Abstract
In theoretical physics, there exist two basic mathematical approaches, algebraic and geometrical methods, which, in most cases, are complementary. In the area of genome sequence analysis, however, algebraic approaches have been widely used, while geometrical approaches have been less explored for a long time. The Z-curve theory is a geometrical approach to genome analysis. The Z-curve is a three-dimensional curve that represents a given DNA sequence in the sense that each can be uniquely reconstructed given the other. The Z-curve, therefore, contains all the information that the corresponding DNA sequence carries. The analysis of a DNA sequence can then be performed through studying the corresponding Z-curve. The Z-curve method has found applications in a wide range of areas in the past two decades, including the identifications of protein-coding genes, replication origins, horizontally-transferred genomic islands, promoters, translational start sides and isochores, as well as studies on phylogenetics, genome visualization and comparative genomics. Here, we review the progress of Z-curve studies from aspects of both theory and applications in genome analysis.
Keywords: GC profile, Gene finding, Genomic island, Replication origin, Z-curve.
1. INTRODUCTION
In theoretical physics, there exist two basic mathematical approaches, algebraic and geometrical methods, which, in most cases, are complementary. In the area of genome studies, however, algebraic approaches, such as Markov chain models and hidden Markov chain models, have been widely used, while geometrical approaches have been less explored for a long time. The Z-curve theory is a geometric approach to genome analysis.
The Z-curve is a 3-dimensional curve that represents a given DNA sequence in the sense that each can be uniquely reconstructed given the other [1-3]. The Z-curve, therefore, contains all the information that the corresponding DNA sequence carries. The analysis of a DNA sequence can then be performed through studying the corresponding Z-curve.
Historically, various methods for the graphical representation of DNA sequences were proposed, such as the H curve [4] and the 2-dimensional DNA walk [5]. It has been shown that most of these methods are, in fact, special cases of the Z-curve, and an extensive comparison between the Z-curve and other representations was detailed in reference [2]. One of the advantages of the Z-curve is its intuitiveness, enabling global and local compositional features of genomes to be grasped quickly in a perceivable form. The methodology of
the Z-curve is a suitable platform on which other methods, such as statistics, can be integrated to address bioinformatics questions. The Z-curve method [1, 2] has found many applications in genome analysis since its initiation two decades ago. Here, we review the progress of the Z-curve studies from aspects of both theory and applications in genome research.
2. PART-1: THEORY OF THE Z-CURVE
2.1. Symmetry of Four DNA Bases and its Geometric Representation
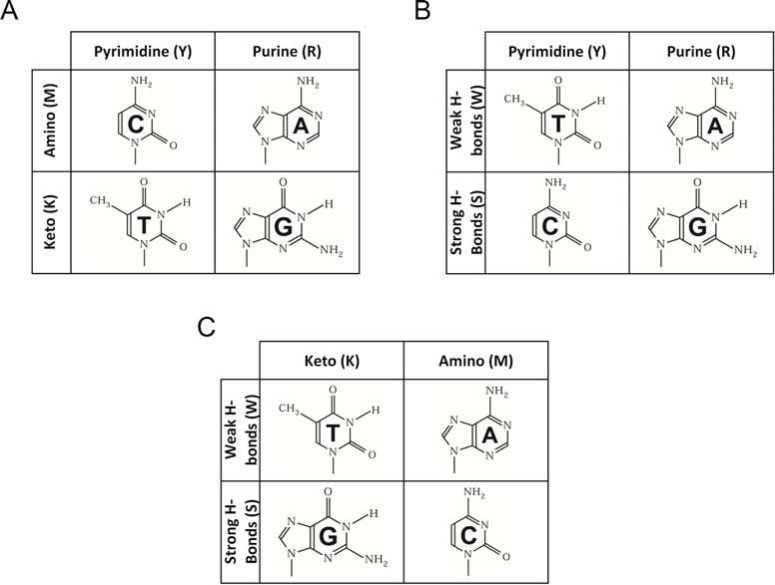
The DNA sequence is composed of 4 kinds of nucleotides, adenine, cytosine, guanine and thymine, denoted by A, C, G and T, respectively. The number of possible combinations when taking 2 bases at a time from 4 bases is 6. The 6 combinations are: R (A/G) and Y (C/T); M (A/C) and K (G/T); W (A/T) and S (G/C), where R, Y, M, K, W and S represent the bases of puRine, pYrimidine, aMino, Keto, Weak hydrogen bonds and Strong hydrogen bonds, respectively, according to the NC-IUB recommendation [6]. The chemical structures of the four bases are shown in (Fig. 1), illustrating the symmetry among the four bases. According to different criteria, the four bases can be classified into two categories.
Fig. (1).
Chemical structures of four DNA bases, displaying the basic symmetry.
(i) Criterion 1, according to the chemical structure of having single or double rings
(ii) Criterion 2, according to the chemical structure of having an amino or keto group
(iii) Criterion 3, according to the structure of the double helix forming two or three hydrogen bonds in the Watson-Crick pair
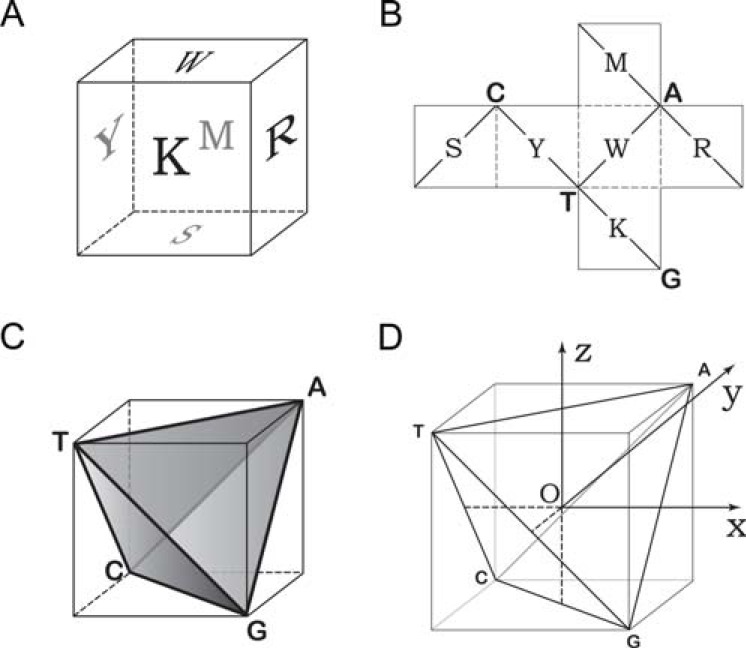
We seek to find some geometrical representation for the above symmetry. If a 2-dimensional (plane) graph is adopted, we find that the symmetry can be represented by (Fig. 1A-C). If a 3-dimensional graph is adopted, the regular cube, as shown in (Fig. 2A), seems to be the unique choice to represent the symmetry. Each face of the cube is assigned to one, and only one, of the six characters: R, Y, M, K, W and S, thereby keeping the rule that R and Y, M and K, as well as W and S are on opposite sides. To prepare (Fig. 2A), readers may cut (Fig. 2B) and fold it along the dashed lines. Diagonals of the regular cube form a regular tetrahedron ACGT, as shown in (Fig. 2C). Assigning one of A, C, G and T to each vertex of the tetrahedron as shown in (Fig. 2C) is not arbitrary. Note that the vertex A of the tetrahedron is also the vertex of the cube, at which three faces of the cube, R, M and W, are crossed. The intersection base of R (A/G), M (A/C) and W(A/T) is A. Similar assignments can be applied to the vertices C, G and T, as shown in (Fig. 2C and D).
Fig. (2).
The coordinate system based on the regular tetrahedron. A) a cube displaying the basic symmetry: R/Y, M/K and S/W symmetry; B) an extended plot for the cube. C) a cube and its inscribed tetrahedron; D) a coordinate system is set up to establish the Z-curve theory.
To further the study, a coordinate system needs to be established (Fig. 2D). The line connecting the middle point of an edge and that of the opposite edge of the tetrahedron is called the middle line. There are a total of three middle lines in a tetrahedron, crossing at the center O, and they are perpendicular to each other. A Cartesian coordinate system OXYZ can be set up by using the three middle lines, as shown in (Fig. 2D).
Thus, the cube-tetrahedron geometric entity established here correctly reflects the symmetry of the four DNA bases.
2.2. The DNA Group
A regular tetrahedron is a geometric entity of high symmetry. All possible rotational motions which keep the tetrahedron fixed in the space form a group, called a tetrahedron group or T-group. As shown in (Fig. 2D), a tetrahedron group consists of 12 operational elements, which are described below.
I, i.e., the identity operation;
Rx, Ry and Rz, i.e., the 180° rotation along x, y and z axes, respectively;
RA, RC, RG and RT, i.e., the 120° rotation along AO, CO, GO and TO axes, respectively;
R2A, R2C, R2G and R2T, i.e., the 240° rotation along AO, CO, GO and TO axes, respectively.
A point with coordinates x, y and z will be transformed accordingly under the operational elements of the T-group. For example,
The transforms of x, y and z under the 12 operations of the T-group are listed in (Table 1). The four elements I, Rx, Ry and Rz form an invariant subgroup of the T-group, which is isomorphic to the Klein-4 group, or K4 group. The K4 group and its cosets exhaust the T-group. Therefore, all the 12 elements of the T-group can be divided into four classes, which are (I), (Rx,Ry, Rz), (RA, RC, RG, RT) and (R2A, R2C, R2G, R2T).
Table 1.
Twelve Elements of the DNA Group (A4 Group or the Tetrahedron Group).
| Element | A4 Group | Tetrahedron Group |
|---|---|---|
| I | A C G T | x y z |
| Rx | G T A C | x -y -z |
| Ry | C A T G | -x y -z |
| Rz | T G C A | -x -y z |
| RA | A T C G | z x y |
| RC | G C T A | z -x -y |
| RG | T A G C | -z -x y |
| RT | C G A T | -z x -y |
| R2A | A G T C | y z x |
| R2C | T C A G | -y -z x |
| R2G | C T G A | -y z -x |
| R2T | G A C T | y -z -x |
On the other hand, the set of all possible permutations of four objects forms a symmetric group, denoted by S4. Among the 24 elements of the symmetric group S4, the set of all 12 even permutations forms an invariant subgroup of S4, referred to as the alternative group of order 4, denoted by A4. The DNA group is defined as a particular A4 group, in which the permuted objects are the four DNA bases A, C, G and T. According to the group theory, the T-group and A4 group are isomorphic with each other. From the perspective of the abstract group, the T-group and the A4 group are the same group, because they have the same group structure and matrix representation. The four bases A, C, G and T are assigned to the four vertices of the tetrahedron, as shown in (Fig. 2D).
The four characters A, C, G and T will be transformed accordingly under the 12 operational elements of the DNA group or the A4 group. For example,
Biologically, the transform Rx is called transition, whereas the transform Ry and Rz are called transversion. Here Rz is termed as the complementary transform.
We have previously established that the T-group and the A4 group are the same group [3], and thus their elements should have one-to-one corresponding relations, as shown in (Table 1). Both the A4 group and the T-group are called the DNA group, which forms the basis of the Z-curve theory.
