Abstract
Metabolic profiling of urine provides a fingerprint of personalized endogenous metabolite markers that correlate to a number of factors such as gender, disease, diet, toxicity, medication, and age. It is important to study these factors individually, if possible to unravel their unique contributions. In this study, age-related metabolic changes in children of age 12 years and below were analyzed by 1H NMR spectroscopy of urine. The effect of age on the urinary metabolite profile was observed as a distinct age-dependent clustering even from the unsupervised principal component analysis. Further analysis, using partial least squares with orthogonal signal correction regression with respect to age, resulted in the identification of an age-related metabolic profile. Metabolites that correlated with age included creatinine, creatine, glycine, betaine/TMAO, citrate, succinate, and acetone. Although creatinine increased with age, all the other metabolites decreased. These results may be potentially useful in assessing the biological age (as opposed to chronological) of young humans as well as in providing a deeper understanding of the confounding factors in the application of metabolomics.
Keywords: age, human urine, metabolite profiling, metabolomics, metabonomics, nuclear magnetic resonance, orthogonal signal correction, principal component analysis
INTRODUCTION
Understanding the hereditary and epidemiological effects of aging is of tremendous importance for a variety of reasons, including the management of health, disease, and prolonged life span. Chronological age alone does not predict a person’s susceptibility to the consequences of aging, as a vast number of effects such as stress, dietary choices, environmental factors, daily activities, and medication affect the so-called ‘biological age’. A large number of parameters have been proposed to assess the biological age of an individual (1). Reliable, sensitive, and specific molecular markers could provide valuable neurobiological and physiological information on age-related changes. These markers might allow new insights into the mechanisms of maturation and aging and raise the hope for accurately assessing an individual’s anatomical status at any given time. Such markers might also lead to the identification of innovative treatments for delaying or reversing abnormal molecular changes. A number of investigations on age-related molecular changes such as gene expression profiles, enzymatic and small molecular changes have been made to date on animals and humans in an effort to establish biomarkers of aging (1–8).
Metabolic profiling provides a new approach and opportunity to explore the metabolic effects of many conditions in complex biological systems. The field of metabolic profiling, including metabolomics and metabonomics, has rapidly developed over the last decade, with successful applications in various research areas including toxicology, disease diagnosis, functional genomics, pharmacology, foods and nutrition, etc. (9–21). The most commonly employed analytical techniques used for metabolic profiling include nuclear magnetic resonance (NMR) and mass spectrometry (MS) (19,22,23). High resolution 1H NMR spectroscopy provides quantitative and reproducible information with little sample preparation, and hence it is widely used to build metabolic profiles in diverse metabolic studies (12,14,16,24–29).
It is well known that the metabolites in urine provide a finger-print for each individual, containing significant amounts of information about age, gender, lifestyle, dietary intake, and disease history (30–38). In most metabolomic studies, high specificity is one of the basic requirements for isolating the desired variables reliably. For this purpose, several signal correction algorithms, including orthogonal signal correction (OSC) and orthogonal projection to latent structures (O-PLS), were developed to remove the chemical and thermal noise, as well as other variables that are not relevant to the biological variation of interest (39–43).
To date, several studies have focused on profiling the metabolites associated with age. While a majority of these investigations have used animal models (36,37,44,45), two age effect studies on adult humans (>19 years of age) were made employing 1H NMR of urine (5,6). Slupsky et al. studied the effect of age on urinary profiles using two groups (young and old), with 40 years as the cut-off point. Carnitine, 3-hydroxyisovalerate, creatinine, alanine, and trigonelline were found to differ significantly between the younger and older age groups (5). Psihogios et al. investigated urine metabolite changes in human subjects ranging from 23 to 74 years of age. They reported several metabolites, including trimethylamine-N-oxide (TMAO), citrate, phenylalanine, creatinine, and hippurate to be linked with the age (6). A study on creatinine levels surveyed a large US population with ages ranging from 6 to 70 years and reported a gradual increase in urinary creatinine concentration up to somewhere between 20 and 29 years of age and a decrease thereafter (46).
In the present study, with a view toward widening our understanding of age-associated metabolic changes, we have studied urine samples of children from newborn to 12 year olds using 1H NMR-based metabolomics methods. In addition to providing complementary evidence on aging, the biochemical variations in urine derived from this study, using unsupervised and supervised statistical methods, provide information on the global age profile in children as they mature.
EXPERIMENTAL METHODS
Sample collection
Urine samples from 55 children, with no diagnosed disease, were collected at the J.W. Riley Children’s Hospital (Indianapolis, IN). Table 1 provides a brief demographic summary of the study subjects. Samples were de-identified in compliance with a protocol approved by the IRB committees at both Purdue University and Indiana University School of Medicine, stored under dry ice, transported to Purdue University, and then frozen at −80°C until they were used for NMR spectroscopy experiments.
Table 1.
Summary of sample numbers according to age
| Age range | Number of normal samples |
|---|---|
| 0–1 year | 23 |
| 2–4 years | 12 |
| 5–12 years | 20 |
| Total from all age ranges | 55 |
1H NMR spectroscopy
After thawing, 640 μL of urine was mixed with 80 μL phosphate buffer (1M; pH 7.0), 80 μL D2O (used as a lock solvent), and a 2 μL solution of 3-(trimethylsilyl) propionic-(2,2,3,3-d4) acid sodium salt (TSP) (20 mM in D2O; used as the chemical shift reference). Each sample was vortexed for 60 s, centrifuged for 10 min at 7000 RPM, and then a 650 μL aliquot was transferred to a standard 5 mm NMR tube for analysis.
