Abstract
Chlamydia trachomatis infections are a global health problem. This obligate intracellular bacterial pathogen comprises lymphogranuloma venereum (L1–L3), ocular (A–C) and genital (D–K) serovars. Although genetically similar, each serovar group differs in disease severity and tissue tropism through mechanisms that are not well understood. It is clear that host genetic differences also play a role in chlamydial disease outcome and key host polymorphisms are beginning to emerge from both human and experimental animal studies. In this review, we will highlight pathogen and host genes that link genetic diversity, disease severity and tissue tropism. We will also use this information to provide new insights that may be helpful in developing improved management strategies for these important pathogens.
Keywords: Chlamydia trachomatis, disease severity, gene polymorphisms, genetic variation, genital tract infections, tissue tropism
Chlamydiae are strict obligate intracellular pathogens that depend on eukaryotic host cells to complete their unique biphasic developmental cycle [1]. Chlamydia trachomatis is one of several chlamydial species that cause disease in humans. It is by far the most significant in terms of public health concerns, causing hundreds of millions of cases of human genital tract (serovars D–K) or ocular (serovars A–C) diseases throughout the world. C. trachomatis genital infections are global and cause substantial morbidity, especially in women [2]. Endemic C. trachomatis ocular disease is restricted to the poorest communities, affecting people with little or no healthcare and resulting in end-stage blinding trachoma [3]. Interestingly, different C. trachomatis serovars can infect and survive in diverse host niches represented by different tissue tropism causing a wide spectrum of diseases in humans (Figure 1) [4]. For example, genital serovars favor genital tract epithelial cells while ocular serovars infect conjunctival epithelial cells, and the lymphogranuloma venereum (LGV) serovars infect macrophages and spread systemically through lymph nodes [5]. There are more than 100 completed genome sequences for C. trachomatis strains archived and publically available in online databases at the National Center for Biotechno logy Information (NCBI) [201], Sanger Institute [202] or European Molecular Biology Laboratory (EMBL) [203], reflecting each of the three disease wbiovars (serovar groups based on pathotype). All sequenced genomes demonstrated similar size (1.04–1.05 Mb), nucleotide sequence similarity (>99% identical) and nearly identical synteny [4,6,7,201,203]. Given that a 1% difference per 1 million base pairs represents approximately 10 kb of variability, and that differences in genes that define C. trachomatis virulence and tissue tropism are disproportionately large [8], it may not be surprising that genetic variability is key to understanding chlamydial virulence differences. In women, lower genital tract infection may or may not be symptomatic and may or may not spread to the upper genital tract. Spread of the infection may or may not result in upper genital tract complications such as pelvic inflammatory diseases [9]. In trachoma endemic areas, the majority of individuals in affected communities becomes infected. Despite this similar exposure rate, only a minority of infected individuals develop severe long-term consequences of acute ocular infection. A number of variables may play roles in defining those at risk for complications, including the presence of different strains circulating within the community, the pathogen burden of each infected individual and polymorphisms in host genetic risk factors [10-19]. The intent of this review is to provide insights for both pathogen virulence factors and host reactivity in defining effectors that contribute to chlamydial disease severity. It is, however, important to note that the full genetic diversity in C. trachomatis is not yet adequately described for any regulatory or epigenetic changes that may lead to a change of phenotype or adaptation to a niche. Current technology allows for the effective management of large ‘omics’ data sets that can be manipulated using systems-based approaches to sort through both pathogen and host genetic variation to determine how genetic and gene expression differences impact disease severity and tissue tropism. A number of system-based approaches, including comparative sequencing, cell culture systems, in vivo infections, epidemiologic studies and mathematical modeling have been recently reviewed [20]. These types of approaches are required to help understand complex traits and epistatic interactions in order to gain a more comprehensive understanding of host and pathogen factors that impact the outcome of chlamydial infections. In this review, it is our purpose to indicate how current information in these areas might help us understand how genetic variability in both the pathogen and the host contribute to disease severity and tissue tropism with a focus on C. trachomatis and the human host.
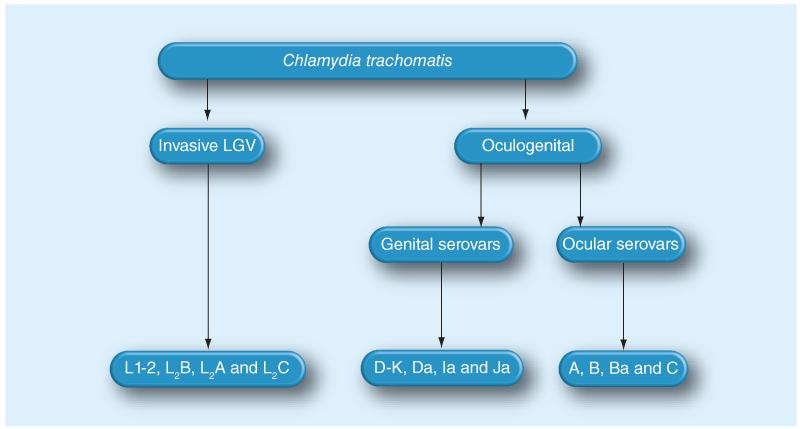
Figure 1. Classification of Chlamydia trachomatis based on tissue tropism.
Chlamydia trachomatis is divided into the oculogenital and LGV biovars. The ocular (A–C) and genital (D–K) serovars infect conjuctival and genital epithelia, respectively, while LGV (L1–L3) spreads systemically in macrophages and other host cell types via lymph nodes. LGV: Lymphogranuloma venerum.
Genetic variation in C. trachomatis
A number of chlamydial virulence factors, such as the chlamydial protease activity factor (CPAF/CT858) and the GroEL (CT110)–GroES (CT111) operon, apparently do not undergo genetic variability and therefore will not be considered here. However, the fittest genome invariably will be selected in niche-specific ways [21]. This is certainly true for both prokaryotic and eukaryotic pathogens, and their capability for DNA exchange is central to virulence gene and fitness trait acquisition [21,22]. While host genetics may contribute to disease severity [12-17,23-29] the pathogen must also successfully evolve to survive in the hostile host environment (Figure 2), and C. trachomatis has developed a number of ways to adapt within host intracellular niches. Prokaryotes acquire beneficial new genetic traits via several standard mechanisms. Point mutations may be selected that encode for effectors with improved functional attributes [30]. Phage transduction, transformation or conjugation enable acquisition by horizontal gene transfer [31]. Gene duplication expands families of related genes with differing functions and expression patterns [32,33]. All of these prominently contribute to bacterial gene variability [22,34].
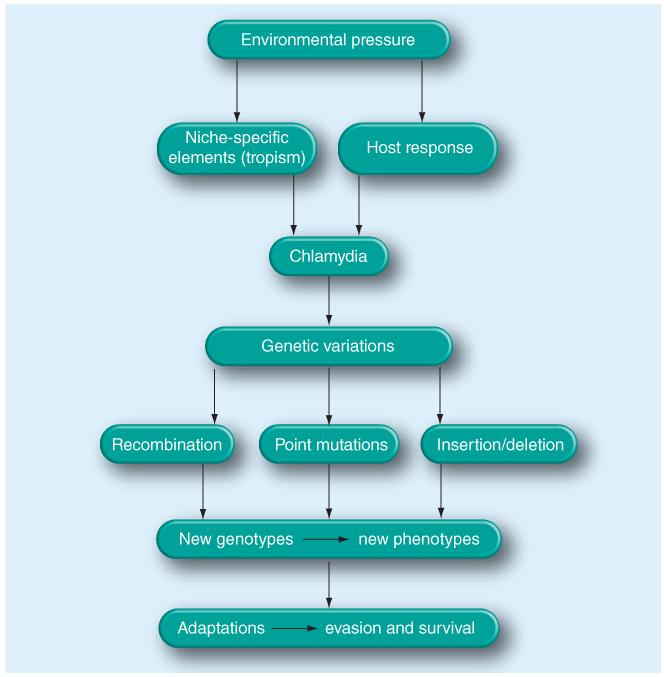
Figure 2. Genetic variation as an adjustment to environmental changes.
Genetic variation allows Chlamydia trachomatis to exploit diverse niches within a host (tissue tropism) and to avoid, escape or resist host responses. Consequently, new Chlamydia genotypes arise within the population and the potential for strain selection with increased virulence is possible.
Certainly, the serovar-defining major outer membrane protein (MOMP) is a prime example of point mutation accumulation resulting in genetic variants, especially in the surface-exposed segments of this molecule [35]. There has also been strong bioinformatics evidence after whole-genome sequencing that chlamydiae have undergone a number of gene duplication events resulting in the creation of several families of proteins important for intracellular survival in cell culture and possibly in disease severity differences [36]. These include the nine poly morphic membrane protein pmp loci, and a large family of secreted inc loci. Chlamydia also undergo genetic recombination [4]. Recombination in the laboratory using mixed infection models occurs readily [37], and several examples of recombined clinical isolates have been described that may impact disease severity. A 30-kb region between ctl393 and ctl417 found in the E/SW2 strain (Swedish New Variant) exactly matched the sequence for C. trachomatis D/UW3/CX [6,38,39]. In another study, an isolate from rectal epithelia was found to be a recombinant of C. trachomatis serovar D and LGV resulting in a hypervirulent strain (L2C) causing severe hemorrhagic proctitis [40]. Recombination was also reported for the cervical isolate Ds/2923. This isolate was serotyped as a D strain, but otherwise the genome was found to be more similar to serovars E and F [41]. Other reports of recombination include the rs2 gene (ct680) upstream of ompA [42], and the mosaic strain B/D, which was isolated from a patient with trachoma, and a B/Da strain isolated from an STD patient [43,44]. Insertion and deletion events have been reported in Chlamydia [40,41,45-48]. One example of this is the emergence of the new variant of C. trachomatis (nvCT) in Sweden, which harbors a 377-bp deletion in addition to a 44-bp duplication in its cryptic plasmid [46-48].
There are a number of C. trachomatis gene products that exhibit variability and may modulate disease severity (Figure 3). The major categories of variable chlamydial gene products and their roles in disease severity, if known or suspected, are described below and summarized in Table 1.
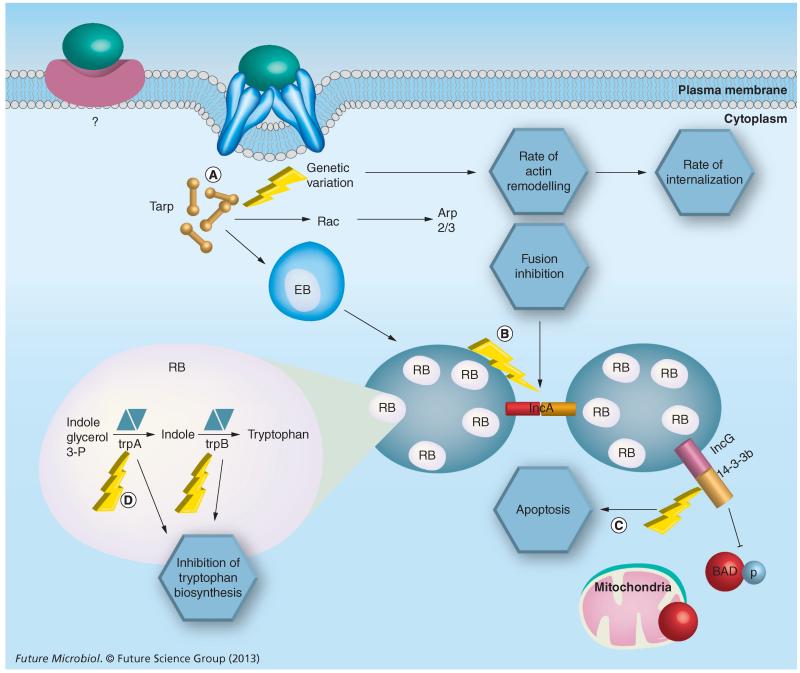
Figure 3. Consequences of genetic variation for key Chlamydia trachomatis virulence factors.