2.3. The Z-transform Formulas
Let the occurrence frequencies of the four bases, A, C, G and T in a DNA sequence be denoted by a, c, g and t, respectively. The normalized condition reads
| (1) |
indicating that among the four real numbers a, c, g and t, only three of them are independent.
Suppose that X, Y and Z are the coordinates of a point P in the coordinate system shown in (Fig. 2D), which can be expressed by a linear combination of the four frequencies a, c, g and t, as follows
| (2) |
where a11, a12, …, c13, c14 are real coefficients. Eqs. (2) can be re-written as a matrix form
| (3) |
The coordinates of the four vertices of the regular tetrahedron A, C, G and T are already known, and shown in (Table 2). Based on the 12 numbers in (Table 2), the 12 coefficients can be uniquely determined, and eqs. (3) becomes
Table 2.
Coordinates of the 4 Vertices of the Regular Tetrahe-dron ACGTa.
| Coordinates | Vertices | |||
|---|---|---|---|---|
| A | C | G | T | |
| X | - | - | ||
| Y | - | - | ||
| Z | - | - | ||
Refer to Fig. 2 (d) for the original coordinate system, where the height of the tetrahe-dron is 1. Consequently, the edge length of the tetrahedron is , and the edge length of the cube is .
| (4a) |
Equivalently, eqs. (4) may be re-written as
| (4b) |
Eqs. (4) are called the Z-transform formulas, which were first derived in 1991 by a totally different way [1]. The Z-transform formulas transform the four base frequencies into three coordinates of a point (called a mapping point) in a three-dimensional space. As previously indicated [1], for convenience, we introduced the reduced coordinate system x, y and z
| (5) |
such that
| (6a) |
In what follows, we always use the Z-transform formulas based on the reduced coordinate system eqs. (6), unless otherwise indicated. Equivalently, eqs. (6a) can be also re-written as a matrix form
| (6b) |
Letting
| (7) |
and
| (8a) |
Equivalently, eqs. (6b) may be re-written with a simplified form
| (9) |
The reverse equation of eqs. (6) is
| (10) |
It is shown that regardless of the values of x, y and z,. In fact, it was shown in 1991 that the mapping point P (x, y, z), corresponding to a, c, g and t, is always situated within the tetrahedron ACGT shown in (Fig. 2D) [1].
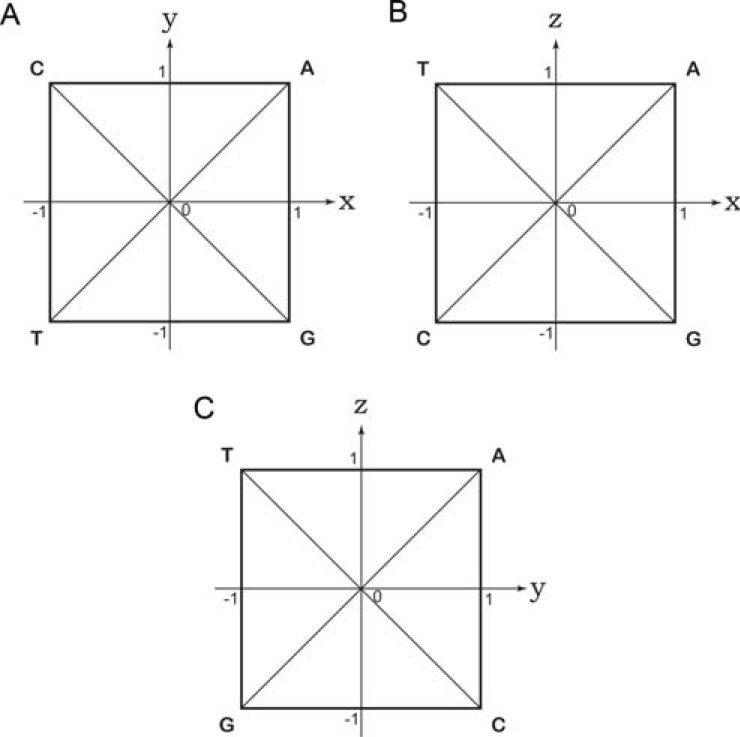
To provide a clear visualization, the tetrahedron and the mapping points within it are projected onto some coordinate planes. Referring to (Fig. 2C and D), note that the tetrahedron ACGT has 4 vertices A, C, G and T, and six edges AC, AG, AT, CG, CT and GT. Interestingly, the projection of six edges onto any coordinate plane forms a regular square and two diagonal lines within the square, where the projection of four vertices of the tetrahedron forms four vertices of the square, as shown in (Fig. 3A, B and C), for the x-y, x-z and y-z planes, respectively. Note that (Fig. 3A, B and C) are in accordance with (Fig. 1A, B and C), respectively. It should be noted that A, C, G and T are sometimes used to denote DNA bases, while the same symbols can represent vertices of the tetrahedron or squares. Refer to (Fig. 3A) first. Projections of four edges AG, AC, CT and GT form the four sides of the square, whereas those of AT and GC form the two diagonal lines of the square. Based on the Z-transform formulas eqs. (6), the base composition of a DNA sequence, i.e., the values of a, c, g and t, can be visualized by observing the position of the mapping point in the square. For example, if the DNA sequence has only one kind of base, say, A, then a = 1, c = g = t = 0. The corresponding mapping point is situated at the vertex A in (Fig. 2A). Similar results for (Fig. 3A) are summarized as follows.
Fig. (3).
Projection of the 3-D coordinates onto planes. The projection of the 3-D coordinate system onto the A) x-y, B) x-z and C) y-z planes.
Similar deductions for the annotation of (Fig. 3B and C) are left out for readers who might be interested in doing so.
2.4. Linear Representation of the DNA Group
Based on the reduced coordinate system, the coordinates of the four vertices A, C, G and T can be represented by
| (11) |
We previously established the linear representation of the DNA group, i.e., the tetrahedron group or the alternative group A4 in 1997 [3]. For readers’ convenience, here we re-write the result (see eqs. (6) in [3] as follows:
| (12) |
This matrix representation depicts correct relationships among the 12 elements of the DNA group. For example, a rotation of 180° along the x-axis in (Fig. 2D), followed by another similar rotation, leads to the original state, i.e.,
| (13) |
That is to say, the matrix representation not only results in a one-to-one correspondence among elements of the DNA group, but also correctly reflects their relations based on the multiplication of matrices.
In the following, we show that the transform matrix (3x4) eq. (8a) and its variants also constitute a one-to-one representation to each element of the DNA group. For this purpose, eq. (8a) can be re-written as
| (8b) |
where (A), (C), (G) and (T) are denoted by eqs. (11). Referring to (Table 2), we find that the order of the four nucleotides above correspond to the element I of the A4 group. Similarly, its 11 variants can be derived, and are listed as follows
| (14) |
where ZI = Z. Based on the 12 Z matrices, we have
| (15) |
Note that each Zi corresponds to each Ri by a way of one-to-one correspondence. Therefore, the Z matrix also constitutes a representation of the DNA group in this sense. However, it is not an ordinary representation of the DNA group, because the similar multiplication relation such as eq. (13) does not exist among the Z matrices.
It should also be noted that the Z-transform formulas eqs. (6), which transform the nucleotide frequencies into the coordinates of a point in a three-dimensional space, are unique and invariant under the operations of the DNA group. The Z-transform formulas shown in eqs. (6) represent the unique set of equations which reflect the inherent features of the DNA group. The Z-transform formulas are the core of the Z-curve theory.
2.5. The Z-transform Formulas for Studying Correlations of Multiple Nucleotides
To extract features of a given DNA sequence, in addition to considering occurrence frequencies of a single nucleotide, correlations of multiple nucleotides should also be considered. Therefore, the Z-transform formulas should be extended to consider the correlations of multiple nucleotides.
The case of a single nucleotide
| (16) |
Eqs. (16) are equivalent to eqs. (9). Here we have 3 (= 3x4°) parameters.
The case of di-nucleotides
| (17) |
where p(HA) represents occurrence frequencies of the di-nucleotide HA, and so forth. Here we have 12 (=3x41)parameters.
The case of tri-nucleotides
| (18) |
where p(HIA) represents occurrence frequencies of the tri-nucleotide HIA, and so forth. Here we have 48 (=3x42) parameters.
The case of tetra-nucleotides
| (19) |
where p(HIJA) represents occurrence frequencies of the four-nucleotide HIJA, and so forth. Here we have 192 (=3x43) parameters.
The case of penta-nucleotides
| (20) |
where p(HIJKA) represents occurrence frequencies of the five-nucleotide HIJKA, and so forth. Here we have 768 (=3x44) parameters.
The case of hexa-nucleotides
| (21) |
where p(HIJKLA) represents occurrence frequencies of the six-nucleotide HIJKLA, and so forth. Here we have 3072 (=3x45) parameters.
To calculate the occurrence frequencies of multiple nucleotides for a given DNA sequence, we use a moving window with size = 1, 2, 3, 4, 5 and 6. Starting from the first nucleotide or base, move the sliding window rightward one base at a time, and then the frequencies can be calculated. Substitute the frequencies into eqs. (16) to (21), and then the Z-curve parameters can be obtained. For some applications, eq. (16), i.e., 3 parameters are sufficient. However, in some cases, more parameters are needed. The space spanned by the 3 parameters is denoted by V1, and similarly we have V2, V3, V4, V5 and V6, respectively, corresponding to eqs. (16) to (21). Usually, the direct sum among different spaces is needed. For most applications, there are six possible choices
| (22) |
where the symbol ⊕ represents the direct sum of two spaces. The dimensions of the spaces V1 to V6 are 3, 15 (= 3+12), 63 (= 15+48), 255 (= 63+192), 1023 (= 255+768) and 4095 (= 1023+3072), respectively. Generally, for the space, the dimension is
2.6. Quadratic Form of x, y and z
Starting from eq. (9)
| (9) |
We have
| (23) |
where “T” means the transpose operation of a matrix. Furthermore, we find
| (24) |
Simple derivation shows that
| (25) |
where S is defined by
| (26) |
S, named as “genome order index” [7], is useful for designing a fast genome segmentation algorithm [8, 9]. We also observed that for most genomes
| (27) |
Eq. (27) has a clear geometrical explanation. The surface of the inscribed sphere is described by the equation
| (28) |
Therefore, S<1/3 implies that the mapping point is within the inscribed sphere [7].
2.7. The Z-curve
One of the most important applications of the Z-transform formulas is to derive the equation of the Z-curve. Consider a DNA sequence with N bases that are inspected one base at a time. From the first base to the nth base, compute accumulative numbers of the bases A, C, G and T, denoted by An, Cn, Gn and Tn, respectively. Based on the Z-transform formulas eqs (6), we find
| (29) |
Multiplied by n to both hands of eq. (29), and letting
| (30) |
we have
| (31a) |
or equivalently
| (31b) |
which was first derived in 1994 by an entirely different method [2]. It should be noted that An, Cn, Gn and Tn are the cumulative occurrence numbers of A, C, G and T, respectively, in the sub-sequence from the 1st base to the nth base in the sequence with length N. We define A0=C0=G0=T0=0, therefore, x0 = y0 = z0 = 0. The Z-curve is defined as the connection of the nodes P0 (x0, y0, z0), , P2 (x2, y2, z2), …, PN(xN, yN, zN) one by one sequentially with straight lines. The connection results in a curve with a zigzag shape, hence the name Z-curve. Note that the Z-curve always starts from the origin of the three-dimensional coordinate system. Once the coordinates xn, yn and zn (n = 1, 2, …, N) of a Z-curve are given, the corresponding DNA sequence can be reconstructed uniquely from the so-called inverse Z-transform formulas
| (32) |
where the normalized relation of An + Cn + Gn + Tn = n is used.