1H NMR experiments were carried out on a Bruker DRX 500 MHz spectrometer equipped with a 5 mm TXI cryoprobe. 1D spectra were recorded by suppressing the strong water signal using a standard WATERGATE pulse sequence from the Bruker library. For each spectrum, 64 transients were collected using 32 000 data points and a 6000 Hz spectral width. An exponential weighting function corresponding to 0.3 Hz line broadening was applied to the free induction decay before applying Fourier transformation. All spectra were phased and baseline corrected using Bruker’s XWINNMR software, and referenced relative to the chemical shift of TSP (0.0 ppm).
Data analysis
Each NMR spectrum was reduced to 800 frequency bins of equal width using the R statistical package (version 2.2.1). Spectral regions within the range of 0.2 to 10 ppm were used after deleting the region containing residual water and urea signals (4.5 to 6.0 ppm). The data were imported into Matlab software (Mathworks, MA) installed with the PLS toolbox (Eigenvector Research, Inc, version 4.0) for PCA and PLS modeling. NMR spectral variables were normalized to total integral and mean-centered prior to all statistical analyses. For PLS modeling, each individual child’s age was used as the one-component Y matrix. OSC-PLS modeling analysis was performed by choosing the OSC filter in the preprocessing step prior to PLS regression. Y-matrix data (age) were mean-centered and the regressions were cross-validated using the leave-one-out process to avoid over-fitting. The leave-one-out cross validation step was also performed before the OSC filter to examine the validation procedure.
In addition, univariate analysis was performed by applying the unpaired Student’s t-test to several metabolites of interest that were identified by the multivariate analysis. The NMR intensities of these metabolites were normalized and mean-centered, and then p-values were calculated using two age groups, newborn and 10–12 year olds (n =6 and 8, respectively).
RESULTS AND DISCUSSION
NMR spectral analysis
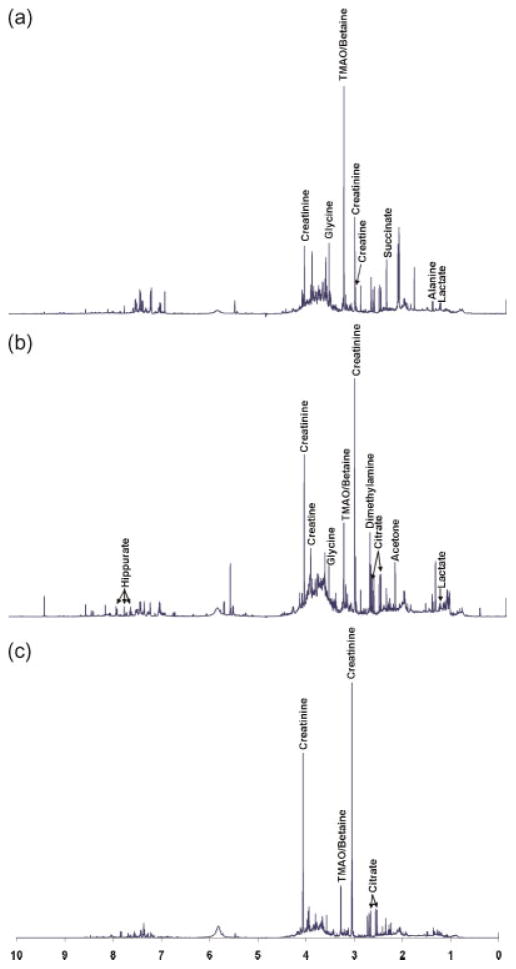
1H NMR spectra of representative urine samples from a newborn, one year old, and a 10-year-old child are shown in Figure 1. As noted in Figure 1, the dominant metabolites identified in the majority of the urine samples include hippurate (7.83, 7.64, 7.55 ppm), lactate (4.12, 1.33 ppm), creatinine (4.07, 3.05 ppm), creatine (3.94, 3.04 ppm), glycine (3.57 ppm), taurine (3.43, 3.27 ppm), trimethylamine oxide (TMAO)/betaine (3.26 ppm), dimethylamine (2.74 ppm), citrate (2.67, 2.56 ppm), succinate (2.42 ppm), acetone (2.24 ppm), and alanine (1.48 ppm). Identification of these metabolites is based on several previous studies (47–51) and KnowItAll software (Bio-Rad Laboratories, Inc., Hercules, CA).
Figure 1.
Typical 1H NMR spectra of normal urine samples from children with different ages: (a) newborn, (b) 1 year old, and (c) 10 years old.
Though the urine spectra are complex due to a number of confounding effects including age, gender, diet, etc., it is evident by visual inspection that the intensity of creatinine increases with age. Other metabolites such as TMAO/betaine and citrate also showed a large variation with age.
Statistical analysis
To unravel global differences associated with age in the complex urine spectra, the data were analyzed using both unsupervised (PCA) and supervised (OSC-PLS) statistical analysis methods.