Genetic variation (lightning bolts) results in serovar-specific differences in invasiveness, evasion of host responses and tissue tropism. (A) Tarp variation alters the number of actin-binding domains and the rate of internalization. (B) Variation in IncA results in nonfusogenic inclusions. (C) IncG, a result of gene duplication within the Inc-family, interacts with the host protein 14-3-3β preventing initiation of apoptosis by the host cell. (D) Mutation in the partial trp operon results in nonfunctional tryptophan synthase unique to ocular Chlamydia trachomatis isolates. ‘?’ indicates that a ‘receptor’ or binding partner of Chlamydia EBs is not fully known and may be different between different chlamydial species. EB: Elementary body; RB: Reticulate body.
Table 1.
Effects of gene diversity in chlamydial proteins on disease severity and tissue tropism.
| Gene family | Gene name | Locus | Function/role | Effect on tissue tropism | Potential effect on disease severity |
|---|---|---|---|---|---|
| Omp family | omp A | ct681 | Most prominent outer membrane protein; porin; antigen-variants |
Classification of Chlamydia trachomatis serovars into A–K and LGV |
No correlation between ompA and disease severity demonstrated |
|
| |||||
| Pmp family | pmp A | ct412 | Type V secreted proteins: some members (D, E, G and H) are expressed on the EB surface. PmpD, D and G elicit a strong inflammatory response. All of these proteins likely play a role in phase variation |
Unknown | High variability of pmp genes by SNPs, gene duplication and indel mutations may be significant in contributing to disease severity |
| pmpB | ct413 | Specific amino acid substitutions distinguish LGV from non-LGV; difference in C. trachomatis E and F to other genital serovars |
|||
| pmpC | ct414 | Specific amino acid substitutions distinguish LGV from non-LGV; difference in C. trachomatis E and F to other genital serovars |
|||
| pmpD | ct812 | Specific amino acid substitutions distinguish LGV from non-LGV |
|||
| pmpE | ct869 | Specific nucleotide substitutions segregate ocular from urogenital and LGV; difference in C. trachomatis E and F to other genital serovars |
|||
| pmpF | ct870 | Specific nucleotide substitutions segregate ocular from urogenital and LGV |
|||
| pmpG | ct871 | Specific amino acid substitutions distinguish LGV from non-LGV |
|||
| pmpH | ct872 | Specific nucleotide substitutions segregate ocular from urogenital and LGV; difference in C. trachomatis E and F to other genital serovars |
|||
| pmpl | ct874 | Difference in C. trachomatis E and F to other genital serovars | |||
| tarp | ct456 | Binds to actin, phosphorylated by host kinases; cytoskeleton rearrangement; involved in internalization |
Number of actin-binding sites differentiate LGV and non-LGV; number of actin-binding sites important for tropism? |
Number of actin-binding sites important for disease severity? |
|
|
| |||||
| Inc proteins | incD | ctll 5 | Sphingolipid transport; interacts with CERT |
Mutations in incD specific to ocular and LGV tissue tropism | Unknown |
| incE | ct116 | Unknown | Unknown | Unknown | |
| incF | ct117 | Unknown | Unknown | Unknown | |
| incG | ctll8 | Blocks release of cytochrome C and inhibits apoptosis |
Unknown | Unknown | |
| incA | ct119 | Homotypic fusion of intracellular inclusions |
In LGV the genetic variation of incA may reflect tissue tropism |
Nonfusogenic clinical isolates have less severe clinical signs |
|
| ct229f † | Modulation of inclusion trafficking; binds to Rab4A |
Unknown | Unknown | ||
| incB | ct232 | Unknown | Unknown | Unknown | |
| incC | ct233 | Unknown | Unknown | Unknown | |
| aaxB | ct373 | AAX: arginine decarboxylase proenzyme; inhibits host cell polyamine synthesis, or inhibition of nitric oxide synthesis? |
Unknown | Unknown | |
| aaxC | ct374 | AAX: cytoplasmic arginine:agmatine antiporter; inhibits host cell polyamine synthesis, or inhibition of nitric oxide synthesis? |
Unknown | Unknown | |
| trpA | ct171 | Subunit of Trp synthase, which produces Trp from indole |
Mutations in trpA segregate ocular from genital isolates | Unknown | |
| ct166f | Toxin-like protein; glycosylates Rac1; causing actin reorganization |
Contains serovar specific loci, missing in LGV strains; potential role in invasiveness? |
Potentially plays a role in disease severity (LGV strains lack the cytotoxin) |
||
These loci have not, as yet, been named.
AAX: Arginine:agmatine exchange system; EB: Elementary body; Indel: Insertion and deletion; LGV: Lymphogranuloma venereum; SNP: Single nucleotide polymorphism.
Major outer membrane protein
The MOMP is a surface-exposed protein encoded by ompA (ct681). In cell culture, ompA is one of the earliest genes detected (2–8 h postinfection, depending on serovar) and reaches its highest expression level at 18–24 h postinfection. The protein is part of a chlamydial outer membrane complex. MOMP represents approximately 60% of the total surface-expressed proteins in elementary bodies (EBs). It is postulated that host immune pressure is the trigger for the variability of surface-exposed proteins [20,49]; however, the most successful genital serovars D, E and F carry the least variable ompA gene [35,44,50,51] suggesting that other undefined factors are at play in selecting MOMP variants [35]. Millman et al. [52] and Andreasen et al. [53] proposed that either negative selection removes deleterious mutations by purifying selection or that a selective sweep results in the fixation of a single haplotype. The MOMP-based micro immunofluorescence method for C. trachomatis strain classification was established more than 40 years ago by Wang and Grayston [54]. Subsequent ompA sequencing studies confirmed the microimmunofluorescence test, and showed that MOMP variability is due to the four surface-exposed variable regions (VRI–VRIV) that define 15 C. trachomatis serovars. It is now very clear that MOMP serovars (or more properly MOMP genovars) accurately predict chlamydial disease biovars (A–C: endemic trachoma; D–K: cervicitis/urethritis genital disease; and L1–L3: LGV) [40,49]. Despite this clear MOMP-based disease biovar classification, no correlation between ompA differences and disease severity has been demonstrated, although Andreasen et al. found an association between an A genotype and clinical symptoms of active trachoma and average ocular infection load [53]. Genovars with the most sequence similarities cross biovar designations. Serum-based cross-reactivity studies have shown that genovars from group B (B, Ba, D, E, L1 and L2) are more cross-reactive, as are genovars from group C (C, J, A, H, I, K and L3) [49]. Thus, classification based on chlamydial pathobiology must involve chlamydial gene products other than MOMP, a supposition that was addressed by Harris et al. who demonstrated that variation in ompA did not reflect C. trachomatis disease severity [6].
If MOMP differences do not reflect disease severity differences, then what does the large amount of genetic variation in ompA signify? The ompA gene is one of the most highly variable genes in the chlamydial genome. Variability occurs in over 30% of the coding sequence, whereas the gene of another surface protein, porB/ct713 (porin), exhibits only 1% variability in the gene sequence [55,56]. Several ompA sequencing studies have included up- and down-stream contiguous regions from different geographical clinical isolates and reference strains [35,44,50,56-63]. One of the largest studies characterized 795 C. trachomatis samples from Lisbon, Portugal [35]. The study demonstrated 80 nucleotide variable sites in all the tested isolates that were distributed randomly throughout the whole gene and included both variable and constant regions. This was confirmed in another study in which ompAs from different geographical regions were sequenced [35,64]. More than half of the mutations in ompA were found in the constant region, and 50% of these were silent. By contrast, almost 94% of the mutations in variable regions coded for amino acid changes [35,57]. These studies are not surprising, but they do reaffirm that exposed portions of MOMP exhibit a high degree of variability, presumably owing to the immuno genicity of this molecule.
Type III secretion system
The type III secretion system (T3SS) is used by many pathogens to directly deliver effector proteins into a target host cell [65]. The T3SS injectisome is assembled as a molecular syringe that spans the inner membrane and periplasmic space. C. trachomatis contains all genes coding for proteins necessary for a fully functional T3SS. These genes are found in several gene clusters across the chlamydial genome in contrast to the T3SS of other Gram-negative bacteria where T3SS genes are located in pathogenicity islands [66]. The chlamydial T3SS consists of two main sets of proteins: the injectisome and the translocator proteins. The injectisome is built from 20–25 contact-dependent secretion proteins, while the chlamydial outer membrane protein B (CopB/CT578) forms a pore in the host cell membrane or inclusion membrane. The pore allows an entry point for the needle to facilitate the injection of effector proteins into the host cell cytosol or inclusion membrane [67]. A number of highly polymorphic genes (e.g., CT049 and CT050) are predicted to encode T3SS effectors; however, gene products have not been functionally characterized [42,68]. Several polymorphic T3SS effectors have been characterized, including an actin-recruiting effector and a large group of inclusion membrane proteins.
Translocated actin-recruiting phosphoprotein
Studies by Carabeo et al. demonstrated that C. trachomatis host cell invasion is initiated by cytoskeletal rearrangements forming actin-rich ‘pedestal-like structures’ to facilitate chlamydial entry [69]. The process starts with host-cell attachment of EBs, followed by injection of preloaded effector proteins through the T3SS injectisome [70]. Tarp is one of the very early secreted effector proteins (30 s after host-cell attachment) and induces actin polymerization within 90 s after attachment via the Arp2/3 nucleation site [71] (Figure 3). Biochemical analysis of Tarp revealed N-terminal tyrosine-rich tandem repeats (phosphorylation domain), a proline-rich C-terminal domain and an actin-binding domain [72]. The actin-binding domain is indispensable for actin polymerization since antibodies against this domain inhibit actin nucleation [73].
Tarp genetic variation & disease severity
Sequence analysis of Tarp orthologs from several Chlamydia species reveals a number of changes that are thought to be associated with intracellular chlamydial survival and therefore may be linked to disease severity [74]. Tarp amino acid sequence comparisons show proline-rich and actin-binding domain conservation, although the number of actin-binding domains differs between and within the species [73]. Serovar L2 has one actin-binding domain while serovars A and D have four and three, respectively [73]. It is not clear if the number of actin-binding domains play a role in disease severity and tissue tropism. Multiple actin-binding domains theoretically facilitate faster polymerization due to simultaneous binding of several actin monomers. In serovar L2 Tarp, there are six tyrosine-rich repeats, while serovar A and D harbor only three [70]. However, Chlamydia muridarum, Chlamydophila caviae and Chlamydophila pneumoniae are missing the tyrosine repeats, yet are efficiently taken up by host cells suggesting a dispensable role for this domain for internalization or that alternative entry mechanisms exist not requiring Tarp [70]. In C. trachomatis, phosphorylation of tyrosinerich repeats was found to be mediated by Src family kinases [75]. The tyrosine-phosphorylated residues then bind PI3K and SHC-1 host-cell adaptor proteins, which in turn affects host-cell growth and survival [76].