The three components of the Z-curve, xn, yn and zn, represent three independent distributions, that is, those of purine/pyrimidine (R/Y), amino/keto (M/K) and strong-H bond/weak-H bond (S/W) bases, respectively, and they completely describe the DNA sequence being studied. In the subsequence constituted from the 1st base to the nth bases of the sequence, when purine bases (A/G) are in excess of pyrimidine bases (C/T), xn > 0, otherwise, xn < 0, and when the numbers of purine (A/G) and pyrimidine bases (C/T) are identical, xn = 0. Similarly, when amino bases (A/C) are in excess of keto bases (G/T), yn > 0, otherwise, yn < 0, and when the numbers of amino (A/C) and keto bases (G/T) are identical, yn = 0. Finally, when weak H-bond bases (A/T) are in excess of strong H-bond bases (G/C), zn > 0, otherwise, zn < 0, and when the numbers of (A/T) and (G/C) bases are identical, zn = 0. The xn and yn components are termed RY and MK disparity curves, respectively. Similarly, the AT and GC disparity curves are defined by (xn + yn)/2 and (xn-yn)/2, which shows the excess of A over T and G over C, along the genome. The RY and MK disparity curves, as well as AT and GC disparity curves, can be used to predict replication origins of various genomes.
2.8. The GC Profile
For most genome sequences, Chargaff Parity Rule II holds, i.e.,, where N is the length of a genome or a chromosome. According to eqs (31), we find
| (33) |
Therefore, the curves of zn ~ n are roughly straight lines in this case. To amplify the variations of the straight-line-like curve, the curve of zn ~ n is firstly fitted by a straight line using the least square technique,
| (34) |
where (z, n) is the coordinate of a point on the fitted straight line and k is its slope. We define the z’ curve, where
| (35) |
Therefore, the variations of zn ~ n curve deviated from the straight line, which corresponds to a constant G+C content (see eq. (36) below), are protruded by the z’ curve. One may also use the average slope of the zn ~ n curve to compute k, k = zN / N, where zN is the terminal coordinate of the zn ~ n curve and N is the sequence length. The essence of the z’ curve is to display the variations of the G+C content along a genome or chromosome based on the cumulative count of G and C bases. Let denote the average G+C content within a region Δn in a sequence, it was shown that [10].
| (36) |
where is the average slope of the z’ curve within the region Δn. Both quantities of and Δn can be calculated by using the z’ curve. It is clear to see from eq. (36) that a jump in the z’ curve, i.e., , indicates a decrease of G+C content or an increase of A+T content, whereas a drop in the z’ curve, i.e., , indicates an increase of G+C content or a decrease of A+T content. The region Δn is usually chosen to be a fragment of a DNA sequence. The above method to calculate G+C content is called the windowless technique [10].
The GC profile is defined as, because it is more intuitive in the sense that a jump denotes an increase in GC content. We emphasize the importance of the GC profile for genome studies, because it represents a windowless technique to calculate the G+C content along genome sequences.
2.9. A Segmentation Algorithm Based on the Z-transform
Let n be a point within a DNA sequence of length N, which divides the whole sequence into two parts: the right and left sub-sequences, and then denote frequencies of bases in the right sub-sequence and left sub-sequence by (ar, cr, gr, tr) and (al, c1, g1, t1), respectively. The frequencies are mapped onto two points, PR (xr, yr, zr) and PL(xl, yl, zl), in a 3-D space, where
| (37) |
The square of Euclidean distance between the two points is denoted by D, where
| (38) |
Substituting eqs. (37) into eq. (38), we have
| (39) |
where C is a constant. Note that D is a function of n. Suppose that when Then the point is called a compositional segmentation point [8]. The segmentation algorithm is recursive, i.e., after n* is determined, the same procedure is applied to both the left and right sub-sequences recursively, until D(n) is less than a given threshold. For more details refer to [8].
Eq. (39) can be extended to the case of a binary sequence. For example, by replacing the bases G and C with S, and bases A and T with W, a DNA sequence can be transformed into a binary sequence of S and W. In this case, the algorithm results in compositional segmentation points according to GC content. A software, called GC-Profile, was developed to implement the algorithm for genome segmentation [9].
3. PART-2: APPLICATIONS IN GENOME ANALYSIS
The Z-curve theory has been successfully applied in many different research areas in analyzing genomes of bacteria, archaea, eukaryotes and viruses. The applications include, to name a few, the identifications of protein-coding genes, replication origins, horizontally-transferred genomic islands, isochore structures, genome segmentation points, promoters and translational start sites, as well as studies on nucleosome positioning, DNA curvature profiles, phylogenetics and comparative genomics, in various organisms (Table 3). It is not practical to cover all these areas in detail in a single review, and thus we will only highlight some studies.
Table 3.
A Partial List of Z-curve Applications in Genome Analysis.
| Research areas | Involved Z-Curve Components | Algorithm, Software or Database | Life Domains or Virus | Species |
|---|---|---|---|---|
| Protein-coding gene recognition a | x, y, z, S | Z-curve algorithm [1, 2], Zcurve [12] | Bacteria | Acinetobacter baumannii [17], Variovorax paradoxus [18], Amycolatopsis mediterranei [19], Bacillus thuringiensis [20], Streptomyces tendae [21], Phaeobacter gallaeciensis [22], Desulfobacterium autotrophicum [23], Mycobacterium tuberculosis [24], Magnetospirillum gryphiswaldense [25], Beggiatoa [26] |
| Phage, plasmid | Fosmids of marine Planctomycetes [127], plasmids in the human gut [128], phage Rtp [129] | |||
| Archaea | Archaea of the ANME-1 group [27] | |||
| Eukaryotes | Leptospira interrogans [130], Yeast [11], Short human protein-coding genes [56, 131], Drosophila [55] | |||
| Zcurve_V [14], Zcurve_CoV [15] | Virus, Coronavirus, phages | Prophage [33], Me Tri virus [28], novel human coronaviruses NL63 and HKU1 [34], novel bat coronaviruses [35], bat coronaviruses 1A, 1B and HKU8 [36], novel human coronavirus [37] | ||
| SARS_CoV | Various strains of SARS_CoV [38-53] | |||
| Replication origin identification | AT, GC, MK and RY disparity b | Ori-finder [78], DoriC [132, 133] | Archaea | Methanosarcina mazei[69], Halobacterium species NRC-1[63], Methanocaldococcus jannaschii [68], Sulfolobus acidocaldarius [72], Haloferax volcanii [73], Desulfurococcus kamchatkensis [74], Thermococcus sibiricus [75], Sulfolobus islandicus [76] |
| Bacteria | Moraxella catarrhalis [79], Sorangium cellulosum [80], Microcystis aeruginosa [80], Cyanothece [81], Cupriavidus metallidurans [82], Azolla filiculoides [83], Variovorax paradoxus [18], Corynebacterium pseudotuberculosis [84], [85], Orientia tsutsugamushi [86], Propionibacterium freudenreichii [87], Laribacter hongkongensis [88], Legionella pneumophila [89], Ehrlichia canis [90] | |||
| Phage, plasmid | Streptococcus pneumoniae Virulent Phage Dp-1 [134], R-plasmid pPRS3a from Bacillus cereus [135] | |||
| Genomic island identification | z’ | GC profile [9, 10] | Bacteria | Corynebacterium efficiens [105], Rhodopseudomonas palustris [106], Corynebacterium glutamicum [104], Vibrio vulnificus and Bacillus cereus [103], Agrobacterium tumefaciens, Rolstonia solanacearum, Xanthomonas axonopodis, Xanthomonas campestris, Xylella fastidiosa and Pseudomonas syringae [107], Streptomyces lividans [108], Parachlamydiaceae UWE25 [109], epsilon proteobacteria Sulfurovum and Nitratiruptor [110], Acinetobacter oleivorans [111], Silicibacter pomeroyi [112] |
| Archaea | Haloquadratum walsbyi [136] | |||
| GC content variation, | z’, S | GC profile [9, 10] | Eukaryotes | Human genome: isochores [94, 98, 137] and replication time zones [138]; Isochores for chicken [97], Arabidopsis thaliana [96], mice [95] and pig [99]; DNA curvature profile for Aspergillus fumigatus [100] |
| isochore, genome segmentation | Bacteria | Bifidobacterium longum [139], Streptomyces avermitilis [140], Erwinia amylovora [141], Ralstonia pickettii [142] | ||
| Promoter, translational start sites, nucleosome positioning | x, y, z | Z-curve algorithm [11, 12], GS-finder [113] | Bacteria | Translational start sites [113] and promoters [115] of Escherichia coli and Bacillus subtilis |
| Eukaryotes | Human Pol II promoter [114], Yeast genome for stable and dynamic nucleosome positioning [116] | |||
| Comparative genomics, genome visualization | x, y, z, z’ | Z-curve database [117] | Bacteria, archaea, eukaryotes and viruses | Bacillus cereus [103], Bacillus cereus ATCC 10987 [119], Coronavirus [118], human immunodeficiency virus [120], human [121, 143], E. coli [122], Seven GC-rich bacteria [126], 90 species [1], Aeropyrum pernix K1 [124], Streptomyces coelicolor [125] |
3.1. Identification of Protein-coding Genes
One of the most important applications of the Z-curve theory is gene-finding in various genomes. The principle in using the Z-curve theory to identify protein-coding genes is straightforward. Based on the Z-transform formulas, the occurrence frequencies of 4 bases in a DNA sequence are mapped onto a point in a 3-dimensional (3-D) or 15-, 63-, 255-, 1023- and 4095-D space, depending on the number of correlated bases under consideration (eqs. (16) to (22)). The first application was for gene recognition in the budding yeast genome, where a 3-D space (eqs. (16)) was adopted [11]. However, since the protein coding sequence has 3 phases, the 3 Z-curve parameters are expanded to 9 (3x3 phases) parameters. Adding the genome order index S (eq. (26)) into the set of 9 Z-curve parameters, a 10-D space is spanned by the 10 parameters. It was observed that the mapping points of protein coding sequences and non-coding sequences are distributed in two distinct regions in the 10-D space, although there is minor overlapping [12]. Therefore, the two kinds of points can be discriminated by the Fisher discriminant method, or other classifiers, such as support vector machines.
The Z-curve algorithm was first applied to recognize protein coding genes in the budding yeast (Saccharomyces cerevisiae) genome with an accuracy better than 95%, where the accuracy is defined as the average of sensitivity and specificity [11]. The same algorithm achieved an accuracy rate over 98% in the Vibrio cholerae genome, based on 9 parameters only [13]. The success of the above studies led to the development of a series of ab initio gene-finding software for various species with different numbers of Z-curve parameters.
3.1.1. Gene-finding in Bacterial, Archaeal, Phage and Virus Genomes
Based on 33 Z-curve parameters, we developed ZCURVE 1.0, which is an ab initio gene-finding software for bacterial and archaeal genomes [12]. Based on the 9 Z-curve parameters, ZCURVE_V was developed for identifying protein-coding genes in viral and phage genomes [14]. We also developed the software, ZCURVE_CoV, for gene-finding in coronavirus genomes, with special applications for SARS-coronavirus genomes [15, 16].