Principal component analysis (PCA)
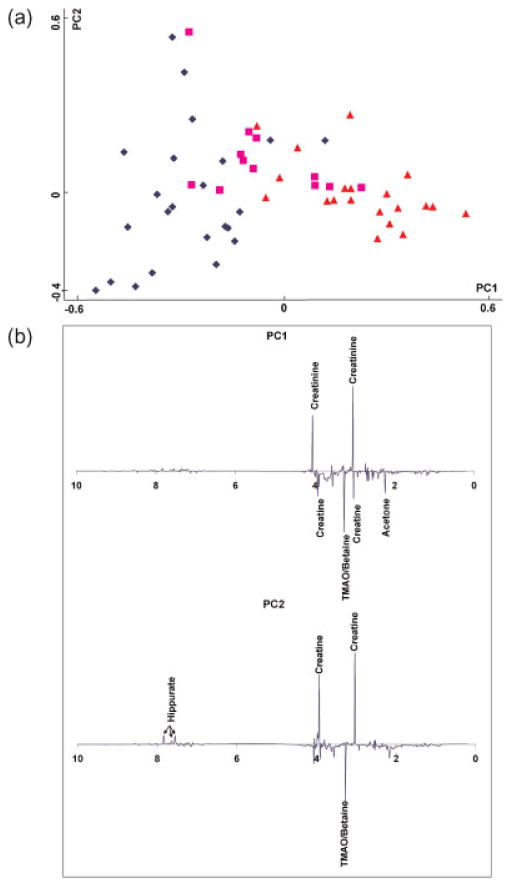
Figure 2 presents the results of PCA of the NMR spectra from 55 normal children’s urine samples. Age ranges are marked with different symbols in the figure. Even from this unsupervised analysis, a separation with respect to age is clearly seen. Older children tend to have higher scores along the PC1 direction; PC1 explains 32.3% of the variance in the NMR data. From the analysis of the PC1 loading plot, a number of metabolites are found to contribute to this separation. Specifically, PC1, the direction in which the samples are separated in relation to age, shows some metabolites such as hippurate, creatinine, creatine, glycine, TMAO/betaine, citrate, succinate, and acetone that differ with age. Some prominent peaks in the PC1 loading plot are marked in Figure 2b.
Figure 2.
(a) PCA score plot of 55 normal urine samples. Blue diamonds: 0–1 year old; pink squares: 2–4 years old; orange triangles: 5–12 years old. (b) PCA loading plots of the 55 normal urine samples.
To further analyze the metabolic changes, p-values between two age groups of <0.5 and 10–12 years old (n =6 and 8, respectively) were calculated for some of the metabolites identi-fied in the PC1 loading plot. Of these, creatinine (p-value =0.0003), TMAO/betaine (p-value =0.0077) and acetone (p-value =0.0348), are significantly altered and confirm the observation that metabolic profiles of pre-adolescent urine change with age.
Age profiling by OSC-PLS modeling
Several factors including age, gender, diet, medications, and environment can contribute to the separation, and it can be difficult to isolate the contributions from these effects by either PCA or PLS methods. In this study, PLS was also performed using NMR spectral regression against age, but this approach did not show much improvement compared to PCA (data not shown). It is therefore important to employ approaches that retain only the effects of one factor such as age by eliminating or reducing the other components. The unique ability of OSC to extract metabolic variations from a particular effect has been proven to be effective in previous studies (41,52,53). In the present study, one component in the OSC filter was chosen to avoid over-fitting of the data. Since samples were collected from subjects with no diagnosed disease, factors other than age did not cause any obvious separation in the PCA score plot (Figure 2a). (Note that gender information was not available for all the subjects, but no obvious separation was noted for samples that could be assigned gender information). Hence, it is reasonable to assume that one component is enough to extract an age-related profile.
To build the OSC-PLS model, age in years was used as the input for the Y-matrix data. The model was constructed based on 3 latent variables (LVs), according to the root-mean-square error of cross validation (RMSECV) curve in the leave-one-out cross validation procedure (54). The values for the important parameters of the OSC-PLS analysis are shown in Table 2. Three LVs were sufficient to understand 100% of the variation in age (cumulative VY) using only 51.6% of the NMR spectral variation (cumulative VX) after one orthogonal component was removed from the X matrix by OSC preprocessing. These results clearly indicate that the OSC-processed NMR spectra are highly correlated with age and the variations caused by other effects are either removed effectively or reduced strongly.
Table 2.
Summary of the OSC-pretreated PLS model of NMR spectra regression against age
| LVs | VX (%) | VX (cuml, %) | VY(%) | VY (cuml, %) |
|---|---|---|---|---|
| 1 | 25.9 | 25.9 | 99.7 | 99.7 |
| 2 | 17.2 | 43.1 | 0.2 | 99.9 |
| 3 | 8.5 | 51.6 | 0.1 | 100 |
VX, VY: variation explained by each LV. VX(cuml), VY(cuml): cumulative variation explained.
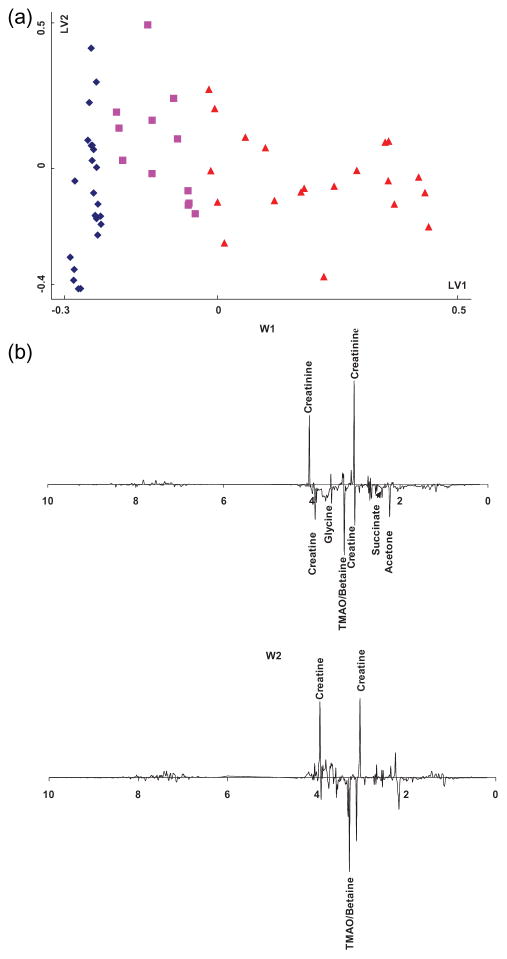
Figure 3a shows the score plot for the OSC pretreated PLS model. Compared to the score plot from PCA (Figure 2a), samples in Figure 3a were better separated without any overlap according to age along the LV1 direction. In this case, LV1 explains 25.9% of total variance in the NMR data (VX), and 99.7% in the age data (VY), as shown in Table 2. Figure 3b shows the weight plots from the OSC-PLS model, where w1 corresponds to the weight of LV1, and similarly for w2. It should be mentioned that the w1 values in the common PLS model are the same as those from the OSC pretreated PLS model (52). The age-related metabolites identified in the weight plots include creatinine, creatine, glycine, betaine/TMAO, citrate, succinate, and acetone. Several compounds with significant peak intensities are marked in the plots (shown in Figure 3b). The fold changes of some of the distinguished metabolites are shown in Table 3.