Genetic variation of inclusion membrane proteins & correlation with disease severity
Inclusion membrane proteins (Incs) are a family of type III secretion-effector proteins that share a common 40–60 amino acid bilobed hydrophobic secondary motif. In LGV strains several inc loci are pseudogenes [77], and although there are 50 or more Incs predicted in C. trachomatis [77,78], only 23 members of this family have been detected in the inclusion membrane [79-81], and only a few Incs have been functionally characterized.
IncA: a fusogenic inclusion membrane protein
IncA (CT119) is required for homotypic fusion of intracellular inclusions [67,82] via the formation of SNARE-like fusogenic intermediates (Figure 3) [69,83]. Genetic variation in different members of the Inc family have been reported in several studies [77,84,85]. The early study of Rockey et al. demonstrated that incA isolated from C. trachomatis clinical isolates harbor several types of mutations that lead to truncated forms of IncA affecting immunofluorescence detection of IncA on inclusion membranes [85]. The absence of IncA was correlated with the formation of multilobed nonfusogenic inclusion bodies [84,86]. One study demonstrated a correlation between disease severity and incA-negative strains [87]. Here, the authors showed that nonfusogenic clinical isolates have less severe clinical signs of infection with low Chlamydia recovery [87]. Variation in incA was also reported in a Dutch population-based study of women with documented chlamydial infection [84]; however, a correlation between disease severity and incA polymorphisms was not possible owing to the limited number of patients enrolled in the study. On the other hand, LGV strains were found to have amino acid substitutions in Inc proteins specific to LGV that are not found in ocular or genital strains. This work demonstrates an example of genetic variation specific to tissue tropism [77]. The polymorphisms specific for LGV strains, although presently unexplained, might reflect host macrophage and lymph node environments unique to LGV.
IncD: a mediator for sphingolipid transport
The IncD locus (CT115) was found to be one of five key genes that helped to segregate ocular (A–C) from genital (D–K) chlamydial genovars [77]. IncD interacts with CERT, which is required for sphingolipid transport. The Pleckstrin homology domain (PH) in CERT was shown to be critical for binding to IncD and subsequent Chlamydia development [88] where knockout of the PH domain resulted in reduced chlamydial growth. Mutations leading to amino acid substitutions specific for ocular and LGV tissue tropism are considered important drivers of C. trachomatis evolution and tissue tropism differences; however, none of these mutations affect the binding domain that interacts with either PH or CERT domains required for ceramide binding, this function appears to be conserved across the species [77,89,90].
IncG: an indirect inhibitor of apoptosis
IncG (CT118) is part of the operon IncD–G where expression can be detected 2 h after internalization [91]. It was found that phosphorylated IncG binds directly to the host phosphoserine binding protein 14–3–3β and hence IncG sequesters it from binding to phospho-BAD and consequently inhibits the release of mitochondrial cytochrome C and induction of apoptosis (Figure 3) [92,93]. This might be a mechanism evolved by Chlamydia to either establish conditions suitable for completing a productive developmental cycle or a chronic infection where host cell viability is crucial for the long-term intimate host–pathogen relationship [94].
CT229: modulation of inclusion trafficking
CT229 is expressed as early as 1 h postinfection and localizes to the inclusion membrane indicating an important role in the early development of the inclusion [95,96]. Inclusion membrane modifications introduced by insertion of Chlamydia proteins occurs as the pathogen-containing vesicle moves to the microtubule organizing center in a dynein-dependent way. Rzomp et al. have demonstrated that CT229 binds to the GTPase Rab4A, which interacts with cytoplasmic dynein [97]. Therefore, CT229 potentially plays a role in the regulation of inclusion trafficking or the fusogenicity of the chlamydial inclusion. Although CT229/Rab interactions are clearly functionally important, other Rab family inclusion membrane interactions dictate host cell vesicle-localization events with distinctions between chlamydial strains, the specific host Rab-binding partner and, therefore, the nature of inclusion trafficking activity.
Inclusion-vesicle trafficking is perceived to be important for the intracellular growth and development of chlamydiae; however, direct links to disease severity are still to be determined.
Other inc genes
Several inc genes (e.g., ct058, ct192 and ct214) are highly expressed in LGV strains and share promoter element motifs, but as with other Incs, their function in disease severity and tissue tropism for LGV strains remains unknown [77].
Pmp autotransporters
In C. trachomatis, the pmps are a family of nine clustered genes (pmp A–I) that encode proteins with molecular weights between 95.5 and 187 kDa [49]. Emergence of this family is likely an example of gene duplications that serve to create functional diversity and possibly environmentally responsive phase variation for chlamydiae. Therefore, this gene family may be significant in contributing to disease severity distinctions between strains. The family members contain two repeated N-terminal motifs, FXXN and GAAL (I, V) [98]. It is likely that Pmps play a pivotal role in Chlamydia biology because the genes represent almost 3% of the genome [49,99]. Bioinformatic analysis predicts that Pmps are most likely members of the type V secretion system autotransporters [100]. They share several conserved autotransporter features, including an N-terminal signal sequence for sec-dependent secretion, a passenger domain and a C-terminal β-barrel allowing transport of the passenger domain to the surface. Some Pmp members are expressed on the EB surface (PmpsD, E, G and H) [49]. They elicit a strong proinflammatory cytokine response from CD4+ cells (PmpB, D and G) or stimulate cytokine production in endothelial cells in an NfκB-pathway dependent manner (PmpB and D) [20].
Genetic variation in pmps: more than just polymorphisms
Genetic variation in pmps has been investigated. Several studies collectively show extensive polymorphisms within this gene family [41,101,102]. For example, pmpB, E, F and H show the highest nonsynonymous to synonymous base-pair change ratios of all chlamydial genes evaluated, suggesting strong selective pressure to retain gene family diversity [78,102]. Variability for pmpB, E, G and H also involves deletions and insertions when compared with a reference (serovar D) strain [102]. For instance, PmpD has a single amino acid deletion. In comparison PmpH, has 10-11 amino acids deleted, and PmpB is conserved in all serovars except Ia, which has an insertion resulting in a protein with the addition of 20 amino acids [102]. A comparison of the pmpC sequence from 12 clinical isolates to a reference strain showed that the gene sequence from seven out of the 12 isolates did not match the reference strain sequences. For example, the clinical isolate Ja/10 has a 32-nucleotide difference compared with the Ja reference strain [101]. Thus, the pmp gene family demonstrates substantive variability acquired via at least three mechanisms (single nucleotide polymorphisms [SNPs], gene duplication and insertion and deletion mutations) and this variability has the potential of contributing to differences in disease severity for C. trachomatis.
Correlation between pmp genetic variation & tissue tropism
Correlation between tissue tropism and genetic variation in pmps has been investigated in several studies [41,77,102]. For example, PmpB, C, D and G in LGV strains harbor specific amino acid substitutions that distinguish them from non-LGV strains [102]. In E and F genital serovars, differences in five Pmps (PmpB, C, D, H and I) are distinct from other genital serovars [102]. Also, computational analysis reveals specific regions in pmpE, F and H associated with high frequency of nucleotide substitutions. These point mutations in pmps result in serovar differences. For example, specific regions in pmpE, F and H segregate ocular serovars from urogenital and LGV serovars. In pmpE, a single midgene region differentiates ocular strains from LGV and urogenital strains [102]. Finally, Jeffery et al. showed that point mutations in both pmpE and F are associated with rectal tropism [41]. In conclusion, point mutations in pmp-specific regions add another factor for strain distinction, tissue tropism and possibly disease severity; however, the mechanisms that confer these differences are still not clear.
The chlamydial cytotoxin
In C. trachomatis, toxin-like genes reside within a 20.3 kb highly polymorphic genomic region called the plasticity zone (PZ) between ycf V (ct152) and dbsB (ct177). The C. trachomatis toxin shares homology with the enzymatic active site of the Clostridium difficile large A and B toxin [45]. Belland et al. reported toxin-mediated cytopathic effects of C. muridarum and C.trachomatis serovar D on HeLa cells [45]. Recently, it has been reported that the C. trachomatis serovar D toxin (CT166) glycosylates the small GTPase Rac1, causing actin reorganization in HeLa cells similar to that seen for the clostridial toxin B [103].
Polymorphisms in the cytotoxin locus
Carlson et al. used comparative DNA–DNA microarray genomic hybridization to compare the genomes of 15 C. trachomatis serovars and found extensive polymorphisms in the cytotoxin locus within the PZ region [104]. The comparison of all serovars demonstrated that each representative strain contained a serovar-specific locus. All genitotropic and oculotropic strains, except serovars J and H, were characterized by a central deletion, while serovars J and H have an apparently intact gene. Furthermore, genital serovars (D–K) possess the N-terminal domains for glucosyltransferase activity and uridine diphosphate–glucose binding, while serovars A and C harbor only the uridine diphosphate–glucose binding domain. LGV serovars (L1–L3) lack both domains [104]. Somboonna et al. showed that the hypervirulent recombinant L2C strain acquired the functional cytotoxin from the D strain, which may account for its reported high level of disease severity [40]. Taken together, these studies provide evidence that cytotoxin polymorphisms may be crucial for biovar classification and likely play a role in disease severity.
Tryptophan synthase genes, an operon that differentiates between ocular & genital strains
C. trachomatis strains possess a partial tryptophan operon (trpRBA; ct169–ct171) [105]. A novel tryptophan synthesis operon (trpRBAFCD) is found in C. caviae, C. felis and C. pecorum, but is absent in other chlamydial species (C. muridarum, C. pneumoniae and C. abortus) [105-109]. A potential role for the tryptophan operon was described in C. trachomatis [105]. In tryptophan-deficient medium, growth of C. trachomatis was inhibited. Growth of genital, but not ocular serovars could be restored after the addition of indole. In addition, the growth inhibition of C. trachomatis after tryptophan depletion by IFN-γ, which induces the tryptophan decyclizing enzyme indoleamine-2,3-dioxygenase, was rescued by addition of indole in genital, but not ocular serovars [110]. Since C. trachomatis carries a truncated version of the operon (trpRBA), containing only the genes for the α- and β-subunit of the tryptophan synthase encoded in the PZ it requires indole to produce tryptophan (Figure 3) [20]. Ocular serovars (A, Ba and C) uniformly contain a frame-shift mutation (nucleotide position 531) in trpA resulting in an early stop codon and a truncated, nonfunctional protein [110]. Thus, trpA (ct171) mutations differentiate ocular from genital serovars. It is well established that unless a genomic function is actively protected by selection, it will accumulate deleterious mutations and will cease to be functional. It is proposed by Caldwell and colleagues that genital serovars retain a functional partial trp operon because indole compounds are available (probably due to the genital microbiota) and tryptophan, an essential amino acid for chlamydiae, is required for productive growth [110]. Thus, retaining the capacity to generate tryptophan from indole possibly helps to ensure successful transmission [111-115]. Ocular C. trachomatis serovars are similar to genital serovars in that they require tryptophan to complete their developmental cycle [110,115]. Since ocular strains uniformly have trp operon mutations resulting in loss of function (Figures 3 & 4) these genes therefore must be nonessential for Chlamydia survival in the conjunctival epithelium. This could be because tryptophan is plentiful in the conjunctival microenvironment, indole-producing flora, although perhaps present [116,117], are in a separate niche from chlamydiae, or that indole is otherwise unavailable.