The above set of gene-finding software has been widely used in various laboratories worldwide. For example, ZCURVE 1.0 has been used for annotating protein-coding genes in many newly sequenced bacterial genomes, such as those of Acinetobacter baumannii [17], Variovorax paradoxus [18], Amycolatopsis mediterranei [19], Bacillus thuringiensis [20], Streptomyces tendae [21], Phaeobacter gallaeciensis [22], Desulfobacterium autotrophicum [23], Mycobacterium tuberculosis [24], Magnetospirillum gryphiswaldense [25] and Beggiatoa [26]. ZCURVE 1.0 was also used for annotating archaeal genomes, e.g., archaea of the ANME-1 group [27] (Table 3).
For some genomes, e.g., those of the bacterium Mycobacterium tuberculosis H37Ra [24] and Me Tri virus [28], ZCURVE 1.0 was the only software used for genome annotation, more frequently, however, results of ZCURVE 1.0 were combined with those of others, such as Glimmer [29] and Genmark [30]. For instance, ZCURVE 1.0 is integrated into meta-gene-finding tool YACOP [31] and GARSA [32]. It is noteworthy that ZCURVE 1.0 is especially suitable for genomes with high GC contents, e.g., GC content > 56% [12]. Likewise, ZCURVE_V and ZCURVE_CoV have been widely used for annotating protein-coding genes in newly sequenced genomes of viruses, coronaviruses [28, 33-37] and SARS coronaviruses [38-53].
3.1.2. Gene-finding in Eukaryotic Genomes
Algorithms based on the Z-curve theory have been used for recognizing protein coding genes in a number of eukaryotic genomes, e.g., the budding yeast genome [11], Leptospira interrogans genome [54] and Drosophila genomes [55]. The Z-curve algorithm has also been used in recognizing short coding sequences of human genes [56]. The algorithm based on the 189 Z-curve parameters was shown to be the most accurate among those tested for a given database, with the second one being an algorithm based on the Markov chain of order five [56], and the result was later confirmed by an independent study [57]. Recognition of exons and introns of human genes was also studied by using the Z-curve method [58].
3.1.3. Gene-finding Using the Fast Fourier Transform (FFT) Technique
The standard genetic code defines a mapping between a codon and an amino acid. According to this mapping, protein coding regions are divided into a series of tri-nucleotides (codon or triplet), resulting in a period-3 property in coding regions. Therefore, it is possible to find coding regions by exploring the 3-periodicity of DNA sequences. Consequently, the first step is to transform the DNA sequence into a digital sequence or signal, and the Z-curve is especially suitable for this purpose.
According to eqs. (31), . Applying the FFT to , respectively, we are able to detect the 3-periodicity in the FFT power spectrum for each of the three numerical sequences. To increase the sensitivity, a lengthen-shuffle FFT algorithm was proposed for finding protein coding regions [59]. For example, the method was used to detect introns in the C. elegance chromosome III [60], and was later improved by using an adaptive filter to predict the exons in DNA sequences [61]. The relationship between the Z-curve and the Fourier transform for DNA sequence classification was studied in details [62].
3.2. Prediction of Replication Origins
3.2.1. Prediction of Replication Origins of Archaeal Genomes
Bacterial and eukaryotic genomes contain single and multiple replication origins, respectively. It was once a mystery whether archaea could have multiple oriCs.
Using the Z-curve method, we firstly predicted three oriCs as well as their precise locations for Sulfolobus solfataricus [63], and the prediction was consistent with later experimental evidence [64-67].
The archaeon Methanococcus jannaschii was the first to have its genome sequenced, however, its oriCs were notoriously difficult to locate by both theoretical and experimental methods. The Z-curve method predicted 2 oriCs [68] that were supported by later experimental evidence [66]. Similarly, we predicted a single oriC in the genome of Methanosarcina mazei [69] and 2 oriCs in the genome of A. pernix [70], which were also supported by experimental evidence [71]. The Z-curve method has been commonly used for annotating newly sequenced archaeal genomes, such as those of Sulfolobus acidocaldarius [72], Haloferax volcanii [73], Desulfurococcus kamchatkensis [74], Thermococcus sibiricus [75], and Sulfolobus islandicus [76].
3.2.2. Prediction of Replication Origins in Bacterial Genomes
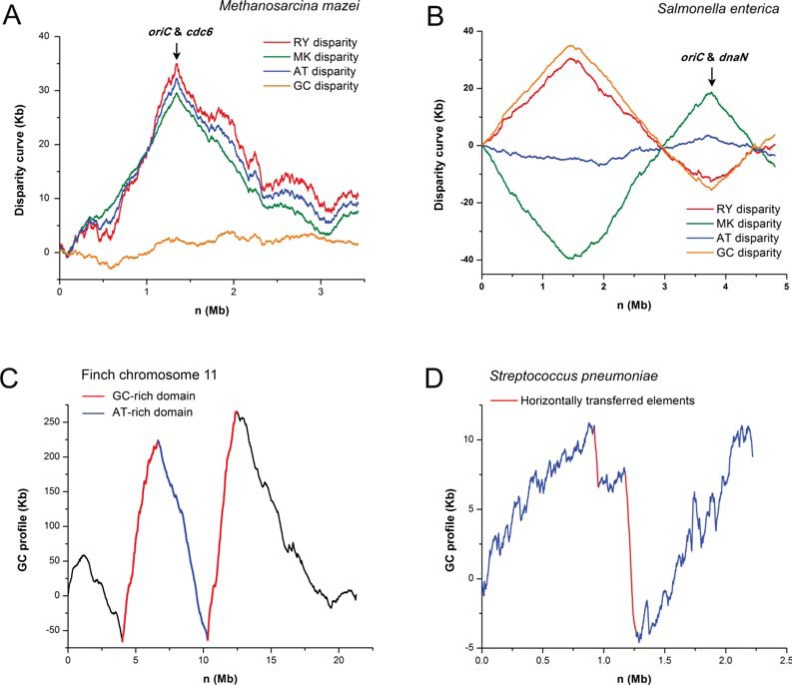
The Z-curve method is an effective technique that detects the asymmetrical nucleotide distribution around replication origins. The Z-curve contains all the information of its corresponding DNA sequence, and therefore the GC-skew [77] is a special case of the Z-curve. Thus the Z-curve can reveal nucleotide asymmetry that is not detectable by GC skew [70]. For instance, RY, MK and AT disparity curves show an oriC in the archaeon Methanosarcina mazei Tuc01 (Fig. 4A), while RY, MK, and GC disparity curves show an oriC in the bacterium Salmonella enterica tr. CT18 (Fig. 4B).
Fig. (4).
The Z-curve reveals features of archaeal, bacterial and eukaryotic genomes. The Z-curve shows replication origins in genomes of A) the archaeon Methanosarcina mazei Tuc01 and B) the bacterium Salmonella enterica subsp. Typhi str. CT18. The Z-curve shows C) the domain structure in chromosome 11 of finch, and D) horizontally-transferred genomic elements in the genome of Streptococcus pneumoniae ATCC 700669.
Ori-Finder, an integrated in silico method to predict oriC regions of bacterial genomes, has been developed, based on the Z-curve method, along with distributions of DnaA box patterns, indicator genes, and phylogenetic relationships [78]. Ori-finder has become a commonly used annotation tool for identifying oriCs in newly sequenced archaeal and bacterial genomes, e.g., those of Moraxella catarrhalis [79], Sorangium cellulosum [80], Microcystis aeruginosa [80], Cyanothece [81], Cupriavidus metallidurans [82], Azolla filiculoides [83], Variovorax paradoxus [18], Corynebacterium pseudotuberculosis [84, 85], Orientia tsutsugamushi [86], Propionibacterium freudenreichii [87], Laribacter hongkongensis [88], Legionella pneumophila [89], and Ehrlichia canis [90] (Table 3).
3.3. Studies of Genome Domain Structures
G+C content is an important characteristic of genome sequences. In the human genome, based on density gradient ultra-centrifugation experiments, it was found that long domains of relatively homogenous G+C content exist, and these domains are referred to as isochores [91, 92]. Traditionally, the G+C content along the genome is calculated using an overlapping or non-overlapping sliding window technique, based on which, however, isochores are hard to identify [93]. We developed a windowless technique in G+C content calculation, the GC profile [9, 10], which was used to study isochore structures in genomes of human [94], mouse [95], Arabidopsis thaliana [96] and chicken [97]. Based on the GC profile, the technique of wavelet multi-resolution analysis was used to identify isochore boundaries in the human genome [98]. For instance, a clear domain structure is revealed by the GC profile in chromosome 11 of finch (Fig. 4C). Other groups also used GC-Profile to study isochores in the pig genome [99] and to assess DNA curvature profiles for Aspergillus fumigatus [100].
3.4. Identification of Horizontally-transferred Genomic Islands in Bacterial Genomes
It is generally accepted that horizontal gene transfer (HGT) plays an important role throughout the genome evolution of prokaryotes, because HGT alters the genotype of a bacterium, and could potentially lead to new traits [101]. Genomic islands (GIs) contain clusters of horizontally transferred genes and therefore, identification of horizontally-transferred GIs is an important biological issue. Because the GC profile is sensitive to changes in GC content, it is a powerful tool in identifying GIs [102].
Based on the method of GC profile, GIs in many bacteria have been identified, e.g., Bacillus cereus [103], Corynebacterium glutamicum [104], Corynebacterium efficiens [105], Vibrio vulnificus CMCP6 [104], and Rhodopseudomonas palustris [106]. For instance, it was once believed that R. palustris does not have GIs, but analysis based on the GC profile identified 3 GIs that help explain how this bacterium survives in a versatile environment [106]. Corynebacterium efficiens can grow and produce glutamate at temperature above 40°C; unexpectedly, however, an aspartate kinase is less thermostable. This kinase gene is located in a GI that we identified, and this result suggests an explanation for its being less thermostable, i.e., the adaptive mutations have not occurred extensively due to the recent HGT [105]. For instance, horizontally transferred elements in Streptococcus pneumoniae ATCC 700669 can be clearly shown by the GC profile (Fig. 4D). The GC profile method has also been used for identification of GIs in other genomes, e.g., those of plant pathogens [107], Streptomyces lividans [108], Parachlamydiaceae UWE25 [109], epsilon proteobacteria Sulfurovum and Nitratiruptor [110], Acinetobacter oleivorans [111], and Silicibacter pomeroyi [112].
3.5. Identification of Promoters, Translation Start Sites and Nucleosome Positioning
Based on the behavior of the Z-curve near the bacterial gene translation start sites (TSS), a self-training method was proposed to find TSS with high accuracy [113]. It is likely that methods based on the same principle can also be used to recognize TSS in archaea and eukaryotes as well. Indeed, the Z-curve method was used to recognize human Pol II promoters [114] and promoters for bacterial genomes [115]. The positioning of nucleosomes, an elementary structural unit in eukaryotic chromatin, is pivotal in regulating many cellular processes, such as gene transcription. The Z-curve algorithm has been used to construct a genome-wide dynamic nucleosome positioning map for the budding yeast [116].