Figure 3.
(a) Score plot of OSC-pretreated PLS modeling of 1H NMR spectra from 55 normal human urine samples. Symbols indicate the same sample types as in Figure 2. (b) Weight plots of OSC-pretreated PLS modeling of 1H NMR spectra from the 55 normal human urine samples.
Table 3.
Fold change of some of the metabolites with age
| Metabolite | Fold change* | |
|---|---|---|
| 2–4 yrs | 5–12 yrs | |
| Acetone | −0.31 | −2.34 |
| Succinate | −1.33 | −1.95 |
| Betaine/TMAO | −1.49 | −1.86 |
| Creatinine | +0.32 | +2.34 |
| Creatine | +4.3 | −3.7 |
| Glycine | −0.58 | −2.50 |
| Citrate | −0.50 | −2.45 |
Fold change relative to 0–1 yr age group. Negative sign indicates decrease with age and the positive sign indicates increase with age.
The relatively large variation of hippurate with age for adults has been reported previously (6). However, in the loading plots from PCA (Figure 2b) in this study, the higher intensities of peaks from hippurate appear in the PC2 direction, which mainly carries variations caused by effects other than age. Compared to Figure 2b, the importance of hippurate in the weight plots of OSC-PLS model (Figure 3b) is further reduced. The present results indicate that hippurate concentrations may vary less with age during childhood development.
A number of other metabolites such as TMAO, citrate, glycine, acetone, and succinate also contribute to the OSC-PLS weight plots. Hippurate has been shown to be related to kidney function (55). Glycine has been implicated in many physiological phenomena as a neurotransmitter or biosynthetic intermediate (56). TMAO has been reported as an osmolyte in muscle tissues of marine organisms (57,58), and it is involved in the detoxification process in the human body (59). Betaine plays an important role in the health of the cardiovascular system and has been used widely in treating vascular, hepatic, and liver diseases (60). It has also been reported that acetone has anticonvulsant effects in animal seizure models (61). Citrate and succinate are involved in several important metabolic pathways (62).
Changes in the concentrations of at least some of the aforementioned metabolites with age have been observed earlier by others (6,63,64). For example, association of TMAO and citrate with age has recently been reported by Psihogios et al. (6) Variation of certain amino acids including glycine with age was reported by Stambaugh et al. in 1963 (63). Organic acid analyses in a Turkish pediatric population revealed that citrate and succinate decrease with age, which agrees with our observations (64). However, definitive biological roles linking these metabolites with age are yet to be established. Potentially important biological functions of these compounds may help explain the appearance of these compounds in the weight plots of the OSC-PLS model as they relate to age profiling.
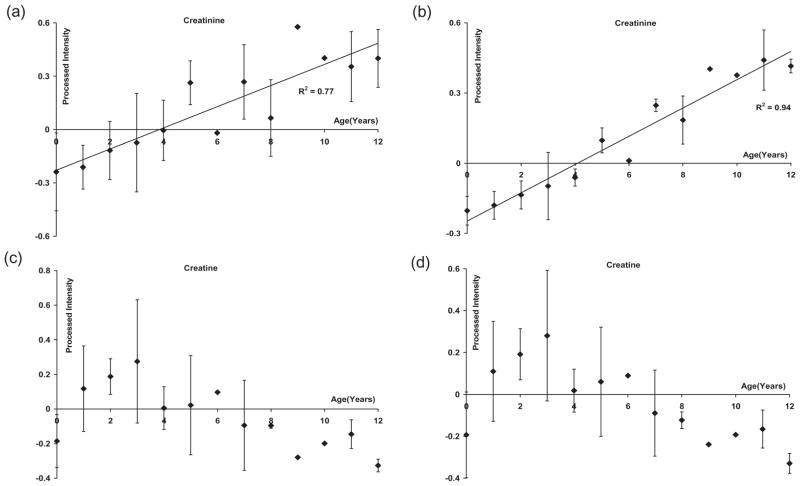
Plots of the average intensity (with the standard deviation) versus age are shown in Figure 4 for two typical prominent metabolites: creatinine and creatine. It can be clearly seen that while creatinine increases with age, creatine decreases after an initial increase. For creatinine, the trend was similar for the data before and after OSC pretreatment, although the correlation improved upon OSC pretreatment. For example, the correlation coefficient (R2) increased from 0.77 before OSC pretreatment to 0.94 after OSC pretreatment, although there were a few sizable deviations. As seen in Figure 4b, the change in intensity of the creatinine signal after the OSC filter arises exclusively from the change in age. This increase of creatinine is in accordance with the expected trend from earlier studies (46). In addition, the urinary concentration of creatinine is highly correlated with skeletal muscle mass which grows significantly during childhood (65).
Figure 4.
Plots of the average intensities and standard deviations for each age group in the 55 normal samples: (a) creatinine without OSC filter, (b) creatinine with OSC filter, (c) creatine without OSC filter, and (d) creatine with OSC filter. A linear fit was used for the data in (a) and (b).
On the other hand, the decrease in creatine with age, especially after 3 years of age, can be understood by the fact that, in the muscle tissue, creatine gets converted into creatinine (66), and this increased creatinine is often correlated with a decrease in creatine levels. For creatine, the difference between the plots (shown in Figures 4c and d) obtained from the data before and after OSC pretreatment was not appreciable, indicating that creatine concentration is less affected by factors other than age.