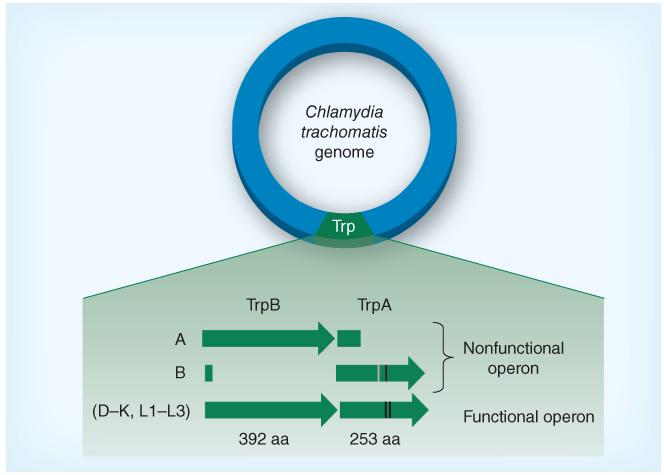
Figure 4. Trp synthase in different Chlamydia trachomatis serovars.
All ocular serovars sequenced thus far harbor a nonfunctional trpRBA operon, whereas trpRBA is functional for all sequenced genital serovars. For ocular serovars, mutations may result either in a truncated TrpA or in a nonsynonymous point mutation that results in incorporation of an amino acid that inhibits function. Point mutations have been described for serovars E, F, G and Ia for trpA (e.g., change at C177Y) and serovars L1–L3 (change at Q178E); however, these mutations do not impact TrpA function. Nonsynonymous point mutations are represented by black lines while deletion mutations and their effect on TrpA are represented by the gray line. Not drawn to scale.
Arginine decarboxylase
Chlamydia import arginine from the host cell by the aaxABC gene products. aaxA (ct372) encodes an outer-membrane transporter, aaxB (ct373) encodes arginine decarboxylase pro-enzyme catalyzing the decarboxylation of arginine to agmantine, and aaxC (ct374) encodes a cytoplasmic membrane arginine:agmatine antiporter [118,119]. Some C. trachomatis clinical isolates express a functional arginine decarboxylase [41], but a number of loss-of-function SNPs have been reported in this operon [120]. C. trachomatis serovar L2/434 has a nonsense point mutation in aaxB resulting in an early stop codon. Replacement of the stop codon with its original amino acid restores the full-length protein and normal function when expressed in Escherichia coli [120]. C. trachomatis serovar D/UW-3 possesses a full length but nonfunctional AaxB protein. Protein function is restored in surrogates by replacing arginine 115 with the original glycine [120]. Chlamydia also have a putative ABC arginine transporter; therefore, it has been suggested that the Aax system may inhibit host cell polyamine synthesis, elevate the intracellular pH or inhibit host nitric oxide synthesis [119]. In this case, it is possible that C. trachomatis D/UW-3 and L2/434 have either developed alternative pathways or are deficient in the perceived role for this operon in promoting chlamydial intracellular survival.
Role of host genetics in C. trachomatis disease severity
Role of host genetics in genital C. trachomatis disease severity
Although pathogen genetic variability has a large impact on disease outcome, host genetics also likely play a role in the interaction between the pathogen and the host. The additive, synergistic or antagonistic effects of bacterial and host factors often determine pathogenicity, disease severity and outcome of infections, including those caused by C. trachomatis [12-17,23-28,62]. For example, not all women infected with genital C. trachomatis develop clinical complications such as tubal factor infertility (TFI).
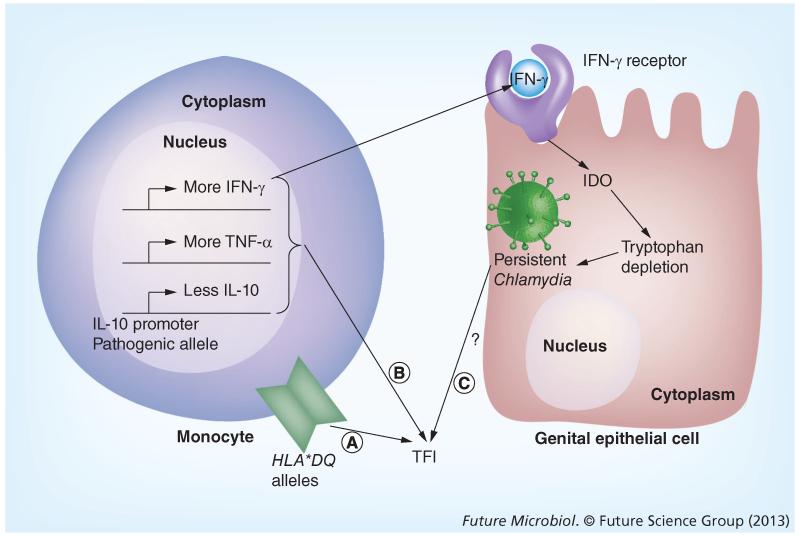
Pathogen burden and reinfection status are undoubtedly contributing variables to disease outcomes; however, several studies have investigated the correlation between host genetic factors and chlamydial disease severity. Kinnunen et al. investigated the correlation between cytokine polymorphisms and C. trachomatis-associated TFI (Figure 5) [23]. These investigators demonstrated that Finnish women attending an infertility clinic and diagnosed with TFI harbored specific HLA DQ alleles (HLA DQA1*0102 and HLA DQB1*0602) as well as SNPs in the IL-10 promoter (IL-10-1082AA genotype) more frequently than control subjects. In another study with a larger cohort of patients, both the IL-10-1082AA promoter allele and the TNF-α-308 promoter allele were found to be associated with a high risk of severe TFI [27]. Furthermore, Ohman et al. found that an IL-10 promoter with an adenosine residue at position 1082 (1082A) was associated not only with decreased IL-10 production, but also with an increase in IFN-γ and TNF-α production from peripheral blood mononuclear cells stimulated in vitro with C. trachomatis, compared with individuals with the 1082G version of the IL-10 promoter [26]. In other studies of genital tract infections with C. trachomatis in women, no association was found between TFI and polymorphisms in TLR4 and its coreceptor CD14, IL-1β and IL-1RN [121,122]. However, women diagnosed with TFI were found to express the variant allele B of mannose-binding lectin-2, while the wildtype allele A was associated with an absence of disease complications [29].
Figure 5. Contribution of host factors to Chlamydia trachomatis disease severity.
Influence of genetic variation in host epithelial and immune cells on Chlamydia trachomatis infection and potential TFI induction. (A) Specific HLA*DQ alleles are correlated to TFI induction. (B) TFI patients carry an IL-10 promoter polymorphism. Cells from these individuals tend to produce less IL-10, more IFNγ and more TNF-α when leukocytes are stimulated in vitro. (C) IFN-γ induces IDO, which degrades tryptophan, an essential amino acid for both Chlamydia and the host cell. This may lead to dampening of the immune system, from a productive to a nonproductive chlamydial growth phenotype, which has been suggested to be associated with chronic infection and increased disease severity. IDO: Indoleamine 2,3-dioxygenase; TFI: Tubal factor infertility.
Role of host genetics in ocular C. trachomatis disease severity
Host genetic factors also likely play a role in trachoma, the leading cause of infectious preventable blindness worldwide [13-17]. In endemic areas, the infection starts early in life, individuals are re-infected multiple times and ultimately develop conjuctival scarring and trichiasis (in-turning of eye lashes) later in life. Although this complex multifactorial disease undoutedly has multiple host and pathogen risk factors associated with severity, several interesting studies have provided links to severity and host inflammatory response genes. Natividad et al. found a specific haplotype (H-RISK) associated with high risk of trichiasis and scarring trachoma in a Gambian population [17]. The haplotype consists of four SNPs (IL10 −3575A, IL10 −1082C, IL10 −592G and IL10 +5009) spanning upstream and downstream regions of the IL-10 gene [17]. Furthermore, this allele was found to be associated with a relative increase in IL-10 gene expression [15]. A previous study showed that TNF-α in tears is important in the pathogenesis of scarring trachoma [123] The study also showed an association between trachoma and the promotor alleles TNFA-308A and TNFA-238A. MMP-9 is another factor that plays a role in trachoma pathogenesis. A nonsynonymous mutation resulted in a single nucleotide polymorphism in MMP-9 (Q279R) and was found to be associated with low risk of trachoma sequelae [13].
Experimental studies of host genetics & disease severity
It is clear that key events controlling disease outcome for chlamydial infections involve more than simply the repertoire of pathogen virulence gene variants and the nature of the host response to infection. Live-attenuated chlamydial strains, cured of the 8 kb plasmid, have been developed. These strains can elicit host protective responses in the absence of disease pathology, presumably due to downregulation of virulence gene expression [124,125]. However, for chlamydial strains circulating in communities, population-based studies suggest that polymorphisms involving the inflammatory response to infection may be key host determinants of disease severity, irrespective of the pathogen strain and the repertoire of virulence factor genes encoded by the pathogen. Qualitative and quantitative differences in host inflammation also seem to be important in experimental infection of animal models of chlamydial disease [121]. Genome-wide association studies in humans have provided some key insights for disease-provoking host responses; however, these types of investigations also have limitations. These studies require large cohorts of patients, large sequence data sets and lack causal conclusions.
The use of genetically modified animal models have been useful in assessing host responses that modulate chlamydial disease severity. Most published studies have focused on single selected host genes to investigate disease severity outcomes. The use of knockout mice, for example, has helped to provide insights concerning the importance of immune-regulated cytokines (e.g., IFN-γ), a variety of inflammatory proteins (e.g., MMPs), chemokines (e.g., CXCRs) and elements of the adaptive response important for resolving infections [126-132]. In some cases, (e.g., type I IFN−/− mice) the absence of an immune-regulated cytokine promoted improved adaptive immunity [133]. This targeted approach is sometimes limited by host response redundancy and compensating mechanisms to overcome the targeted deficiency, and can also be considered biased in the sense that host factors are specifically selected by the investigator, which may limit the relevance based on the deficiency selected and whether or not it actually plays a role in development of severe chlamydial disease. Any approach chosen to sort out host factors important to disease outcome has limitations; however, the use of sets of genetically diverse advanced recombinant inbred mice, have proven to be valuable tools in forward genetic studies to help relate host susceptibility, resistance and disease severity phenotypes to specific genetic loci with a high degree of penetrance. The mouse genome, although 95% similar to the human genome [134], exhibits critical differences in innate and acquired responses. This fact may limit the degree of correlation for inflammatory responses in mice and humans, although the cited study was only on the use of endotoxin and therefore may have its own set of interpretation limitations [135]. Other types of differences exist between responses in mice and in humans. For example, in a C. psittaci 6BC systemic mouse infection model, forward genetics approaches identified a locus on chromosome 11 (1.5 Mb) responsible for susceptibility and resistance to the infection. This locus comprised 18 genes including p47 GTPases (Irgb10, Irgm2 and Iigp2) [136]. These GTPases exhibit cell autonomous effects on chlamydial growth, influence inflammatory cell recruitment to the site of disease [137], and are important in resolving LGV [138,139] and C. muridarum infections in mice [140]. However, this family of interferon-induced GTPases is not present in humans and therefore a host genetic correlation with direct translational potential was lacking in these studies. Despite limitations of animal models regarding reproducing results that correlate with human disease, use of advanced recombinant inbred mice have been shown to be of benefit and relevance to human disease [136,141,142]. These genetically diverse murine strain sets may prove to be of value in the study of genital, respiratory and ocular chlamydial infections.
Therapeutics against C. trachomatis infection
There are several avenues for development of therapeutics and management of chlamydial infections. One way is to target the pathogen arm such as the usage of antibiotics or targeting virulence factors such as type III secretion effectors or the cytotoxin with small inhibitors especially designed for these proteins. The other way is to target the host side by dampening the inflammatory response or by upregulating protective immunity via vaccination. Although antibiotic resistance is problematic in C. suis [143,144], the emergence of antibiotic resistance in C. trachomatis is not yet a threat to limit the effectiveness of postexposure antibiotic use.
