;34(Web Server issue):W686–691. doi: 10.1093/nar/gkl040. [DOI] [PMC free article] [PubMed] [Google Scholar]