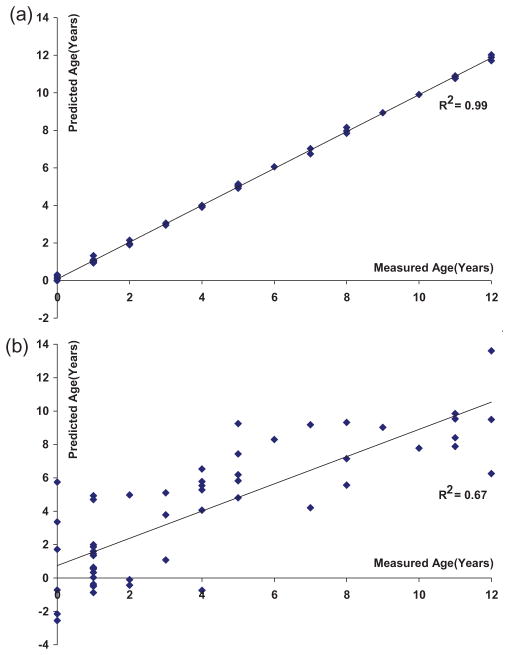
The leave-one-out cross validation procedure used for the OSC-PLS regression was also examined to evaluate the predictive ability of the model. During cross validation, one sample at a time was used as a test sample for the prediction while the rest defined the training set. In this way, each sample was used as a test sample for cross validation. Figure 5a shows the prediction plot in the cross validation using 3 LVs with the leave-one-out step performed after OSC. A very good fit was achieved (R2 =0.99; Figure 5a), indicating the robustness of the prediction model that can be achieved under ideal conditions for the present sample set. However, Figure 5b shows the prediction plot in the cross validation with the same parameters used in Figure 5a except that the leave-one-out step was performed before OSC. The prediction errors are quite large for several samples in Figure 5b, with a residual standard deviation of 2.3 years. One possibility is that these errors may be due to a difference between ‘chronological’ and ‘biological’ ages, but this hypothesis will need to be proven with further detailed studies. Nevertheless, a linear trend between the measured and predicted ages is noticeable (R2 =0.67). The effectiveness of the OSC pretreatment to retain the age-associated metabolic variations is clearly noticeable from the comparison of Figures 5a and b, however, one must be careful not to over-fit the data set. In particular, it will be important to validate these findings in additional samples.
Figure 5.
Results of cross validation in the OSC-pretreated PLS modeling of 1H NMR spectra from the 55 normal human urine samples using 3 LVs, with the leave-one-out cross validation performed: (a) after OSC, and (b) before OSC.
CONCLUSIONS
Age is an important variable that contributes to the metabolic profile of body fluids, and thus establishing that metabolic profiles linked to age have numerous applications. In addition to being able to predict the biological age of a subject based on the metabolic profile, the knowledge of an age-related metabolic profile can make the interpretation of data for other effects such as disease easier, which is important in biomarker discovery for disease diagnosis or prognosis. This study presents the metabolic variation in the pre-adolescent age group, and the data analysis using OSC-PLS regression clearly explains the variance of the samples with respect to age, indicating the potential of such an approach for obtaining a useful and reliable profile. Metabolites, such as creatinine, creatine, glycine, betaine/TMAO, citrate, succinate, and acetone were identified as key molecules with high correlation to age. Further development of the model utilizing a large number of samples is needed to substantiate these findings and to obtain more biological insight into age-related metabolic variations.
Supplementary Material
Acknowledgments
Contract/grant sponsor: NIH Roadmap Initiative on Metabolomics Technology; NIH/NIDDK 3 R21 DK070290-01.
Contract/grant: Purdue University/Discovery Park and the Indiana University School of Medicine collaborative grant.
Abbreviations used
- LVs
latent variables
- MS
mass spectrometry
- O-PLS
orthogonal projection to latent structures
- OSC-PLS
orthogonal signal correction-projection to latent
- PLS
partial least square
- RMSECV
root-mean-square error of cross validation
- TMAO
trimethylamine-N-oxide
Footnotes
Supporting information may be found in the online version of this article.