3.6. Visualization of DNA Sequences, Comparative Genomics and the Z-curve Database
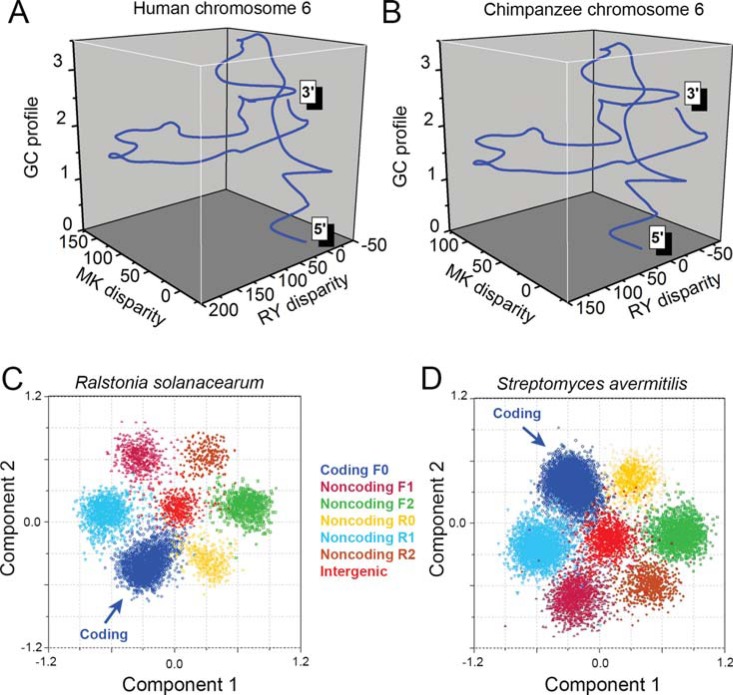
One of the aims for developing the Z-curve theory is to visualize DNA sequences. By using the Z-curve, features of related DNA sequences can be grasped quickly in a perceivable form [1, 2]. Therefore, we constructed the Z-curve database (www.zcurve.net), which contains Z-curves for currently available genomes, online Z-curve drawing tools and other Z-curve related software [117]. For instance, human chromosome 6 and chimpanzee chromosome 6 are homologous, and they apparently have similar Z-curve patterns (Fig. 5A and B). A typical example is the visualization of the genomes of related SARS-coronaviruses. Based on the 3-D coordinates of the corresponding Z-curves, the phylogenetic tree was constructed and was found to be in agreement with that based on sequence alignment [118]. Comparative genomics based on the GC profile was used to identify genomic islands [103, 119].
Fig. (5).
Genomic nucleotide composition features revealed by the Z-curve method. 3-D Z-curves for human chromosome 6 (A) and chimpanzee chromosome 6 (B). The 2 homologous chromosomes show similar Z-curves. To show global nucleotide composition patterns, Z-curves have been smoothed for 50,000 times by using the B-spline function. An ORF-flower phenomenon is revealed by the Z-curve method in genomes with high GC content. All open reading frames are mapped onto a 9-dimensional space using the Z-curve method, and protein-coding ORFs are located in a distinct region, compared with non-coding ORFs and intergenic sequences. Shown are principal component analysis for the genomes of Ralstonia solanacearum GMI1000 (C) and Streptomyces avermitilis MA 4680 (D). F0, F1, F2, R0, R1, and R2 stand for reading frames of protein-coding, forward 1, forward 2, non-coding reverse 0, reverse 1 and reverse 2, respectively.
According to eqs. (6), the base composition of a DNA sequence can be represented by a point in a 3-D space, thus providing an intuitive method to display base compositions. This method was used to study the codon usage in the genomes of AIDS virus [120], human [121], E. coli [122], Vibrio cholerae [123], Aeropyrum pernix K1 [124], Streptomyces coelicolor A3(2) [125] and seven GC-rich bacteria [126]. In prokaryotic genomes with high-GC content, coding ORFs and non-coding ORFs are located in distinct regions in a 9-dimensional space revealed by the Z-curve method, forming a flower-like pattern (Fig. 5C and D).
4. SUMMARY
The three components of the Z-curve, x, y and z, which display distributions of purine/pyrimidine (R/Y), amino/keto (M/K) and strong-H bond/weak-H bond (S/W) bases, respectively, are independent, and completely describe the DNA sequence. The x and y components are related to the disparities of RY, MK, AT and GC bases, and can therefore be used to identify oriC regions in prokaryotic and eukaryotic genomes. The component z is related to G+C content, and can therefore be used to identify domain structures of eukaryotic genomes and genomic islands of prokaryotic genomes. The set of all three components can be used in identifications of protein-coding genes, promoters, translational start sites or in other bioinformatics issues. Generally, further applications are expected to benefit from the use of functions based on the three components, i.e., f (x, y, z), with potential integration of other parameters.
In conclusion, the methodology of the Z-curve provides a geometrical approach to analyzing genomic DNA sequences. Considerable progress in applying the Z-curve method has been achieved, and the Z-curve theory provides a solid basis for future developments.
ACKNOWLEDGEMENTS
The authors cordially thank the students and staff in the laboratory for assistance during the course of Z-curve studies.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Zhang CT, Zhang R. Analysis of distribution of bases in the coding sequences by a diagrammatic technique. Nucleic Acids Res. 1991;19 (22):6313–6317. doi: 10.1093/nar/19.22.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang R, Zhang CT. Z-curves, an intuitive tool for visualizing and analyzing the DNA sequences. J. Biomol. Struct. Dyn. 1994;11 (4):767–782. doi: 10.1080/07391102.1994.10508031. [DOI] [PubMed] [Google Scholar]
- 3.Zhang CT. A symmetrical theory of DNA sequences and its applications. J. Theor. Biol. 1997;187 (3):297–306. doi: 10.1006/jtbi.1997.0401. [DOI] [PubMed] [Google Scholar]
- 4.Hamori E, Ruskin J. H curves, a novel method of representation of nucleotide series especially suited for long DNA sequences. J. Biol. Chem. 1983;258 (2):1318–1327. [PubMed] [Google Scholar]
- 5.Lobry JR. A simple vectorial representation of DNA sequences for the detection of replication origins in bacteria. Biochimie. 1996;78 (5):323–326. doi: 10.1016/0300-9084(96)84764-x. [DOI] [PubMed] [Google Scholar]
- 6.Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985;13(9):3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang CT, Zhang R. A nucleotide composition constraint of genome sequences. Comput. Biol. Chem. 2004;28(2):149–153. doi: 10.1016/j.compbiolchem.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Zhang CT, Gao F, Zhang R. Segmentation algorithm for DNA sequences. Phys. Rev. E. Stat. Nonlin Soft Matter Phys. 2005;72 (4 Pt 1):041917. doi: 10.1103/PhysRevE.72.041917. [DOI] [PubMed] [Google Scholar]
- 9.Gao F, Zhang CT. GC-Profile: a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 2006;34(Web Server issue):W686–691. doi: 10.1093/nar/gkl040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang CT, Wang J, Zhang R. A novel method to calculate the G+C content of genomic DNA sequences. J. Biomol. Struct. Dyn. 2001;19 (2):333–341. doi: 10.1080/07391102.2001.10506743. [DOI] [PubMed] [Google Scholar]
- 11.Zhang CT, Wang J. Recognition of protein coding genes in the yeast genome at better than 95% accuracy based on the Z-curve. Nucleic Acids Res. 2000;28 (14):2804–2814. doi: 10.1093/nar/28.14.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo FB, Ou HY, Zhang CT. ZCURVE: a new system for recognizing protein-coding genes in bacterial and archaeal genomes. Nucleic Acids Res. 2003;31 (6):1780–1789. doi: 10.1093/nar/gkg254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Zhang CT. Identification of protein-coding genes in the genome of Vibrio cholerae with more than 98% accuracy using occurrence frequencies of single nucleotides. Eur. J. Biochem. 2001;268(15):4261–4268. doi: 10.1046/j.1432-1327.2001.02341.x. [DOI] [PubMed] [Google Scholar]
- 14.Guo FB, Zhang CT. ZCURVE_V: a new self-training system for recognizing protein-coding genes in viral and phage genomes. BMC Bioinform. 2006;7:9. doi: 10.1186/1471-2105-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen LL, Ou HY, Zhang R, Zhang CT. ZCURVE_CoV: a new system to recognize protein coding genes in coronavirus genomes, and its applications in analyzing SARS-CoV genomes. Biochem. Biophys. Res. Commun. 2003;307(2):382–388. doi: 10.1016/S0006-291X(03)01192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao F, Ou HY, Chen LL, Zheng WX, Zhang CT. Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes. FEBS Lett. 2003;553(3):451–456. doi: 10.1016/S0014-5793(03)01091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao F, Wang Y, Liu YJ, Wu XM, Lv X, Gan YR, Song SD, Huang H. Genome sequence of Acinetobacter baumannii MDR-TJ. J. Bacteriol. 2011;193(9):2365–2366. doi: 10.1128/JB.00226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han JI, Choi HK, Lee SW, Orwin PM, Kim J, Laroe SL, Kim TG, O'Neil J, Leadbetter JR, Lee SY, Hur CG, Spain JC, Ovchinnikova G, Goodwin L, Han C. Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J. Bacteriol. 2011;193(5):1183–1190. doi: 10.1128/JB.00925-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao W, Zhong Y, Yuan H, Wang J, Zheng H, Wang Y, Cen X, Xu F, Bai J, Han X, Lu G, Zhu Y, Shao Z, Yan H, Li C, Peng N, Zhang Z, Zhang Y, Lin W, Fan Y, Qin Z, Hu Y, Zhu B, Wang S, Ding X, Zhao GP. Complete genome sequence of the rifamycin SV-producing Amycolatopsis mediterranei U32 revealed its genetic characteristics in phylogeny and metabolism. Cell Res. 2010;20(10):1096–1108. doi: 10.1038/cr.2010.87. [DOI] [PubMed] [Google Scholar]
- 20.He J, Shao X, Zheng H, Li M, Wang J, Zhang Q, Li L, Liu Z, Sun M, Wang S, Yu Z. Complete genome sequence of Bacillus thuringiensis mutant strain BMB171. J. Bacteriol. 2010;192(15):4074–4075. doi: 10.1128/JB.00562-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez P, Hornung A, Welzel K, Unsin C, Wohlleben W, Weber T, Pelzer S. Isolation of the lysolipin gene cluster of Streptomyces tendae Tu 4042. Gene. 2010;461(1-2):5–14. doi: 10.1016/j.gene.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Zech H, Thole S, Schreiber K, Kalhofer D, Voget S, Brinkhoff T, Simon M, Schomburg D, Rabus R. Growth phase-dependent global protein and metabolite profiles of Phaeobacter gallaeciensis strain DSM 17395, a member of the marine Roseobacter-clade. Proteomics. 2009;9(14):3677–3697. doi: 10.1002/pmic.200900120. [DOI] [PubMed] [Google Scholar]