There are two concerns related to antibiotics and the management of chlamydial infections. The first is accurate identification of those with chlamydial infections, who are often asymptomatic [145-147]. There is equal risk of developing untoward complications of infection, especially genital tract infection in women, irrespective of whether acute infection is symptomatic or asymptomatic [148]. Therefore, to identify all of those infected with Chlamydia and at risk of complications, blanket screening programs – although costly and manageable in only a limited number of countries – need to be effectively implemented when feasible. The second concern is that in areas where screening and treatment are most effective, the incidence of infection is not diminishing, but rather is increasing [204]. The ‘arrested immunity’ hypothesis is one possibility that has been posited to account for this observation [149], although if arrested immunity is important, it apparently does not impact disease complications, which continue to fall even with increasing prevalence of lower genital tract infection [150,151].
If arrested immunity contributes to the maintenance of a high level of susceptible individuals in the population, then other intervention strategies must continue to be considered. Certainly, development of a safe and effective vaccine is first on the list of alternatives; however, chlamydiae are highly successful pathogens and development of testable candidate vaccines has yet to occur, and safety due to untoward complications of vaccination as a result of heightened re-exposure pathology may be a concern.
One additional alternative management strategy might be considered, especially if host reactivity can be convincingly established as a contributing factor to disease severity. The development and use of compounds that can safely and effectively reduce host responses that contribute to development of disease might prove to be beneficial in limiting the severity of chlamydial infection complications. This strategy could be as simple as developing management strategies that take maximum advantage of beneficial side effects of antibiotic regimens including interfering with MMPs [152] and dampening NFκB-mediated inflammation [153], or as sophisticated as targeting more individual host-specific responses as personalized medicine becomes a reality.
Conclusion
Genetic variation is a method by which pathogens have evolved new phenotypes to evade host responses in a dynamic and niche-specific manner. Several types of genetic variation are found in C. trachomatis that impact variability and expression of virulence factors, as described in this review. These strategies have been shown to promote chlamydial intracellular survival, greatly impact disease severity and are the basis for distinct chlamydial disease biovars and differences in host tissue tropism. Host genetic factors also play a role in disease severity where there is genetic variation in a wide range of pathogen-elicited response genes ranging from MMPs, TLRs, cytokines, cytokine promoters and receptors, to MHCs; all of which have been shown to contribute to disease severity and its control. Therefore, the overall picture of chlamydial disease severity is the outcome of the interaction between host and pathogen factors, which must be considered in a systems-based manner to gain a complete understanding of the process of chlamydial pathogenesis and the management of chlamydial infectious diseases.
Future perspective
The advent of genome sequencing, systems biology and bioinformatics has advanced the field of Chlamydia research to enable identification and function of chlamydial virulence factors. These approaches have also provided refined insights of host responses that are linked to either beneficial or detrimental outcomes. The identification of signature chlamydial genetic variants and key host polymorphisms is likely to provide highly specific diagnostic tools for identifying highly virulent strains of chlamydiae and individuals who are at risk for developing complications of their chlamydial infections. This type of information may also provide a basis for improved management of chlamydial infections in the era of personalized medicine.
Executive summary.
Background
-
■
Chlamydia trachomatis is a major health burden worldwide in humans.
-
■
The two C. trachomatis biovars show large differences in disease severity and tissue tropism despite less than 1% genetic differences.
Genetic variation in Chlamydia
-
■
Genetic variations by gene duplication, insertion/deletions and point mutations in the form of single nucleotide polymorphisms in pathogen factors are the means of evading the immune response and adaptation of C. trachomatis to diverse environments.
-
■
A high degree of mutations in ompA altering the variable region in major outer membrane protein and mutations in pmp genes causes differences in the immune response to these surface proteins.
-
■
Gene duplication of inc genes results in different functions of the Inc proteins within the Inc family.
-
■
The presence of the chlamydial cytotoxin and the type III secretion effector Tarp allow invasion of serovar/biovar-specific eukaryotic cells as a means of tissue tropism.
-
■
Metabolic enzymes such as gene products of the tryptophan operon and the arginine decarboxylase allow a response to environmental changes triggered by the immune response.
Role of host genetics in C. trachomatis disease severity
-
■
Differences in host genetics affect susceptibility and resistance to C. trachomatis.
-
■
Genetic variability in pathways and proteins important for the immune system, such as antigen-presenting proteins, cytokines and metalloproteinases, affect host response to C. trachomatis.
-
■
The utilization of recombinant inbred mice allows identification of important host genes that affect susceptibility to C. trachomatis and the study of their role during infection.
Therapeutics
-
■
The use of effective therapeutics is limited by the proper identification of infected individuals.
-
■
Arrested immunity in populations with effective screening and treatment programs leads to an increase in the number of susceptible individuals providing impetus for vaccine development.
-
■
Interfering with host responses that contribute to disease severity opens new research opportunities for investigating chlamydial infections.
Conclusion
-
■
Genetic variations in C. trachomatis enable the pathogen to evade and survive the host immune response, compete with other bacteria or grow in a suitable environment within the host; while genetic variations in the host alter susceptibility to C. trachomatis and the ability of the host to clear the pathogen.
-
■
Understanding the connection between genetic variation on the pathogen and host side will lead to a greater understanding of the pathogenicity of C. trachomatis and provide new targets to develop specific and novel therapeutics for C. trachomatis treatment.
Future perspective
-
■
The identification of genetic variations in the host and pathogen as well as understanding their role in disease severity during C. trachomatis infections will be important in the future to develop novel therapeutics, to understand pathogenicity of C. trachomatis and to design new diagnostic methods.
Footnotes
Financial & competing interests disclosure
Work on chlamydial immunity and host factors involved in disease severity is supported by an NIH public health service grant, AI 019782 (GI Byrne) and a Department of Defense award W81XWH-09-01-0391 (GI Byrne). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Abdelrahman YM, Belland RJ. The chlamydial developmental cycle. FEMS Microbiol. Rev. 2005;29(5):949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Mylonas I. Female genital Chlamydia trachomatis infection: where are we heading? Arch. Gynecol. Obstet. 2012;285(5):1271–1285. doi: 10.1007/s00404-012-2240-7. [DOI] [PubMed] [Google Scholar]
- 3.WHO WHO weekly epidemiological record. 2012;87:161–168. [Google Scholar]
- 4.Stephens RS, Myers G, Eppinger M, Bavoil PM. Divergence without difference: phylogenetics and taxonomy of Chlamydia resolved. FEMS Immunol. Med. Microbiol. 2009;55(2):115–119. doi: 10.1111/j.1574-695X.2008.00516.x. [DOI] [PubMed] [Google Scholar]
- 5.Schachter J. Chlamydia intracellular biology, pathogenesis, and immunity. In: Stephens R, editor. American Society for Microbiology. Washington, DC, USA: 1999. pp. 139–177. [Google Scholar]
- 6.Harris SR, Clarke IN, Seth-Smith HM, et al. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat. Genet. 2012;44(4):413–419. S1. doi: 10.1038/ng.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seth-Smith HM, Harris SR, Skilton RJ, et al. Whole-genome sequences of Chlamydia trachomatis directly from clinical samples without culture. Genome Res. 2013;23(5):855–866. doi: 10.1101/gr.150037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collingro A, Tischler P, Weinmaier T, et al. Unity in variety – the pan-genome of the Chlamydiae. Mol. Biol. Evol. 2011;28(12):3253–3270. doi: 10.1093/molbev/msr161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottlieb SL, Martin DH, Xu F, Byrne GI, Brunham RC. Summary: the natural history and immunobiology of Chlamydia trachomatis genital infection and implications for chlamydia control. J. Infect. Dis. 2010;201(Suppl. 2):S190–S204. doi: 10.1086/652401. [DOI] [PubMed] [Google Scholar]
- 10.Atik B, Skwor TA, Kandel RP, et al. Identification of novel single nucleotide polymorphisms in inflammatory genes as risk factors associated with trachomatous trichiasis. PLoS ONE. 2008;3(10):e3600. doi: 10.1371/journal.pone.0003600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conway DJ, Holland MJ, Campbell AE, et al. HLA class I and II polymorphisms and trachomatous scarring in a Chlamydia trachomatis-endemic population. J. Infect. Dis. 1996;174(3):643–646. doi: 10.1093/infdis/174.3.643. [DOI] [PubMed] [Google Scholar]
- 12.Mozzato-Chamay N, Mahdi OS, Jallow O, Mabey DC, Bailey RL, Conway DJ. Polymorphisms in candidate genes and risk of scarring trachoma in a Chlamydia trachomatis-endemic population. J. Infect. Dis. 2000;182(5):1545–1548. doi: 10.1086/315891. [DOI] [PubMed] [Google Scholar]
- 13.Natividad A, Cooke G, Holland MJ, et al. A coding polymorphism in matrix metalloproteinase 9 reduces risk of scarring sequelae of ocular Chlamydia trachomatis infection. BMC Med. Genet. 2006;7:40. doi: 10.1186/1471-2350-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Natividad A, Hanchard N, Holland MJ, et al. Genetic variation at the TNF locus and the risk of severe sequelae of ocular Chlamydia trachomatis infection in Gambians. Genes Immun. 2007;8(4):288–295. doi: 10.1038/sj.gene.6364384. [DOI] [PubMed] [Google Scholar]
- 15.Natividad A, Holland MJ, Rockett KA, et al. Susceptibility to sequelae of human ocular Chlamydial infection associated with allelic variation in IL10 cis-regulation. Hum. Mol. Genet. 2008;17(2):323–329. doi: 10.1093/hmg/ddm310. [DOI] [PubMed] [Google Scholar]
- 16.Natividad A, Hull J, Luoni G, et al. Innate immunity in ocular Chlamydia trachomatis infection: contribution of IL8 and CSF2 gene variants to risk of trachomatous scarring in Gambians. BMC Med. Genet. 2009;10:138. doi: 10.1186/1471-2350-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Natividad A, Wilson J, Koch O, et al. Risk of trachomatous scarring and trichiasis in Gambians varies with SNP haplotypes at the interferon-gamma and interleukin-10 loci. Genes Immun. 2005;6(4):332–340. doi: 10.1038/sj.gene.6364182. [DOI] [PubMed] [Google Scholar]