References
- 1.Jackson SHD, Weale MR, Weale RA. Biological age – what is it and can it be measured? Arch Gerontol Geriatr. 2003;36:103–115. doi: 10.1016/s0167-4943(02)00060-2. [DOI] [PubMed] [Google Scholar]
- 2.Ingram DK, Nakamura E, Smucny D, Roth GS, Lane MA. Strategy for identifying biomarkers of aging in long-lived species. Exp Gerontol. 2001;36:1025–1034. doi: 10.1016/s0531-5565(01)00110-3. [DOI] [PubMed] [Google Scholar]
- 3.Randerath K, Zhou GD, Hart RW, Turturro A, Randerath E. Biomarkers of aging-correlation of DNA I-compound levels with median lifespan of calorically restricted and ad libitum fed rats and mice. Mutat Res. 1993;295:247–263. doi: 10.1016/0921-8734(93)90024-w. [DOI] [PubMed] [Google Scholar]
- 4.Vranckx R, Savu L, Lambert N, Vermeil-De-Conchard G, Grosse R, Mourey MS, Corman B. Plasma proteins as biomarkers of the aging process. Am J Physiol Regul Integr Comp Physiol. 1995;37:536–548. doi: 10.1152/ajpregu.1995.268.2.R536. [DOI] [PubMed] [Google Scholar]
- 5.Slupsky CM, Rankin KN, Wagner J, Fu H, Chang D, Weljie AM, Saude EJ, Lix B, Adamko DJ, Shah S, Greiner R, Sykers BD, Marrie TJ. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal Chem. 2007;79:6995–7004. doi: 10.1021/ac0708588. [DOI] [PubMed] [Google Scholar]
- 6.Psihogios NG, Gazi IF, Elisaf MS, Seferiadis KI, Bairaktari ET. Gender-related and age-related urinalysis of healthy subjects by NMR-based metabonomics. NMR Biomed. 2008;21:195–207. doi: 10.1002/nbm.1176. [DOI] [PubMed] [Google Scholar]
- 7.Kochhar S, Jacobs DM, Ramadan Z, Berruex F, Fuerhoz A, Fay LB. Probing gender-specific metabolism differences in humans by nuclear magnetic resonance-based metabonomics. Anal Biochem. 2006;352:274–281. doi: 10.1016/j.ab.2006.02.033. [DOI] [PubMed] [Google Scholar]
- 8.Kristal BS, Shurubor YI, Kaddurah-Daouk R, Matson WR. Metabolomics in the study of aging and caloric restriction. Methods Mol Biol. 2007;371:393–409. doi: 10.1007/978-1-59745-361-5_25. [DOI] [PubMed] [Google Scholar]
- 9.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discovery. 2002;1:153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 10.Allen J, Davey HM, Broadhurst D, Heald JK, Rowland JJ, Oliver SG, Kell DB. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat Biotechnol. 2003;21:692–696. doi: 10.1038/nbt823. [DOI] [PubMed] [Google Scholar]
- 11.Clayton TA, Lindon JC, Cloarec O, Antti H, Charuel C, Hanton G, Provost JP, Net JL, Baker D, Walley RJ, Everett JR, Nicholson JK. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440:1073–1077. doi: 10.1038/nature04648. [DOI] [PubMed] [Google Scholar]
- 12.Odunsi K, Wollman RM, Ambrosone CB, Hutson A, McCann SE, Tammela J, Geisler JP, Miller G, Sellers T, Cliby W, Qian F, Keitz B, Intengan M, Lele S, Alderfer JL. Detection of epithelial ovarian cancer using 1H-NMR-based metabonomics. Int. J Cancer. 2005;113:782–788. doi: 10.1002/ijc.20651. [DOI] [PubMed] [Google Scholar]
- 13.Sandusky P, Raftery D. Use of selective TOCSY NMR experiments for quantifying minor components in complex mixtures: Application to the metabonomics of amino acids in honey. Anal Chem. 2005;77:2455–2463. doi: 10.1021/ac0484979. [DOI] [PubMed] [Google Scholar]
- 14.Pan Z, Gu H, Talaty N, Chen H, Shanaiah N, Hainline BE, Cooks RG, Raftery D. Principal component analysis of urine metabolites detected by NMR and DESI–MS in patients with inborn errors of metabolism. Anal Bioanal Chem. 2007;387:539–549. doi: 10.1007/s00216-006-0546-7. [DOI] [PubMed] [Google Scholar]
- 15.Beger RD, Schnackenberg LK, Holland RD, Li DH, Dragan Y. Metabonomic models of human pancreatic cancer using 1D proton NMR spectra of lipids in plasma. Metabolomics. 2006;2:125–134. [Google Scholar]
- 16.Serkova NJ, Niemann CU. Pattern recognition and biomarker validation using quantitative 1H-NMR-based metabolomics. Expert Rev. Mol. Diagn. 2006;6:717–731. doi: 10.1586/14737159.6.5.717. [DOI] [PubMed] [Google Scholar]
- 17.Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, Fearnside J, Tatoud R, Blanc V, Lindon JC, Mitchell SC, Holmes E, McCarthy MI, Scott J, Gauguier D, Nicholoson JK. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Nat Acad Sci US A. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiehn O. Metabolomics-the link between genotypes and phenotypes. Plant Mol Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 19.Gowda GAN, Zhang S, Gu H, Asiago V, Shanaiah N, Raftery D. Metabolomics-based methods for early disease diagnostics: a review. Exp Rev Mol Diagn. 2008;8:617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu H, Pan Z, Duda C, Mann D, Kissinger C, Rohde C, Raftery D. 1H NMR study of the effects of sample contamination in the metabolomic analysis of mouse urine. J. Pharm. Biomed. Anal. 2007;45:134–140. doi: 10.1016/j.jpba.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 21.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 22.Pan Z, Raftery D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal Bioanal Chem. 2007;387:525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- 23.van-der-Greef J, Smilde AK. Symbiosis of chemometrics and metabolomics: past, present, and future. J Chemom. 2005;19:376–386. [Google Scholar]