- 23.Strittmatter AW, Liesegang H, Rabus R, Decker I, Amann J, Andres S, Henne A, Fricke WF, Martinez-Arias R, Bartels D, Goesmann A, Krause L, Puhler A, Klenk HP, Richter M, Schuler M, Glockner FO, Meyerdierks A, Gottschalk G, Amann R. Genome sequence of Desulfobacterium autotrophicum HRM2, a marine sulfate reducer oxidizing organic carbon completely to carbon dioxide. Environ. Microbiol. 2009;11(5):1038–1055. doi: 10.1111/j.1462-2920.2008.01825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng H, Lu L, Wang B, Pu S, Zhang X, Zhu G, Shi W, Zhang L, Wang H, Wang S, Zhao G, Zhang Y. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS ONE. 2008;3(6):e2375. doi: 10.1371/journal.pone.0002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richter M, Kube M, Bazylinski DA, Lombardot T, Glockner FO, Reinhardt R, Schuler D. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. J. Bacteriol. 2007;189(13):4899–4910. doi: 10.1128/JB.00119-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mussmann M, Hu FZ, Richter M, de Beer D, Preisler A, Jorgensen BB, Huntemann M, Glockner FO, Amann R, Koopman WJ, Lasken RS, Janto B, Hogg J, Stoodley P, Boissy R, Ehrlich GD. Insights into the genome of large sulfur bacteria revealed by analysis of single filaments. PLoS Biol. 2007;5(9):e230. doi: 10.1371/journal.pbio.0050230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyerdierks A, Kube M, Kostadinov I, Teeling H, Glockner FO, Reinhardt R, Amann R. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ. Microbiol. 2010;12(2):422–439. doi: 10.1111/j.1462-2920.2009.02083.x. [DOI] [PubMed] [Google Scholar]
- 28.Tan le V, Ha do Q, Hien VM, van der Hoek L, Farrar J, de Jong MD. Me Tri virus: a Semliki Forest virus strain from Vietnam? J. Gen. Virol. 2008;89(Pt 9):2132–2135. doi: 10.1099/vir.0.2008/002121-0. [DOI] [PubMed] [Google Scholar]
- 29.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27(23):4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes.Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29(12):2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tech M, Merkl R. YACOP: Enhanced gene prediction obtained by a combination of existing methods. In Silico Biol. 2003;3(4):441–451. [PubMed] [Google Scholar]
- 32.Dávila AMR, Lorenzini DM, Mendes PN, Satake TS, Sousa GR, Campos LM, Mazzoni CJ, Wagner G, Pires PF, Grisard EC, Cavalcanti MCR, Campos MLM. GARSA: genomic analysis resources for sequence annotation. Bioinform. 2005;21(23):4302–4303. doi: 10.1093/bioinformatics/bti705. [DOI] [PubMed] [Google Scholar]
- 33.Lan SF, Huang CH, Chang CH, Liao WC, Lin IH, Jian WN, Wu YG, Chen SY, Wong HC. Characterization of a new plasmid-like prophage in a pandemic Vibrio parahaemolyticus O3:K6 strain. Appl. Environ. Microbiol. 2009;75(9):2659–2667. doi: 10.1128/AEM.02483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pyrc K, Berkhout B, van der Hoek L. The novel human coronaviruses NL63 and HKU1. J. Virol. 2007;81(7):3051–3057. doi: 10.1128/JVI.01466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang XC, Zhang JX, Zhang SY, Wang P, Fan XH, Li LF, Li G, Dong BQ, Liu W, Cheung CL, Xu KM, Song WJ, Vijaykrishna D, Poon LL, Peiris JS, Smith GJ, Chen H, Guan Y. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006;80(15):7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu DK, Peiris JS, Chen H, Guan Y, Poon LL. Genomic characterizations of bat coronaviruses (1A, 1B and HKU8) and evidence for co-infections in Miniopterus bats. J. Gen. Virol. 2008;89(Pt 5):1282–1287. doi: 10.1099/vir.0.83605-0. [DOI] [PubMed] [Google Scholar]
- 37.van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10(4):368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan YJ, Goh PY, Fielding BC, Shen S, Chou CF, Fu JL, Leong HN, Leo YS, Ooi EE, Ling AE, Lim SG, Hong W. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 2004;11(2):362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang HZ, Zhang H, Kemnitzer W, Tseng B, Cinatl J Jr, Michaelis M, Doerr HW, Cai SX. Design and synthesis of dipeptidyl glutaminyl fluoromethyl ketones as potent severe acute respiratory syndrome coronovirus (SARS-CoV) inhibitors. J. Med. Chem. 2006;49(3):1198–1201. doi: 10.1021/jm0507678. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Gui C, Luo X, Yang Q, Günther S, Scandella E, Drosten C, Bai D, He X, Ludewig B, Chen J, Luo H, Yang Y, Yang Y, Zou J, Thiel V, Chen K, Shen J, Shen X, Jiang H. Cinanserin Is an Inhibitor of the 3C-Like Proteinase of Severe Acute Respiratory Syndrome Coronavirus and Strongly Reduces Virus Replication In Vitro. J. Virol. 2005;79(11):7095–7103. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen S, Hu T, Zhang J, Chen J, Chen K, Ding J, Jiang H, Shen X. Mutation of Gly-11 on the Dimer Interface Results in the Complete Crystallographic Dimer Dissociation of Severe Acute Respiratory Syndrome Coronavirus 3C-like Protease. J. Biol. Chem. 2008;283(1):554–564. doi: 10.1074/jbc.M705240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S, Luo H, Chen L, Chen J, Shen J, Zhu W, Chen K, Shen X, Jiang H. An overall picture of SARS coronavirus (SARS-CoV) genome-encoded major proteins: structures, functions and drug development. Curr. Pharm. Des. 2006;12(35):4539–4553. doi: 10.2174/138161206779010459. [DOI] [PubMed] [Google Scholar]
- 43.Fan K, Ma L, Han X, Liang H, Wei P, Liu Y, Lai L. The substrate specificity of SARS coronavirus 3C-like proteinase. Biochem. Biophy. Res. Commun. 2005;329(3):934–940. doi: 10.1016/j.bbrc.2005.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang S, Shen H, Wang J, Tay FPL, Liu DX. Functional and Genetic Studies of the Substrate Specificity of Coronavirus Infectious Bronchitis Virus 3C-Like Proteinase. J. Virology. 2010;84(14):7325–7336. doi: 10.1128/JVI.02490-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang SG, Shen H, Wang J, Tay FPL, Liu DX. Proteolytic processing of polyproteins 1a and 1ab between non-structural proteins 10 and 11/12 of Coronavirus infectious bronchitis virus is dispensable for viral replication in cultured cells. Virol. 2008;379(2):175–180. doi: 10.1016/j.virol.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goetz DH, Choe Y, Hansell E, Chen YT, McDowell M, Jonsson CB, Roush WR, McKerrow J, Craik CS. Substrate Specificity Profiling and Identification of a New Class of Inhibitor for the Major Protease of the SARS Coronavirus†,‡. Biochem. 2007;46(30):8744–8752. doi: 10.1021/bi0621415. [DOI] [PubMed] [Google Scholar]
- 47.Han YS, Chang GG, Juo CG, Lee HJ, Yeh SH, Hsu JTA, Chen X. Papain-like protease 2 (PLP2) from severe acute respiratory syndrome coronavirus (SARS-CoV): Expression, purification, characterization, and inhibition. Biochem. 2005;44(30):10349–10359. doi: 10.1021/bi0504761. [DOI] [PubMed] [Google Scholar]
- 48.Joseph JS, Saikatendu KS, Subramanian V, Neuman BW, Brooun A, Griffith M, Moy K, Yadav MK, Velasquez J, Buchmeier MJ, Stevens RC, Kuhn P. Crystal Structure of Nonstructural Protein 10 from the Severe Acute Respiratory Syndrome Coronavirus Reveals a Novel Fold with Two Zinc-Binding Motifs. J. Virol. 2006;80(16):7894–7901. doi: 10.1128/JVI.00467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joseph JS, Saikatendu KS, Subramanian V, Neuman BW, Buchmeier MJ, Stevens RC, Kuhn P. Crystal Structure of a Monomeric Form of Severe Acute Respiratory Syndrome Coronavirus Endonuclease nsp15 Suggests a Role for Hexamerization as an Allosteric Switch. J. Virol. 2007;81(12):6700–6708. doi: 10.1128/JVI.02817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiemer L, Lund O, Brunak S, Blom N. Coronavirus 3CLpro proteinase cleavage sites: possible relevance to SARS virus pathology. BMC Bioinform. 2004;5:72–72. doi: 10.1186/1471-2105-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin C-W, Tsai C-H, Tsai F-J, Chen P-J, Lai C-C, Wan L, Chiu H-H, Lin K-H. Characterization of trans- and cis-cleavage activity of the SARS coronavirus 3CLpro protease: basis for the in vitro screening of anti-SARS drugs. FEBS Lett. 2004;574(1-3):131–137. doi: 10.1016/j.febslet.2004.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sydnes MO, Hayashi Y, Sharma VK, Hamada T, Bacha U, Barrila J, Freire E, Kiso Y. Synthesis of glutamic acid and glutamine peptides possessing a trifluoromethyl ketone group as SARS-CoV 3CL protease inhibitors. Tetrahedron. 2006;62(36):8601–8609. doi: 10.1016/j.tet.2006.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tian X, Lu G, Gao F, Peng H, Feng Y, Ma G, Bartlam M, Tian K, Yan J, Hilgenfeld R, Gao GF. Structure and Cleavage Specificity of the Chymotrypsin-Like Serine Protease (3CLSP/nsp4) of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV). J. Mol. Biol. 2009;392(4):977–993. doi: 10.1016/j.jmb.2009.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren S-X, Fu G, Jiang X-G, Zeng R, Miao Y-G, Xu H, Zhang Y-X, Xiong H, Lu G, Lu L-F, Jiang H-Q, Jia J, Tu Y-F, Jiang J-X, Gu W-Y, Zhang Y-Q, Cai Z, Sheng H-H, Yin H-F, Zhang Y, Zhu G-F, Wan M, Huang H-L, Qian Z, Wang S-Y, Ma W, Yao Z-J, Shen Y, Qiang B-Q, Xia Q-C, Guo X-K, Danchin A, Saint Girons I, Somerville RL, Wen Y-M, Shi M-H, Chen Z, Xu J-G, Zhao G-P. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422(6934):888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- 55.Lin MF, Deoras AN, Rasmussen MD, Kellis M. Performance and scalability of discriminative metrics for comparative gene identification in 12 Drosophila genomes. PLoS Comput. Biol. 2008;4(4):e1000067. doi: 10.1371/journal.pcbi.1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao F, Zhang CT. Comparison of various algorithms for recognizing short coding sequences of human genes. Bioinfor- m. 2004;20(5):673–681. doi: 10.1093/bioinformatics/btg467. [DOI] [PubMed] [Google Scholar]
- 57.Saeys Y, Rouzé P, Van de Peer Y. In search of the small ones: improved prediction of short exons in vertebrates, plants, fungi and protists. Bioinform. 2007;23(4):414–420. doi: 10.1093/bioinformatics/btl639. [DOI] [PubMed] [Google Scholar]
- 58.Wu Y, Liew AW-C, Yan H, Yang M. Classification of short human exons and introns based on statistical features. Phys. Rev. E. 2003;67(6):061916. doi: 10.1103/PhysRevE.67.061916. [DOI] [PubMed] [Google Scholar]
- 59.Yan M, Lin ZS, Zhang CT. A new fourier transform approach for protein coding measure based on the format of the Z-curve. Bioinform. 1998;14(8):685–690. doi: 10.1093/bioinformatics/14.8.685. [DOI] [PubMed] [Google Scholar]
- 60.Rushdi A, Tuqan J. In Acoustics, Speech and Signal Processing 2006. ICASSP 2006 Proceedings. 2006 IEEE International Conference on. 2006;2 pp:II–II. [Google Scholar]
- 61.Ma B, Zhu Y-S. In Bioinformatics and Biomedical Engineering 2007. ICBBE 2007. The 1st International Conference on. 2007:188–191. [Google Scholar]