- 18.White AG, Bogh J, Leheny W, et al. HLA antigens in Omanis with blinding trachoma: markers for disease susceptibility and resistance. Brit. J. Ophthalmol. 1997;81(6):431–434. doi: 10.1136/bjo.81.6.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abbas M, Bobo LD, Hsieh YH, et al. Human leukocyte antigen (HLA)-B, DRB1, and DQB1 allotypes associated with disease and protection of trachoma endemic villagers. Invest. Ophthalmol. Vis. Sci. 2009;50(4):1734–1738. doi: 10.1167/iovs.08-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan M, Bavoil P, editors. Intracellular Pathogens I: Chlamydiales. American Society for Microbiology; Washington, DC, USA: 2012. [Google Scholar]
- 21.Vos M. Why do bacteria engage in homologous recombination? Trends Microbiol. 2009;17(6):226–232. doi: 10.1016/j.tim.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Didelot X, Maiden MC. Impact of recombination on bacterial evolution. Trends Microbiol. 2010;18(7):315–322. doi: 10.1016/j.tim.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinnunen AH, Surcel HM, Lehtinen M, et al. HLA DQ alleles and interleukin-10 polymorphism associated with Chlamydia trachomatis-related tubal factor infertility: a case-control study. Hum. Reprod. 2002;17(8):2073–2078. doi: 10.1093/humrep/17.8.2073. [DOI] [PubMed] [Google Scholar]
- 24.Murillo LS, Land JA, Pleijster J, Bruggeman CA, Pena AS, Morre SA. Interleukin-1B (IL-1B) and interleukin-1 receptor antagonist (IL-1RN) gene polymorphisms are not associated with tubal pathology and Chlamydia trachomatis-related tubal factor subfertility. Hum. Reprod. 2003;18(11):2309–2314. doi: 10.1093/humrep/deg436. [DOI] [PubMed] [Google Scholar]
- 25.Ohman H, Bailey R, Natividad A, et al. Effect of IL12A and IL12B polymorphisms on the risk of Chlamydia trachomatis-induced tubal factor infertility and disease severity. Hum. Reprod. 2012;27(7):2217–2223. doi: 10.1093/humrep/des136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohman H, Tiitinen A, Halttunen M, et al. IL-10 polymorphism and cell-mediated immune response to Chlamydia trachomatis. Genes Immun. 2006;7(3):243–249. doi: 10.1038/sj.gene.6364293. [DOI] [PubMed] [Google Scholar]
- 27.Ohman H, Tiitinen A, Halttunen M, Lehtinen M, Paavonen J, Surcel HM. Cytokine polymorphisms and severity of tubal damage in women with Chlamydia-associated infertility. J. Infect. Dis. 2009;199(9):1353–1359. doi: 10.1086/597620. [DOI] [PubMed] [Google Scholar]
- 28.Ohman H, Tiitinen A, Halttunen M, Paavonen J, Surcel HM. Cytokine gene polymorphism and Chlamydia trachomatis- specific immune responses. Hum. Immunol. 2011;72(3):278–282. doi: 10.1016/j.humimm.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 29.Sziller I, Babula O, Hupuczi P, et al. Mannose-binding lectin (MBL) codon 54 gene polymorphism protects against development of pre-eclampsia, HELLP syndrome and pre-eclampsia-associated intrauterine growth restriction. Mol. Hum. Reprod. 2007;13(4):281–285. doi: 10.1093/molehr/gam003. [DOI] [PubMed] [Google Scholar]
- 30.Tokuriki N, Stricher F, Serrano L, Tawfik DS. How protein stability and new functions trade off. PLoS Comput. Biol. 2008;4(2):e1000002. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dreiseikelmann B. Translocation of DNA across bacterial membranes. Microbiol. Rev. 1994;58(3):293–316. doi: 10.1128/mr.58.3.293-316.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raes J, van De Peer Y. Gene duplication, the evolution of novel gene functions, and detecting functional divergence of duplicates in silico. Appl. Bioinformatics. 2003;2(2):91–101. [PubMed] [Google Scholar]
- 33.Zhang J. Evolution by gene duplication: an update. Trends Ecol. Evol. 2003;18(6):292–298. [Google Scholar]
- 34.Serres MH, Kerr AR, Mccormack TJ, Riley M. Evolution by leaps: gene duplication in bacteria. Biol. Direct. 2009;4:46. doi: 10.1186/1745-6150-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nunes A, Borrego MJ, Nunes B, Florindo C, Gomes JP. Evolutionary dynamics of ompA, the gene encoding the Chlamydia trachomatis key antigen. J. Bacteriol. 2009;191(23):7182–7192. doi: 10.1128/JB.00895-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voigt A, Schofl G, Saluz HP. The Chlamydia psittaci genome: a comparative analysis of intracellular pathogens. PLoS ONE. 2012;7(4):e35097. doi: 10.1371/journal.pone.0035097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suchland RJ, Jeffrey BM, Sandoz KM, Stamm WE, Rockey DD. Generation of recombinant C. trachomatis strains for associating individual genes with known phenotypes. Proceedings of the 12th International Symposium on Human Chlamydial Infections. International Chlamydia Symposium; San Fransisco, CA, USA. 2010. [Google Scholar]
- 38.Seth-Smith HM, Harris SR, Persson K, et al. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics. 2009;10:239. doi: 10.1186/1471-2164-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Unemo M, Seth-Smith HM, Cutcliffe LT, et al. The Swedish new variant of Chlamydia trachomatis: genome sequence, morphology, cell tropism and phenotypic characterization. Microbiology. 2010;156(Pt 5):1394–1404. doi: 10.1099/mic.0.036830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somboonna N, Wan R, Ojcius DM, et al. Hypervirulent Chlamydia trachomatis clinical strain is a recombinant between lymphogranuloma venereum (L(2)) and D lineages. MBio. 2011;2(3):e00045–11. doi: 10.1128/mBio.00045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41■■.Jeffrey BM, Suchland RJ, Quinn KL, Davidson JR, Stamm WE, Rockey DD. Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infect. Immun. 2010;78(6):2544–2553. doi: 10.1128/IAI.01324-09. Describes how comparative genome analysis with new sequencing techniques can be used to investigate the correlation of polymorphisms with tissue tropism and chlamydial disease severity.
- 42.Gomes JP, Bruno WJ, Nunes A, et al. Evolution of Chlamydia trachomatis diversity occurs by widespread interstrain recombination involving hotspots. Genome Res. 2007;17(1):50–60. doi: 10.1101/gr.5674706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dean D, Stephens RS. Identification of individual genotypes of Chlamydia trachomatis from experimentally mixed serovars and mixed infections among trachoma patients. J. Clin. Microbiol. 1994;32(6):1506–1510. doi: 10.1128/jcm.32.6.1506-1510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Millman K, Black CM, Johnson RE, et al. Population-based genetic and evolutionary analysis of Chlamydia trachomatis urogenital strain variation in the United States. J. Bacteriol. 2004;186(8):2457–2465. doi: 10.1128/JB.186.8.2457-2465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belland RJ, Scidmore MA, Crane DD, et al. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl Acad. Sci. USA. 2001;98(24):13984–13989. doi: 10.1073/pnas.241377698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herrmann B. A new genetic variant of Chlamydia trachomatis. Sex Transm. Infect. 2007;83(4):253–254. doi: 10.1136/sti.2007.026260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ripa T, Nilsson P. A variant of Chlamydia trachomatis with deletion in cryptic plasmid: implications for use of PCR diagnostic tests. Euro. Surveill. 2006;11(11):E061109.2. doi: 10.2807/esw.11.45.03076-en. [DOI] [PubMed] [Google Scholar]
- 48.Ripa T, Nilsson PA. A Chlamydia trachomatis strain with a 377-bp deletion in the cryptic plasmid causing false-negative nucleic acid amplification tests. Sex Transm. Dis. 2007;34(5):255–256. doi: 10.1097/OLQ.0b013e31805ce2b9. [DOI] [PubMed] [Google Scholar]
- 49.Byrne GI. Chlamydia trachomatis strains and virulence: rethinking links to infection prevalence and disease severity. J. Infect. Dis. 2010;201(Suppl. 2):S126–S133. doi: 10.1086/652398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jurstrand M, Falk L, Fredlund H, et al. Characterization of Chlamydia trachomatis omp1 genotypes among sexually transmitted disease patients in Sweden. J. Clin. Microbiol. 2001;39(11):3915–3919. doi: 10.1128/JCM.39.11.3915-3919.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lysen M, Osterlund A, Rubin CJ, Persson T, Persson I, Herrmann B. Characterization of ompA genotypes by sequence analysis of DNA from all detected cases of Chlamydia trachomatis infections during 1 year of contact tracing in a Swedish county. J. Clin. Microbiol. 2004;42(4):1641–1647. doi: 10.1128/JCM.42.4.1641-1647.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Millman KL, Tavare S, Dean D. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J. Bacteriol. 2001;183(20):5997–6008. doi: 10.1128/JB.183.20.5997-6008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andreasen AA, Burton MJ, Holland MJ, et al. Chlamydia trachomatis ompA variants in trachoma: what do they tell us? PLoS Negl. Trop. Dis. 2008;2(9):e306. doi: 10.1371/journal.pntd.0000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang SP, Grayston JT. Immunologic relationship between genital TRIC, lymphogranuloma venereum, and related organisms in a new microtiter indirect immunofluorescence test. Am. J. Ophthalmol. 1970;70(3):367–374. doi: 10.1016/0002-9394(70)90096-6. [DOI] [PubMed] [Google Scholar]
- 55.Brunelle BW, Sensabaugh GF. The ompA gene in Chlamydia trachomatis differs in phylogeny and rate of evolution from other regions of the genome. Infect. Immun. 2006;74(1):578–585. doi: 10.1128/IAI.74.1.578-585.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stothard DR, Boguslawski G, Jones RB. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect. Immun. 1998;66(8):3618–3625. doi: 10.1128/iai.66.8.3618-3625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brunelle BW, Sensabaugh GF. Nucleotide and phylogenetic analyses of the Chlamydia trachomatis ompA gene indicates it is a hotspot for mutation. BMC Res. Notes. 2012;5:53. doi: 10.1186/1756-0500-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dean D, Millman K. Molecular and mutation trends analyses of omp1 alleles for serovar E of Chlamydia trachomatis. Implications for the immunopathogenesis of disease. J. Clin. Invest. 1997;99(3):475–483. doi: 10.1172/JCI119182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee G, Park J, Kim B, Kim SA, Yoo CK, Seong WK. OmpA genotyping of Chlamydia trachomatis from Korean female sex workers. J. Infect. 2006;52(6):451–454. doi: 10.1016/j.jinf.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 60.Lister NA, Tabrizi SN, Fairley CK, Smith A, Janssen PH, Garland S. Variability of the Chlamydia trachomatis omp1 gene detected in samples from men tested in male-only saunas in Melbourne, Australia. J. Clin. Microbiol. 2004;42(6):2596–2601. doi: 10.1128/JCM.42.6.2596-2601.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mossman D, Beagley KW, Landay AL, et al. Genotyping of urogenital Chlamydia trachomatis in regional New South Wales, Australia. Sex Transm. Dis. 2008;35(6):614–616. doi: 10.1097/OLQ.0b013e31816b1b80. [DOI] [PubMed] [Google Scholar]
- 62.Ngandjio A, Clerc M, Fonkoua MC, et al. Screening of volunteer students in Yaounde (Cameroon, central Africa) for Chlamydia trachomatis infection and genotyping of isolated C. trachomatis strains. J. Clin. Microbiol. 2003;41(9):4404–4407. doi: 10.1128/JCM.41.9.4404-4407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pedersen LN, Kjaer HO, Moller JK, Orntoft TF, Ostergaard L. High-resolution genotyping of Chlamydia trachomatis from recurrent urogenital infections. J. Clin. Microbiol. 2000;38(8):3068–3071. doi: 10.1128/jcm.38.8.3068-3071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64■.Nunes A, Nogueira PJ, Borrego MJ, Gomes JP. Adaptive evolution of the Chlamydia trachomatis dominant antigen reveals distinct evolutionary scenarios for B- and T-cell epitopes: worldwide survey. PLoS ONE. 2010;5(10):e13171. doi: 10.1371/journal.pone.0013171. Gives a great overview of the genetic variation in the ompA gene and highlights differences between the conserved and variable regions.