- 24.Chen H, Pan Z, Talaty N, Raftery D, Cooks RG. Combining desorption electrospray ionization mass spectrometry and nuclear magnetic resonance for differential metabolomics without sample preparation. Rapid Commun Mass Spectrom. 2006;20:1577–1584. doi: 10.1002/rcm.2474. [DOI] [PubMed] [Google Scholar]
- 25.Crockford DJ, Holmes E, Lindon JC, Plumb RS, Zirah S, Bruce SJ, Rainville P, Stumpf CL, Nicholosn JK. Statistical heterospectroscopy, an approach to the integrated analysis of NMR and UPLC-MS data sets: Application in metabonomic toxicology studies. Anal Chem. 2006;78:363–371. doi: 10.1021/ac051444m. [DOI] [PubMed] [Google Scholar]
- 26.Engelke UFH, Liebrand-Van-Sambeek MLF, De-Jong JGN, Leroy JG, Morava E, Smeitink JAM, Wevers RA. N-acetylated metabolites in urine: proton nuclear magnetic resonance spectroscopic study on patients with inborn errors of metabolism. Clin Chem. 2004;50:58–66. doi: 10.1373/clinchem.2003.020214. [DOI] [PubMed] [Google Scholar]
- 27.Zhang S, Gowda GAN, Asiago V, Shanaiah N, Barbas C, Raftery D. Quantitative NMR-based metabolic profiling in diabetic rats. Anal Biochem. 2008;383:76–84. doi: 10.1016/j.ab.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Constantinou MA, Papakonstantinou E, Benaki D, Spraul M, Shulpis K, Koupparis MA, Mikros E. Application of nuclear magnetic resonance spectroscopy combined with principal component analysis in detecting inborn errors of metabolism using blood spots: a metabonomic approach. Anal Chim Acta. 2004;511:303–312. [Google Scholar]
- 29.Assfalg M, Bertini I, Colangiuli D, Luchinat C, Shafer H, Schutz B, Spraul M. Evidence of different metabolic phenotypes in humans. Proc Nat Acad Sci US A. 2008;105:1420–1424. doi: 10.1073/pnas.0705685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phipps AN, Stewart J, Wright B, Wilson ID. Effect of diet on the urinary excretion of hippuric acid and other dietary-derived aromatics in rat. A complex interaction between diet, gut microflora and substrate specificity. Xenobiotica. 1998;28:527–537. doi: 10.1080/004982598239443. [DOI] [PubMed] [Google Scholar]
- 31.Ramadan Z, Jacobs D, Grigorov M, Kochhar S. Metabolic profiling using principal component analysis, discriminant partial least squares, and genetic algorithms. Talanta. 2006;68:1683–1691. doi: 10.1016/j.talanta.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 32.Remer T, Boye KR, Hartmann MF, Wudy SA. Urinary markers of adrenarche: reference values in healthy subjects, aged 3–18 years. J Clin Endocrinol Metab. 2005;90:2015–2021. doi: 10.1210/jc.2004-1571. [DOI] [PubMed] [Google Scholar]
- 33.Valongo C, Cardoso ML, Domingues P, Almeida L, Verhoeven N, Salomons G, Jakobs C, Vilarinho L. Age related reference values for urine creatine and guanidinoacetic acid concentration in children and adolescents by gas chromatography-mass spectrometry. Clin Chim Acta. 2004;348:155–161. doi: 10.1016/j.cccn.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 34.Stella C, Beckwith-Hall B, Cloarec O, Holmes E, Lindon JC, Powell J, van-der-Ouderaa F, Bingham S, Cross AJ, Nicholson JK. Susceptibility of human metabolic phenotypes to dietary modulation. J Proteome Res. 2006;5:2780–2788. doi: 10.1021/pr060265y. [DOI] [PubMed] [Google Scholar]
- 35.Lindon JC, Holmes E, Nicholson JK. Pattern recognition methods and applications in biomedical magnetic resonance. Prog Nucl Magn Reson Spectrosc. 2001;39:1–40. [Google Scholar]
- 36.Williams RE, Lenz EM, Lowden JS, Rantalainen M, Wilson ID. The metabonomics of aging and development in the rat: an investigation into the effect of age on the profile of endogenous metabolites in the urine of male rats using 1H NMR and HPLC-TOF MS. Mol. Biosyst. 2005;1:166–175. doi: 10.1039/b500852b. [DOI] [PubMed] [Google Scholar]
- 37.Williams RE, Lenz EM, Rantalainen M, Willson ID. The comparative metabonomics of age-related changes in the urinary composition of male Wistar-derived and Zucker (fa/fa) obese rats. Mol Biosyst. 2006;2:193–202. doi: 10.1039/b517195d. [DOI] [PubMed] [Google Scholar]
- 38.Bell JD, Sadler PJ, Morris VC, Levander OA. Effect of aging and diet on proton NMR spectra of rat urine. Magn Reson Med. 1991;17:414–422. doi: 10.1002/mrm.1910170213. [DOI] [PubMed] [Google Scholar]
- 39.Wold S, Antti H, Lindgren F, Ohman J. Orthogonal signal correction of near-infrared spectra. Chemom Intell Lab Syst. 1998;44:175–185. [Google Scholar]
- 40.Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS) J Chemom. 2002;16:119–128. [Google Scholar]
- 41.Gavaghan CL, Wilson ID, Nicholson JK. Physiological variation in metabolic phenotyping and functional genomic studies: use of orthogonal signal correction and PLS-DA. FEBS Lett. 2002;530:191–196. doi: 10.1016/s0014-5793(02)03476-2. [DOI] [PubMed] [Google Scholar]
- 42.Rantalainen M, Cloarec O, Beckonert O, Wilson ID, Jackson D, Tonge R, Rowlinson R, Rayner S, Nickson J, Wilkinson RW, Mills JD, Trygg J, Nicholson JK, Holmes E. Statistically integrated metabonomic-proteomic studies on a human prostate cancer xenograft model in mice. J Proteome Res. 2006;5:2642–2655. doi: 10.1021/pr060124w. [DOI] [PubMed] [Google Scholar]
- 43.Mao HL, Xu M, Wang B, Wang HM, Deng XM, Lin DH. Evaluation of filtering effects of orthogonal signal correction on metabonomic analysis of healthy human serum 1H NMR spectra. Acta Chim Sinica. 2007;65:152–158. [Google Scholar]