- 62.Law NF, Cheng KO, Siu WC. On relationship of Z-curve and Fourier approaches for DNA coding sequence classification. Bioinform. 2006;1(7):242–246. doi: 10.6026/97320630001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang R, Zhang CT. Multiple replication origins of the archaeon Halobacterium species NRC-1. Biochem. Biophys. Res. Commun. 2003;302(4):728–734. doi: 10.1016/s0006-291x(03)00252-3. [DOI] [PubMed] [Google Scholar]
- 64.Robinson NP, Dionne I, Lundgren M, Marsh VL, Bernander R, Bell SD. Identification of Two Origins of Replication in the Single Chromosome of the Archaeon Sulfolobus solfataricus. Cell. 2004;116(1):25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- 65.Robinson NP, Bell SD. Origins of DNA replication in the three domains of life. The FEBS J. 2005;272(15):3757–3766. doi: 10.1111/j.1742-4658.2005.04768.x. [DOI] [PubMed] [Google Scholar]
- 66.Lundgren M, Bernander R. Archaeal cell cycle progress. Curr. Opin. Microbiol. 2005;8(6):662–668. doi: 10.1016/j.mib.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 67.Soppa J. From genomes to function: haloarchaea as model organisms. Microbiol. 2006;152(3):585–590. doi: 10.1099/mic.0.28504-0. [DOI] [PubMed] [Google Scholar]
- 68.Zhang R, Zhang CT. Identification of replication origins in the genome of the methanogenic archaeon, Methanocaldococcus jannaschii. Extremophiles. 2004;8(3):253–258. doi: 10.1007/s00792-004-0385-4. [DOI] [PubMed] [Google Scholar]
- 69.Zhang R, Zhang CT. Single replication origin of the archaeon Methanosarcina mazei revealed by the Z-curve method. Biochem. Biophys. Res. Commun. 2002;297(2):396–400. doi: 10.1016/s0006-291x(02)02214-3. [DOI] [PubMed] [Google Scholar]
- 70.Zhang R, Zhang CT. Identification of replication origins in archaeal genomes based on the Z-curve method. Archaea. 2005;1(5):335–346. doi: 10.1155/2005/509646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson NP, Bell SD. Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes. Proc. Natl. Acad. Sci. 2007;104(14):5806–5811. doi: 10.1073/pnas.0700206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen L, Brügger K, Skovgaard M, Redder P, She Q, Torarinsson E, Greve B, Awayez M, Zibat A, Klenk H-P, Garrett RA. The Genome of Sulfolobus acidocaldarius, a Model Organism of the Crenarchaeota. J. Bacteriol. 2005;187(14):4992–4999. doi: 10.1128/JB.187.14.4992-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Norais C, Hawkins M, Hartman AL, Eisen JA, Myllykallio H, Allers T. Genetic and Physical Mapping of DNA Replication Origins in Haloferax volcanii. PLoS Genet. 2007;3(5):e77. doi: 10.1371/journal.pgen.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ravin NV, Mardanov AV, Beletsky AV, Kublanov IV, Kolganova TV, Lebedinsky AV, Chernyh NA, Bonch-Osmolovskaya EA, Skryabin KG. Complete Genome Sequence of the Anaerobic, Protein-Degrading Hyperthermophilic Crenarchaeon Desulfurococcus kamchatkensis. J. Bacteriol. 2009;191(7):2371–2379. doi: 10.1128/JB.01525-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mardanov AV, Ravin NV, Svetlitchnyi VA, Beletsky AV, Miroshnichenko ML, Bonch-Osmolovskaya EA, Skryabin KG. Metabolic Versatility and Indigenous Origin of the Archaeon Thermococcus sibiricus, Isolated from a Siberian Oil Reservoir, as Revealed by Genome Analysis. App. Environ. Microbiol. 2009;75(13):4580–4588. doi: 10.1128/AEM.00718-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flynn KM, Vohr SH, Hatcher PJ, Cooper VS. Evolutionary Rates and Gene Dispensability Associate with Replication Timing in the Archaeon Sulfolobus islandicus. Genom. Biol. Evol. 2010;2:859–869. doi: 10.1093/gbe/evq068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 1996;13(5):660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 78.Gao F, Zhang CT. Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinform. 2008;9:79. doi: 10.1186/1471-2105-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Vries SP, van Hijum SA, Schueler W, Riesbeck K, Hays JP, Hermans PW, Bootsma HJ. Genome analysis of Moraxella catarrhalis strain BBH18 [corrected] a human respiratory tract pathogen. J. Bacteriol. 2010;192(14):3574–3583. doi: 10.1128/JB.00121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao F, Zhang CT. Origins of replication in Sorangium cellulosum and Microcystis aeruginosa. DNA Res. 2008;15(3):169–171. doi: 10.1093/dnares/dsn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao F, Zhang CT. Origins of replication in Cyanothece 51142. Proc. Natl. Acad. Sci. U. S. A. E125 author reply. 2008;105(52):E126–127. doi: 10.1073/pnas.0809987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, Michaux A, Benotmane MA, Leys N, Vallaeys T, Lapidus A, Monchy S, Medigue C, Taghavi S, McCorkle S, Dunn J, van der Lelie D, Mergeay M. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS ONE. 2010;5(5):e10433. doi: 10.1371/journal.pone.0010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ran L, Larsson J, Vigil-Stenman T, Nylander JA, Ininbergs K, Zheng WW, Lapidus A, Lowry S, Haselkorn R, Bergman B. Genome erosion in a nitrogen-fixing vertically transmitted endosymbiotic multicellular cyanobacterium. PLoS ONE. 2010;5(7):e11486. doi: 10.1371/journal.pone.0011486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trost E, Ott L, Schneider J, Schroder J, Jaenicke S, Goesmann A, Husemann P, Stoye J, Dorella FA, Rocha FS, Soares Sde C, D'Afonseca V, Miyoshi A, Ruiz J, Silva A, Azevedo V, Burkovski A, Guiso N, Join-Lambert OF, Kayal S, Tauch A. The complete genome sequence of Corynebacterium pseudotuberculosis FRC41 isolated from a 12-year-old girl with necrotizing lymphadenitis reveals insights into gene-regulatory networks contributing to virulence. BMC Genom. 2010;11:728. doi: 10.1186/1471-2164-11-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paul D, Bridges SM, Burgess SC, Dandass YS, Lawrence ML. Complete genome and comparative analysis of the chemolithoautotrophic bacterium Oligotropha carboxidovorans OM5. BMC Genom. 2010;11:511. doi: 10.1186/1471-2164-11-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakayama K, Kurokawa K, Fukuhara M, Urakami H, Yamamoto S, Yamazaki K, Ogura Y, Ooka T, Hayashi T. Genome comparison and phylogenetic analysis of Orientia tsutsugamushi strains. DNA Res. 2010;17(5):281–291. doi: 10.1093/dnares/dsq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Falentin H, Deutsch SM, Jan G, Loux V, Thierry A, Parayre S, Maillard MB, Dherbecourt J, Cousin FJ, Jardin J, Siguier P, Couloux A, Barbe V, Vacherie B, Wincker P, Gibrat JF, Gaillardin C, Lortal S. The complete genome of Propionibacterium freudenreichii CIRM-BIA1, a hardy actinobacterium with food and probiotic applications. PLoS ONE. 2010;5(7):e11748. doi: 10.1371/journal.pone.0011748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lau SK, Fan RY, Ho TC, Wong GK, Tsang AK, Teng JL, Chen W, Watt RM, Curreem SO, Tse H, Yuen KY, Woo PC. Environmental adaptability and stress tolerance of Laribacter hongkongensis: a genome-wide analysis. Cell Biosci. 2011;1(1):22. doi: 10.1186/2045-3701-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bryan A, Swanson MS. Oligonucleotides stimulate genomic alterations of Legionella pneumophila. Mol. Microbiol. 2011;80(1):231–247. doi: 10.1111/j.1365-2958.2011.07573.x. [DOI] [PubMed] [Google Scholar]
- 90.Wei W, Guo FB. Strong Strand Composition Bias in the Genome of Ehrlichia canis Revealed by Multiple Methods. Open Microbiol. J. 2010;4:98–102. doi: 10.2174/1874285801004010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Macaya G, Thiery J-P, Bernardi G. An approach to the organization of eukaryotic genomes at a macromolecular level. J. Mol. Biol. 1976;108(1):237–254. doi: 10.1016/s0022-2836(76)80105-2. [DOI] [PubMed] [Google Scholar]
- 92.Cuny G, Soriano P, Macaya G, Bernardi G. The major components of the mouse and human genomes.1. Prepartion basic properties and compositional heterogeneity. Eur. J. Biochem. FEBS . 1981; 115(2):227–233. doi: 10.1111/j.1432-1033.1981.tb05227.x. [DOI] [PubMed] [Google Scholar]
- 93.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 94.Zhang CT, Zhang R. An isochore map of the human genome based on the Z-curve method. Gene. 2003;317(1-2):127–135. doi: 10.1016/s0378-1119(03)00665-6. [DOI] [PubMed] [Google Scholar]
- 95.Zhang CT, Zhang R. Isochore structures in the mouse genome. Genom. 2004;83(3):384–394. doi: 10.1016/j.ygeno.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 96.Zhang R, Zhang CT. Isochore structures in the genome of the plant Arabidopsis thaliana. J. Mol. Evol. 2004;59(2):227–238. doi: 10.1007/s00239-004-2617-8. [DOI] [PubMed] [Google Scholar]
- 97.Gao F, Zhang CT. Isochore structures in the chicken genome. FEBS J. 2006;273(8):1637–1648. doi: 10.1111/j.1742-4658.2006.05178.x. [DOI] [PubMed] [Google Scholar]
- 98.Wen SY, Zhang CT. Identification of isochore boundaries in the human genome using the technique of wavelet multiresolution analysis. Biochem. Biophys. Res. Commun. 2003;311(1):215–222. doi: 10.1016/j.bbrc.2003.09.198. [DOI] [PubMed] [Google Scholar]
- 99.Zhang W, Wu W, Lin W, Zhou P, Dai L, Zhang Y, Huang J, Zhang D. Deciphering heterogeneity in pig genome assembly Sscrofa9 by isochore and isochore-like region analyses. PLoS ONE. 2010;5(10):e13303. doi: 10.1371/journal.pone.0013303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Do JH, Miyano S. The GC and window-averaged DNA curvature profile of secondary metabolite gene cluster in Aspergillus fumigatus genome. Appl. Microbiol. Biotechnol. 2008;80(5):841–847. doi: 10.1007/s00253-008-1638-4. [DOI] [PubMed] [Google Scholar]
- 101.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 102.Charkowski AO. Making sense of an alphabet soup: the use of a new bioinformatics tool for identification of novel gene islands.Focus on "Identification of genomic islands in the genome of Bacillus cereus by comparative analysis with Bacillus anthracis". Physiol. Genom. 2004;16(2):180–181. doi: 10.1152/physiolgenomics.00199.2003. [DOI] [PubMed] [Google Scholar]