- 65.Galan JE, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284(5418):1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 66.Peters J, Wilson DP, Myers G, Timms P, Bavoil PM. Type III secretion a la Chlamydia. Trends Microbiol. 2007;15(6):241–251. doi: 10.1016/j.tim.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 67.Fields KA, Fischer ER, Mead DJ, Hackstadt T. Analysis of putative Chlamydia trachomatis chaperones Scc2 and Scc3 and their use in the identification of type III secretion substrates. J. Bacteriol. 2005;187(18):6466–6478. doi: 10.1128/JB.187.18.6466-6478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jorgensen I, Valdivia RH. Pmp-like proteins Pls1 and Pls2 are secreted into the lumen of the Chlamydia trachomatis inclusion. Infect. Immun. 2008;76(9):3940–3950. doi: 10.1128/IAI.00632-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carabeo RA, Grieshaber SS, Fischer E, Hackstadt T. Chlamydia trachomatis induces remodeling of the actin cytoskeleton during attachment and entry into HeLa cells. Infect. Immun. 2002;70(7):3793–3803. doi: 10.1128/IAI.70.7.3793-3803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clifton DR, Dooley CA, Grieshaber SS, Carabeo RA, Fields KA, Hackstadt T. Tyrosine phosphorylation of the chlamydial effector protein Tarp is species specific and not required for recruitment of actin. Infect. Immun. 2005;73(7):3860–3868. doi: 10.1128/IAI.73.7.3860-3868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiwani S, Alvarado S, Ohr RJ, Romero A, Nguyen B, Jewett TJ. Chlamydia trachomatis Tarp harbors distinct G and F actin binding domains which bundle actin filaments. J. Bacteriol. 2012;195(4):708–716. doi: 10.1128/JB.01768-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jewett TJ, Fischer ER, Mead DJ, Hackstadt T. Chlamydial Tarp is a bacterial nucleator of actin. Proc. Natl Acad. Sci. USA. 2006;103(42):15599–15604. doi: 10.1073/pnas.0603044103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73■.Jewett TJ, Miller NJ, Dooley CA, Hackstadt T. The conserved Tarp actin binding domain is important for chlamydial invasion. PLoS Pathog. 2010;6(7):e1000997. doi: 10.1371/journal.ppat.1000997. Demonstrates the importance of Tarp for chlamydial cell invasion, highlights species-specific differences in the protein and shows the role of the actin binding domain in the species-specific actin-nucleation speed.
- 74.Lutter EI, Bonner C, Holland MJ, et al. Phylogenetic analysis of Chlamydia trachomatis Tarp and correlation with clinical phenotype. Infect. Immun. 2010;78(9):3678–3688. doi: 10.1128/IAI.00515-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jewett TJ, Dooley CA, Mead DJ, Hackstadt T. Chlamydia trachomatis Tarp is phosphorylated by src family tyrosine kinases. Biochem. Biophys. Res. Commun. 2008;371(2):339–344. doi: 10.1016/j.bbrc.2008.04.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mehlitz A, Banhart S, Maurer AP, et al. Tarp regulates early Chlamydia-induced host cell survival through interactions with the human adaptor protein SHC1. J. Cell Biol. 2010;190(1):143–157. doi: 10.1083/jcb.200909095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Almeida F, Borges V, Ferreira R, Borrego MJ, Gomes JP, Mota LJ. Polymorphisms in Inc proteins and differential expression of Inc genes among Chlamydia trachomatis strains correlate with invasiveness and tropism of lymphogranuloma venereum isolates. J. Bacteriol. 2012;194(23):6574–6585. doi: 10.1128/JB.01428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lutter EI, Martens C, Hackstadt T. Evolution and conservation of predicted inclusion membrane proteins in Chlamydiae. Comp. Funct. Genomics. 2012;2012:362104. doi: 10.1155/2012/362104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dehoux P, Flores R, Dauga C, Zhong G, Subtil A. Multi-genome identification and characterization of Chlamydiae-specific type III secretion substrates: the Inc proteins. BMC Genomics. 2011;12:109. doi: 10.1186/1471-2164-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Z, Chen C, Chen D, Wu Y, Zhong Y, Zhong G. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect. Immun. 2008;76(6):2746–2757. doi: 10.1128/IAI.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mital J, Miller NJ, Fischer ER, Hackstadt T. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell Microbiol. 2010;12(9):1235–1249. doi: 10.1111/j.1462-5822.2010.01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spaeth KE, Chen YS, Valdivia RH. The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog. 2009;5(9):e1000579. doi: 10.1371/journal.ppat.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clifton DR, Fields KA, Grieshaber SS, et al. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl Acad. Sci. USA. 2004;101(27):10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pannekoek Y, Spaargaren J, Langerak AA, Merks J, Morre SA, van Der Ende A. Interrelationship between polymorphisms of IncA, fusogenic properties of Chlamydia trachomatis strains, and clinical manifestations in patients in The Netherlands. J. Clin. Microbiol. 2005;43(5):2441–2443. doi: 10.1128/JCM.43.5.2441-2443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rockey DD, Viratyosin W, Bannantine JP, Suchland RJ, Stamm WE. Diversity within inc genes of clinical Chlamydia trachomatis variant isolates that occupy non-fusogenic inclusions. Microbiology. 2002;148(Pt 8):2497–2505. doi: 10.1099/00221287-148-8-2497. [DOI] [PubMed] [Google Scholar]
- 86■.Suchland RJ, Rockey DD, Bannantine JP, Stamm WE. Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect. Immun. 2000;68(1):360–367. doi: 10.1128/iai.68.1.360-367.2000. Describes for the first time that differences in IncA are potentially correlated to species-specific differences and result in nonfusogenic inclusions of some chlamydial species.
- 87.Geisler WM, Suchland RJ, Rockey DD, Stamm WE. Epidemiology and clinical manifestations of unique Chlamydia trachomatis isolates that occupy nonfusogenic inclusions. J. Infect. Dis. 2001;184(7):879–884. doi: 10.1086/323340. [DOI] [PubMed] [Google Scholar]
- 88.Derre I, Swiss R, Agaisse H. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathog. 2011;7(6):e1002092. doi: 10.1371/journal.ppat.1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Borges V, Nunes A, Ferreira R, Borrego MJ, Gomes JP. Directional evolution of Chlamydia trachomatis towards niche-specific adaptation. J. Bacteriol. 2012;194(22):6143–6153. doi: 10.1128/JB.01291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sugiki T, Takeuchi K, Yamaji T, et al. Structural basis for the Golgi association by the pleckstrin homology domain of the ceramide trafficking protein (CERT) J. Biol. Chem. 2012;287(40):33706–33718. doi: 10.1074/jbc.M112.367730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scidmore-Carlson MA, Shaw EI, Dooley CA, Fischer ER, Hackstadt T. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol. Microbiol. 1999;33(4):753–765. doi: 10.1046/j.1365-2958.1999.01523.x. [DOI] [PubMed] [Google Scholar]
- 92.Scidmore MA, Hackstadt T. Mammalian 14-13-3β associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol. Microbiol. 2001;39(6):1638–1650. doi: 10.1046/j.1365-2958.2001.02355.x. [DOI] [PubMed] [Google Scholar]
- 93.Verbeke P, Welter-Stahl L, Ying S, et al. Recruitment of BAD by the Chlamydia trachomatis vacuole correlates with host-cell survival. PLoS Pathog. 2006;2(5):e45. doi: 10.1371/journal.ppat.0020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Byrne GI, Ojcius DM. Chlamydia and apoptosis: life and death decisions of an intracellular pathogen. Nat. Rev. Microbiol. 2004;2(10):802–808. doi: 10.1038/nrmicro1007. [DOI] [PubMed] [Google Scholar]
- 95.Belland RJ, Zhong G, Crane DD, et al. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl Acad. Sci. USA. 2003;100(14):8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol. Microbiol. 2000;37(4):913–925. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- 97.Rzomp KA, Moorhead AR, Scidmore MA. The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrane protein CT229. Infect. Immun. 2006;74(9):5362–5373. doi: 10.1128/IAI.00539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Grimwood J, Stephens RS. Computational analysis of the polymorphic membrane protein superfamily of Chlamydia trachomatis and Chlamydia pneumoniae. Microb. Comp. Genomics. 1999;4(3):187–201. doi: 10.1089/omi.1.1999.4.187. [DOI] [PubMed] [Google Scholar]
- 99.Stephens RS, Kalman S, Lammel C, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282(5389):754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 100.Henderson IR, Lam AC. Polymorphic proteins of Chlamydia spp.-autotransporters beyond the Proteobacteria. Trends Microbiol. 2001;9(12):573–578. doi: 10.1016/s0966-842x(01)02234-x. [DOI] [PubMed] [Google Scholar]
- 101.Gomes JP, Bruno WJ, Borrego MJ, Dean D. Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J. Bacteriol. 2004;186(13):4295–4306. doi: 10.1128/JB.186.13.4295-4306.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gomes JP, Nunes A, Bruno WJ, Borrego MJ, Florindo C, Dean D. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J. Bacteriol. 2006;188(1):275–286. doi: 10.1128/JB.188.1.275-286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thalmann J, Janik K, May M, et al. Actin re-organization induced by Chlamydia trachomatis serovar D-evidence for a critical role of the effector protein CT166 targeting Rac. PLoS ONE. 2010;5(3):e9887. doi: 10.1371/journal.pone.0009887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104■.Carlson JH, Hughes S, Hogan D, et al. Polymorphisms in the Chlamydia trachomatis cytotoxin locus associated with ocular and genital isolates. Infect. Immun. 2004;72(12):7063–7072. doi: 10.1128/IAI.72.12.7063-7072.2004. A thorough comparative analysis of the cytotoxin loci between Chlamydia trachomatis serovars, and highlights the polymorphism of the cytotoxin and a potential role for tissue tropism.
- 105.Fehlner-Gardiner C, Roshick C, Carlson JH, et al. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 2002;277(30):26893–26903. doi: 10.1074/jbc.M203937200. [DOI] [PubMed] [Google Scholar]
- 106.Mojica S, Huot Creasy H, Daugherty S, et al. Genome sequence of the obligate intracellular animal pathogen Chlamydia pecorum E58. J. Bacteriol. 2011;193(14):3690. doi: 10.1128/JB.00454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Read TD, Myers GS, Brunham RC, et al. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 2003;31(8):2134–2147. doi: 10.1093/nar/gkg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thomson NR, Yeats C, Bell K, et al. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 2005;15(5):629–640. doi: 10.1101/gr.3684805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xie G, Bonner CA, Jensen RA. Dynamic diversity of the tryptophan pathway in Chlamydiae: reductive evolution and a novel operon for tryptophan recapture. Genome Biol. 2002;3(9):research0051. doi: 10.1186/gb-2002-3-9-research0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110■.Caldwell HD, Wood H, Crane D, et al. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Invest. 2003;111(11):1757–1769. doi: 10.1172/JCI17993. Describes for the first time the polymorphisms in the tryptophan operon in C. trachomatis and its potential role in tissue tropism between the ocular and genital serovars.