- 44.Plumb RS, Granger JH, Stumpf CL, Johnson KA, Smith BW, Gaulitz S, Wilson ID, Castro-Perez J. A rapid screening approach to metabonomics using UPLC and oa-TOF mass spectrometry: application to age, gender and diurnal variation in normal/Zucker obese rats and black, white and nude mice. Analyst. 2005;130:844–849. doi: 10.1039/b501767j. [DOI] [PubMed] [Google Scholar]
- 45.Moser VC, Padilla S, Hunter DL, Marshall RS, McDaniel KL, Phillips PM. Age- and gender-related differences in the time course of behavioral and biochemical effects produced by oral chlorpyrifos in rats. Toxicol Appl Pharmacol. 1998;149:107–119. doi: 10.1006/taap.1997.8354. [DOI] [PubMed] [Google Scholar]
- 46.Barr DB, Wilder LC, Caudill SP, Gonzalez AJ, Needham LL, Pirkle JL. Urinary creatinine concentrations in the US population: Implications for urinary biologic monitoring measurements. Environ Health Perspect. 2005;113:192–200. doi: 10.1289/ehp.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan WMT. Metabolite profiling by one- and two-dimensional NMR analysis of complex mixtures. Prog Nucl Magn Reson Spectrosc. 1996;28:161–219. [Google Scholar]
- 48.Constantinou MA, Papakonstantinou E, Spraul M, Sevastiadou S, Costalos C, Koupparis MA, Shulpis K, Tsantili-Kakoulidou A, Mikros E. 1H NMR-based metabonomics for the diagnosis of inborn errors of metabolism in urine. Anal. Chim Acta. 2005;542:169–177. [Google Scholar]
- 49.Nicholls AW, Mortishire-Smith RJ, Nicholson JK. NMR spectroscopic-based metabonomic studies of urinary metabolite variation in acclimatizing germ-free rats. Chem Res Toxicol. 2003;16:1395–1404. doi: 10.1021/tx0340293. [DOI] [PubMed] [Google Scholar]
- 50.Griffin JL, Walker LA, Garrod S, Holmes E, Shore RF, Nicholson JK. NMR spectroscopy based metabonomic studies on the comparative biochemistry of the kidney and urine of the bank vole (Clethrionomys glareolus), wood mouse (Apodemus sylvaticus), white toothed shrew (Crocidura suaveolens) and the laboratory rat. Comp Biochem Physiol B, Biochem Mol Biol. 2000;127:357–367. doi: 10.1016/s0305-0491(00)00276-5. [DOI] [PubMed] [Google Scholar]
- 51.Feng JH, Li XJ, Pei FK, Chen X, Li SL, Nie YX. 1H NMR analysis for metabolites in serum and urine from rats administrated chronically with La(NO3)3. Anal. Biochem. 2002;301:1–7. doi: 10.1006/abio.2001.5471. [DOI] [PubMed] [Google Scholar]
- 52.Hauksson JB, Edlund U, Trygg J. NMR processing techniques based on multivariate data analysis and orthogonal signal correction13C CP/MAS NMR spectroscopic characterization of softwood kraft pulp. Magn Reson Chem. 2001;39:267–275. [Google Scholar]
- 53.Eriksson L, Antti H, Gottfries J, Holmes E, Johansson E, Lindgren F, Long I, Lundstedt T, Trygg J, Wold S. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (gpm) Anal Bioanal Chem. 2004;380:419–429. doi: 10.1007/s00216-004-2783-y. [DOI] [PubMed] [Google Scholar]
- 54.Westerhuis JA, Hoefsloot HCJ, Smit S, Vis DJ, Smilde AK, van-Velzen EJJ, van-Duijnhoven JPM, van-Dorsten FA. Assessment of PLSDA cross validation. Metabolomics. 2008;4:81–89. [Google Scholar]
- 55.Aoyama H, Kamiyama Y, Ukikusa M, Ozawa K. Clinical significance of hippurate synthesizing capacity in surgical patients with liver disease: a metabolic tolerance test. J Lab Clin Med. 1986;108:456–460. [PubMed] [Google Scholar]
- 56.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. W.H. Freeman and Company; New York: 2005. [Google Scholar]
- 57.Dos Santos JP, Iobbi-Nivol C, Couillault C, Giordano G, Mejean V. Molecular analysis of the trimethylamine N-oxide (TMAO) reductase respiratory system from a Shewanella species. J Mol Biol. 1998;284:421–433. doi: 10.1006/jmbi.1998.2155. [DOI] [PubMed] [Google Scholar]
- 58.Athawale MV, Dordick JS, Garde S. Osmolyte trimethylamine-N-oxide does not affect the strength of hydrophobic interactions: origin of osmolyte compatibility. Biophys J. 2005;89:858–866. doi: 10.1529/biophysj.104.056671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brittain T. Functional protection of human haemoglobin against protein dissociation. IUBMB Life. 2000;50:131–134. doi: 10.1080/713803694. [DOI] [PubMed] [Google Scholar]
- 60.Craig SAS. Betaine in human nutrition. Am J Clin Nutr. 2004;80:539–549. doi: 10.1093/ajcn/80.3.539. [DOI] [PubMed] [Google Scholar]
- 61.Likhodii SS, Serbanescu I, Cortez MA, Murphy P, Snead OC, Burnham WM. Anticonvulsant properties of acetone, a brain ketone elevated by the ketogenic diet. Ann Neurology. 2003;54:219–226. doi: 10.1002/ana.10634. [DOI] [PubMed] [Google Scholar]
- 62.Berg MJ, Tymoczko LJ, Stryer L. Biochemistry. W.H. Freeman and Company; New York: 2001. [Google Scholar]
- 63.Stambaugh R, Davidson DT, Jr, Elkinston JR. Variation of excretion of certain amino acids with age. Clin Chem. 1963;9(2):210–216. [PubMed] [Google Scholar]
- 64.Guneral F, Bachmann C. Age-related reference values for urinary organic acids in a healthy Turkish pediatric population. Clin Chem. 1994;40(6):862–868. [PubMed] [Google Scholar]
- 65.Heymsfield SB, Lohman TG, Wang Z, Going SB. Human Body Composition. Human Kinetics Publishers; Champaign, Illinois: 2005. [Google Scholar]
- 66.Greenhaff PL. The nutritional biochemistry of creatine. J Nutr Biochem. 1997;8:610–618. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.