- 103.Zhang R, Zhang CT. Identification of genomic islands in the genome of Bacillus cereus by comparative analysis with Bacillus anthracis. Physiol. Genom. 2003;16(1):19–23. doi: 10.1152/physiolgenomics.00170.2003. [DOI] [PubMed] [Google Scholar]
- 104.Zhang R, Zhang CT. A systematic method to identify genomic islands and its applications in analyzing the genomes of Corynebacterium glutamicum and Vibrio vulnificus CMCP6 chromosome I. Bioinform. 2004;20(5):612–622. doi: 10.1093/bioinformatics/btg453. [DOI] [PubMed] [Google Scholar]
- 105.Zhang R, Zhang CT. Genomic islands in the Corynebacterium efficiens genome. Appl. Environ. Microbiol. 2005;71(6):3126–3130. doi: 10.1128/AEM.71.6.3126-3130.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang CT, Zhang R. Genomic islands in Rhodopseudomonas palustris. Nat. Biotechnol. 2004;22(9):1078–1079. doi: 10.1038/nbt0904-1078b. [DOI] [PubMed] [Google Scholar]
- 107.Chen LL. Identification of genomic islands in six plant pathogens. Gene. 2006;374:134–141. doi: 10.1016/j.gene.2006.01.029. [DOI] [PubMed] [Google Scholar]
- 108.Jayapal KP, Lian W, Glod F, Sherman DH, Hu WS. Comparative genomic hybridizations reveal absence of large Streptomyces coelicolor genomic islands in Streptomyces lividans. BMC Genom. 2007;8:229. doi: 10.1186/1471-2164-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Greub G, Collyn F, Guy L, Roten CA. A genomic island present along the bacterial chromosome of the Parachlamydiaceae UWE25, an obligate amoebal endosymbiont, encodes a potentially functional F-like conjugative DNA transfer system. BMC Microbiol. 2004;4:48. doi: 10.1186/1471-2180-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nakagawa S, Takaki Y, Shimamura S, Reysenbach AL, Takai K, Horikoshi K. Deep-sea vent epsilon-proteobacterial genomes provide insights into emergence of pathogens. Proc. Natl. Acad. Sci. U. S. A. 2007;104(29):12146–12150. doi: 10.1073/pnas.0700687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jung J, Madsen EL, Jeon CO, Park W. Comparative genomic analysis of Acinetobacter oleivorans DR1 to determine strain-specific genomic regions and gentisate biodegradation. Appl Environ. Microbiol. 2011;77(20):7418–7424. doi: 10.1128/AEM.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yan DZ, Kang JX, Liu DQ. Genomic analysis of the aromatic catabolic pathways from Silicibacter pomeroyi DSS-3. Ann. Microbiol. 2009;59(4):789–800. [Google Scholar]
- 113.Ou HY, Guo FB, Zhang CT. GS-Finder: a program to find bacterial gene start sites with a self-training method. Int. J. Biochem. Cell. Biol. 2004;36(3):535–544. doi: 10.1016/j.biocel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 114.Yang JY, Zhou Y, Yu ZG, Anh V, Zhou LQ. Human Pol II promoter recognition based on primary sequences and free energy of dinucleotides. BMC Bioinform. 2008;9:113. doi: 10.1186/1471-2105-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Song K. Recognition of prokaryotic promoters based on a novel variable-window Z-curve method. Nucleic Acids Res. 2012;40(3):963–971. doi: 10.1093/nar/gkr795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu X, Liu H, Su J, Lv J, Cui Y, Wang F, Zhang Y. Z-curve theory-based analysis of the dynamic nature of nucleosome positioning in Saccharomyces cerevisiae. Gene. 2013;530(1):8–18. doi: 10.1016/j.gene.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 117.Zhang CT, Zhang R, Ou HY. The Z-curve database: a graphic representation of genome sequences. Bioinform. 2003;19(5):593–599. doi: 10.1093/bioinformatics/btg041. [DOI] [PubMed] [Google Scholar]
- 118.Zheng WX, Chen LL, Ou HY, Gao F, Zhang CT. Coronavirus phylogeny based on a geometric approach. Mol. Phylogenet. Evol. 2005;36(2):224–232. doi: 10.1016/j.ympev.2005.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang R, Zhang CT. Accurate localization of the integration sites of two genomic islands at single-nucleotide resolution in the genome of Bacillus cereus ATCC 10987. Comp. Funct. Genom. 2008;1:451930. doi: 10.1155/2008/451930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chou KC, Zhang CT. Diagrammatization of codon usage in 339 human immunodeficiency virus proteins and its biological implication. AIDS Res Hum Retroviruses. 1992;8(12):1967–1976. doi: 10.1089/aid.1992.8.1967. [DOI] [PubMed] [Google Scholar]
- 121.Zhang CT, Zhan Y. Analysis on the distribution of bases in 1487 human protein coding sequences. J. Theor. Biol. 1994;167(2):161–166. doi: 10.1006/jtbi.1994.1060. [DOI] [PubMed] [Google Scholar]
- 122.Zhang CT, Chou KC. A graphic approach to analyzing codon usage in 1562 Escherichia coli protein coding sequences. J. Mol. Biol. 1994;238(1):1–8. doi: 10.1006/jmbi.1994.1263. [DOI] [PubMed] [Google Scholar]
- 123.Wang J, Zhang CT. Analysis of the codon usage pattern in the Vibrio cholerae genome. J. Biomol. Struct. Dyn. 2001;18(6):872–880. doi: 10.1080/07391102.2001.10506714. [DOI] [PubMed] [Google Scholar]
- 124.Guo FB, Wang J, Zhang CT. Gene recognition based on nucleotide distribution of ORFs in a hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 2004;11(6):361–370. doi: 10.1093/dnares/11.6.361. [DOI] [PubMed] [Google Scholar]
- 125.Ou HY, Guo FB, Zhang CT. Analysis of nucleotide distribution in the genome of Streptomyces coelicolor A3(2) using the Z-curve method. FEBS Lett. 2003;540(1-3):188–194. doi: 10.1016/s0014-5793(03)00263-1. [DOI] [PubMed] [Google Scholar]
- 126.Chen LL, Zhang CT. Seven GC-rich microbial genomes adopt similar codon usage patterns regardless of their phylogenetic lineages. Biochem. Biophys. Res. Commun. 2003;306(1):310–317. doi: 10.1016/s0006-291x(03)00973-2. [DOI] [PubMed] [Google Scholar]
- 127.Woebken D, Teeling H, Wecker P, Dumitriu A, Kostadinov I, Delong EF, Amann R, Glockner FO. Fosmids of novel marine Planctomycetes from the Namibian and Oregon coast upwelling systems and their cross-comparison with planctomycete genomes. ISME J. 2007;1(5):419–435. doi: 10.1038/ismej.2007.63. [DOI] [PubMed] [Google Scholar]
- 128.Jones BV, Marchesi JR. Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nat. Methods. 2007;4(1):55–61. doi: 10.1038/nmeth964. [DOI] [PubMed] [Google Scholar]
- 129.Wietzorrek A, Schwarz H, Herrmann C, Braun V. The genome of the novel phage Rtp, with a rosette-like tail tip, is homologous to the genome of phage T1. J. Bacteriol. 2006;188(4):1419–1436. doi: 10.1128/JB.188.4.1419-1436.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, Xu H, Zhang YX, Xiong H, Lu G, Lu LF, Jiang HQ, Jia J, Tu YF, Jiang JX, Gu WY, Zhang YQ, Cai Z, Sheng HH, Yin HF, Zhang Y, Zhu GF, Wan M, Huang HL, Qian Z, Wang SY, Ma W, Yao ZJ, Shen Y, Qiang BQ, Xia QC, Guo XK, Danchin A, Saint Girons I, Somerville RL, Wen YM, Shi MH, Chen Z, Xu JG, Zhao GP. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422(6934):888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- 131.Song K, Zhang Z, Tong TP, Wu F. Classifier assessment and feature selection for recognizing short coding sequences of human genes. J. Comput. Biol. 2012;19(3):251–260. doi: 10.1089/cmb.2011.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gao F, Luo H, Zhang CT. DoriC 5.: an updated database of oriC regions in both bacterial and archaeal genomes. Nucleic Acids Res. 2013;41(Database issue):D90–93. doi: 10.1093/nar/gks990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gao F, Zhang CT. DoriC: a database of oriC regions in bacterial genomes. Bioinform. 2007;23(14):1866–1867. doi: 10.1093/bioinformatics/btm255. [DOI] [PubMed] [Google Scholar]
- 134.Sabri M, Hauser R, Ouellette M, Liu J, Dehbi M, Moeck G, Garcia E, Titz B, Uetz P, Moineau S. Genome annotation and intraviral interactome for the Streptococcus pneumoniae virulent phage Dp-1. J. Bacteriol. 2011;193(2):551–562. doi: 10.1128/JB.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jain PK, Kush D, Ramachandran S, Verma SK. Isolation and characterization of R-plasmid pPRS3a from Bacillus cereus GC subgroup a PRS3. Int. J. Integr. Biol. Int. J. Integ. Biol. 2011;11(1):1–7. [Google Scholar]
- 136.Dyall-Smith ML, Pfeiffer F, Klee K, Palm P, Gross K, Schuster SC, Rampp M, Oesterhelt D. Haloquadratum walsbyi: limited diversity in a global pond. PLoS ONE. 2011;6(6):e20968. doi: 10.1371/journal.pone.0020968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zheng WX, Zhang CT. Biological implications of isochore boundaries in the human genome. J. Biomol. Struct. Dyn. 2008;25(4):327–336. doi: 10.1080/07391102.2008.10507181. [DOI] [PubMed] [Google Scholar]
- 138.Gao F, Zhang CT. Prediction of replication time zones at single nucleotide resolution in the human genome. FEBS Lett. 2008;582(16):2441–2444. doi: 10.1016/j.febslet.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 139.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. The genome sequence of Bifidobacterium longum subsp.infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U. S. A. 2008;105(48):18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hernandez A, Lopez JC, Santamaria R, Diaz M, Fernandez-Abalos JM, Copa-Patino JL, Soliveri J. Xylan-binding xylanase Xyl30 from Streptomyces avermitilis: cloning, characterization, and overproduction in solid-state fermentation. Int. Microbiol. 2008;11(2):133–141. [PubMed] [Google Scholar]
- 141.McNally RR, Toth IK, Cock PJ, Pritchard L, Hedley PE, Morris JA, Zhao Y, Sundin GW. Genetic characterization of the HrpL regulon of the fire blight pathogen Erwinia amylovora reveals novel virulence factors. Mol. Plant. Pathol. 2012;13(2):160–173. doi: 10.1111/j.1364-3703.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ryan MP, Pembroke JT, Adley CC. Novel Tn4371-ICE like element in Ralstonia pickettii and genome mining for comparative elements. BMC Microbiol. 2009;9:242. doi: 10.1186/1471-2180-9-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang CT. In Visualizing biological information Pickover C.World Scientific Singapore. River Edge N J. . 1995:84–95. [Google Scholar]