- 111.Byrne GI, Lehmann LK, Landry GJ. Induction of tryptophan catabolism is the mechanism for gamma-interferon-mediated inhibition of intracellular Chlamydia psittaci replication in T24 cells. Infect. Immun. 1986;53(2):347–351. doi: 10.1128/iai.53.2.347-351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Carlin JM, Borden EC, Byrne GI. Interferon-induced indoleamine 2,3-dioxygenase activity inhibits Chlamydia psittaci replication in human macrophages. J. Interferon Res. 1989;9(3):329–337. doi: 10.1089/jir.1989.9.329. [DOI] [PubMed] [Google Scholar]
- 113.Kane CD, Vena RM, Ouellette SP, Byrne GI. Intracellular tryptophan pool sizes may account for differences in gamma interferon-mediated inhibition and persistence of chlamydial growth in polarized and nonpolarized cells. Infect. Immun. 1999;67(4):1666–1671. doi: 10.1128/iai.67.4.1666-1671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Leonhardt RM, Lee SJ, Kavathas PB, Cresswell P. Severe tryptophan starvation blocks onset of conventional persistence and reduces reactivation of Chlamydia trachomatis. Infect. Immun. 2007;75(11):5105–5117. doi: 10.1128/IAI.00668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rapoza PA, Tahija SG, Carlin JP, Miller SL, Padilla ML, Byrne GI. Effect of interferon on a primary conjunctival epithelial cell model of trachoma. Invest. Ophthalmol. Vis. Sci. 1991;32(11):2919–2923. [PubMed] [Google Scholar]
- 116.Dong Q, Brulc JM, Iovieno A, et al. Diversity of bacteria at healthy human conjunctiva. Invest. Ophthalmol. Vis. Sci. 2011;52(8):5408–5413. doi: 10.1167/iovs.10-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhou Y, Gao H, Mihindukulasuriya KA, et al. Biogeography of the ecosystems of the healthy human body. Genome Biol. 2013;14(1):R1. doi: 10.1186/gb-2013-14-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Giles TN, Graham DE. Characterization of an acid-dependent arginine decarboxylase enzyme from Chlamydophila pneumoniae. J. Bacteriol. 2007;189(20):7376–7383. doi: 10.1128/JB.00772-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Smith CB, Graham DE. Outer and inner membrane proteins compose an arginine-agmatine exchange system in Chlamydophila pneumoniae. J. Bacteriol. 2008;190(22):7431–7440. doi: 10.1128/JB.00652-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Giles TN, Fisher DJ, Graham DE. Independent inactivation of arginine decarboxylase genes by nonsense and missense mutations led to pseudogene formation in Chlamydia trachomatis serovar L2 and D strains. BMC Evol. Biol. 2009;9:166. doi: 10.1186/1471-2148-9-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Darville T, Hiltke TJ. Pathogenesis of genital tract disease due to Chlamydia trachomatis. J. Infect. Dis. 2010;201(Suppl. 2):S114–S125. doi: 10.1086/652397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang C, Tang J, Geisler WM, Crowley-Nowick PA, Wilson CM, Kaslow RA. Human leukocyte antigen and cytokine gene variants as predictors of recurrent Chlamydia trachomatis infection in high-risk adolescents. J. Infect. Dis. 2005;191(7):1084–1092. doi: 10.1086/428592. [DOI] [PubMed] [Google Scholar]
- 123.Conway DJ, Holland MJ, Bailey RL, et al. Scarring trachoma is associated with polymorphism in the tumor necrosis factor alpha (TNF-alpha) gene promoter and with elevated TNF-alpha levels in tear fluid. Infect. Immun. 1997;65(3):1003–1006. doi: 10.1128/iai.65.3.1003-1006.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kari L, Whitmire WM, Olivares-Zavaleta N, et al. A live-attenuated chlamydial vaccine protects against trachoma in nonhuman primates. J. Exp. Med. 2011;208(11):2217–2223. doi: 10.1084/jem.20111266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.O’Connell CM, Ingalls RR, Andrews CW, Jr, Scurlock AM, Darville T. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J. Immunol. 2007;179(6):4027–4034. doi: 10.4049/jimmunol.179.6.4027. [DOI] [PubMed] [Google Scholar]
- 126.Bode J, Dutow P, Sommer K, et al. A new role of the complement system: C3 provides protection in a mouse model of lung infection with intracellular Chlamydia psittaci. PLoS ONE. 2012;7(11):e50327. doi: 10.1371/journal.pone.0050327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gondek DC, Roan NR, Starnbach MN. T cell responses in the absence of IFN-gamma exacerbate uterine infection with Chlamydia trachomatis. J. Immunol. 2009;183(2):1313–1319. doi: 10.4049/jimmunol.0900295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Imtiaz MT, Distelhorst JT, Schripsema JH, et al. A role for matrix metalloproteinase-9 in pathogenesis of urogenital Chlamydia muridarum infection in mice. Microbes Infect. 2007;9(14-15):1561–1566. doi: 10.1016/j.micinf.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee HY, Schripsema JH, Sigar IM, et al. A role for CXC chemokine receptor-2 in the pathogenesis of urogenital Chlamydia muridarum infection in mice. FEMS Immunol. Med. Microbiol. 2010;60(1):49–56. doi: 10.1111/j.1574-695X.2010.00715.x. [DOI] [PubMed] [Google Scholar]
- 130.Olive AJ, Gondek DC, Starnbach MN. CXCR3 and CCR5 are both required for T cell-mediated protection against C. trachomatis infection in the murine genital mucosa. Mucosal Immunol. 2011;4(2):208–216. doi: 10.1038/mi.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pal S, Schmidt AP, Peterson EM, Wilson CL, de la Maza LM. Role of matrix metalloproteinase-7 in the modulation of a Chlamydia trachomatis infection. Immunology. 2006;117(2):213–219. doi: 10.1111/j.1365-2567.2005.02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang X, Brunham RC. Gene knockout B cell-deficient mice demonstrate that B cells play an important role in the initiation of T cell responses to Chlamydia trachomatis (mouse pneumonitis) lung infection. J. Immunol. 1998;161(3):1439–1446. [PubMed] [Google Scholar]
- 133.Nagarajan UM, Prantner D, Sikes JD, et al. Type I interferon signaling exacerbates Chlamydia muridarum genital infection in a murine model. Infect. Immun. 2008;76(10):4642–4648. doi: 10.1128/IAI.00629-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Waterston RH, Lindblad-Toh K, Birney E, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 135.Seok J, Warren HS, Cuenca AG, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl Acad. Sci. USA. 2013;110(9):3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Miyairi I, Tatireddigari VR, Mahdi OS, et al. The p47 GTPases Iigp2 and Irgb10 regulate innate immunity and inflammation to murine Chlamydia psittaci infection. J. Immunol. 2007;179(3):1814–1824. doi: 10.4049/jimmunol.179.3.1814. [DOI] [PubMed] [Google Scholar]
- 137■■.Miyairi I, Ziebarth J, Laxton JD, et al. Host genetics and chlamydia disease: prediction and validation of disease severity mechanisms. PLoS ONE. 2012;7(3):e33781. doi: 10.1371/journal.pone.0033781. Demonstrates how systems biology and forward genetics using advanced recombinant inbred mice can be utilized to identify host genetic factors that are important for the infection and disease outcome.
- 138.Bernstein-Hanley I, Balsara ZR, Ulmer W, Coers J, Starnbach MN, Dietrich WF. Genetic analysis of susceptibility to Chlamydia trachomatis in mouse. Genes Immun. 2006;7(2):122–129. doi: 10.1038/sj.gene.6364285. [DOI] [PubMed] [Google Scholar]
- 139.Bernstein-Hanley I, Coers J, Balsara ZR, Taylor GA, Starnbach MN, Dietrich WF. The p47 GTPases Igtp and Irgb10 map to the Chlamydia trachomatis susceptibility locus Ctrq-3 and mediate cellular resistance in mice. Proc. Natl Acad. Sci. USA. 2006;103(38):14092–14097. doi: 10.1073/pnas.0603338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nelson DE, Virok DP, Wood H, et al. Chlamydial IFN-gamma immune evasion is linked to host infection tropism. Proc. Natl Acad. Sci. USA. 2005;102(30):10658–10663. doi: 10.1073/pnas.0504198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Abdeltawab NF, Aziz RK, Kansal R, et al. An unbiased systems genetics approach to mapping genetic loci modulating susceptibility to severe streptococcal sepsis. PLoS Pathog. 2008;4(4):e1000042. doi: 10.1371/journal.ppat.1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Boon AC, Debeauchamp J, Hollmann A, et al. Host genetic variation affects resistance to infection with a highly pathogenic H5N1 influenza A virus in mice. J. Virol. 2009;83(20):10417–10426. doi: 10.1128/JVI.00514-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sandoz KM, Rockey DD. Antibiotic resistance in Chlamydiae. Future Microbiol. 2010;5(9):1427–1442. doi: 10.2217/fmb.10.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schautteet K, de Clercq E, Miry C, et al. Tetracycline-resistant Chlamydia suis in cases of reproductive failure on Belgian, Cypriote and Israeli pig production farms. J. Med. Microbiol. 2013;62(Pt 2):331–334. doi: 10.1099/jmm.0.042861-0. [DOI] [PubMed] [Google Scholar]
- 145.Gaydos CA, Howell MR, Pare B, et al. Chlamydia trachomatis infections in female military recruits. N. Engl. J. Med. 1998;339(11):739–744. doi: 10.1056/NEJM199809103391105. [DOI] [PubMed] [Google Scholar]
- 146.Geisler WM, Wang C, Morrison SG, Black CM, Bandea CI, Hook EW., 3rd The natural history of untreated Chlamydia trachomatis infection in the interval between screening and returning for treatment. Sex Transm. Dis. 2008;35(2):119–123. doi: 10.1097/OLQ.0b013e318151497d. [DOI] [PubMed] [Google Scholar]
- 147.Imai H, Nakao H, Shinohara H, et al. Population-based study of asymptomatic infection with Chlamydia trachomatis among female and male students. Int. J. STD AIDS. 2010;21(5):362–366. doi: 10.1258/ijsa.2010.010026. [DOI] [PubMed] [Google Scholar]
- 148.CDC Grand Rounds Chlamydia prevention: challenges and strategies for reducing disease burden and sequelae. MMWR Morb. Mortal. Wkly Rep. 2011;60:370–373. [PubMed] [Google Scholar]
- 149.Brunham RC, Rekart ML. The arrested immunity hypothesis and the epidemiology of chlamydia control. Sex Transm. Dis. 2008;35(1):53–54. doi: 10.1097/OLQ.0b013e31815e41a3. [DOI] [PubMed] [Google Scholar]
- 150.Brunham RC, Rappuoli R. Chlamydia trachomatis control requires a vaccine. Vaccine. 2013;31(15):1892–1897. doi: 10.1016/j.vaccine.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Brunham RC, Rekart M. Considerations on Chlamydia trachomatis disease expression. FEMS Immunol. Med. Microbiol. 2009;55(2):162–166. doi: 10.1111/j.1574-695X.2008.00509.x. [DOI] [PubMed] [Google Scholar]
- 152.Su W, Li Z, Li F, Chen X, Wan Q, Liang D. Doxycycline-mediated inhibition of corneal angiogenesis: an MMP-independent mechanism. Invest. Ophthalmol. Vis. Sci. 2013;54(1):783–788. doi: 10.1167/iovs.12-10323. [DOI] [PubMed] [Google Scholar]
- 153.Bernardino AL, Kaushal D, Philipp MT. The antibiotics doxycycline and minocycline inhibit the inflammatory responses to the Lyme disease spirochete Borrelia burgdorferi. J. Infect. Dis. 2009;199(9):1379–1388. doi: 10.1086/597807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 201.NCBI genome database. www.ncbi.nlm.nih.gov/genome/browse/
- 202.Sanger Institute Chlamydia trachomatis. www.sanger.ac.uk/resources/downloads/bacteria/chlamydia-trachomatis.html.
- 203.European Nucleotide Archive at EMBL. www.ebi.ac.uk/ena.
- 204.CDC Sexually transmitted disease surveillance 2011. www.cdc.gov/std/stats11/Surv2011.pdf.