There is tremendous interest in the use of bioorthogonal reactions for imaging of unnatural building blocks that are metabolically incorporated into biosynthetic pathways. Applications include visualizing glycans, imaging proteins tagged with unnatural amino acids, monitoring cellular proliferation, and tracking lipid analogues.[1] For live-cell imaging applications, the azide–cyclooctyne catalyst-free cycloaddition has been extremely useful in providing a rapid and biocompatible method to label small azide tags with fluorophore.[1a] Recently, there has been growing interest in exploring bioorthogonal cycloadditions involving tetrazines for live-cell imaging applications.[2] Tetrazines have been shown to react rapidly through inverse-electron-demand Diels–Alder reactions with a variety of strained alkenes and alkynes including trans-cyclooctenes, norbornenes, and cyclooctynes. These reactions can be used for live-cell imaging, and tetrazines can quench the fluorescence of commonly used imaging probes such as BODIPY dyes and fluoresceins.[3] This leads to a fluorogenic response after reaction, which can improve signal-to-background, which is particularly useful for intracellular live-cell imaging applications.[1b,4] Though tetrazine cycloadditions would be exciting developments for a wide array of metabolic imaging applications, the large size of both the tetrazine and cycloalkene coupling partners has limited their ability to be incorporated into small bio-active molecules. In response to this challenge, we recently developed small and stable methylcyclopropenes as coupling partners for fluorogenic tetrazines.[5] The molecular weight of these tags rivaled those of azides and were used to fluorogenically image lipids in live mammalian cells. However, we were interested in whether methylcyclopropenes could substitute for azides in metabolic imaging applications with stringent steric constraints. Here we demonstrate that unnatural cyclopropene-mannosamine derivatives can be used to image glycans on live human cancer cell lines.
Tetrazine-based cycloadditions are an emerging class of bioorthogonal reactions that can proceed with rapid rate constants, enable imaging of dienophile tags in live-cells and animals, and be mutually orthogonal to azide–alkyne cycloadditions.[6] Despite these applications, the use of tetrazine cycloadditions has been limited in metabolic imaging. This is due to the size of tetrazines and paired dienophiles such as trans-cyclooctene and norbornene, which are large compared to the azide and alkyne tags commonly used in bioorthogonal chemistry. We recently developed methylcyclopropenes as tetrazine-reactive cellular imaging tags that are comparable to azides in terms of molecular weight (Scheme 1A).[5] Following our work, others have shown that cyclopropene-containing amino acids and cyclopropene-derivatized neuraminic acid analogues can be incorporated into cellular macromolecules and later tagged using photoinitated reactions or tetrazine-biotin–avidin coupling.[7] However, although the experiments demonstrated cellular uptake of the chosen analogues, these systems are well known to tolerate larger substituents.[8] For instance, past work has demonstrated that trans-cyclooctene-containing unnatural amino acids and aryl-azide-containing neuraminic acid analogues can be taken up by cells.[4a,9] Thus, in those studies, the cyclopropene would not be a necessary dienophile for inverse-electron-demand Diels–Alder chemistry.
Scheme 1.
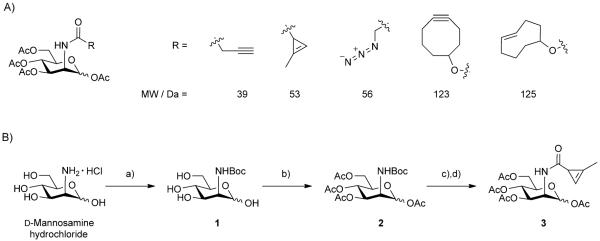
A) Comparison of N-acyl substituents on unnatural mannosamine derivatives. The cyclopropene handle is similar in size to the commonly used azide handle. B) Synthesis of peracetylated Ac4ManNCyc (3). a) 1.0 eq. NaOH, dioxane, sat. NaHCO3, Boc2O; b) pyridine, Ac2O, 20% (2 steps); c) 20% TFA, CH2Cl2; d) CH2Cl2, Et3N, DMAP, 2-methyl-2-cyclopropenyl-1-carbonyl chloride, 60% (2 steps).
Seeking to provide a more stringent test, we attempted to metabolically label human cancer cell lines with an unnatural cyclopropene-mannosamine derivative. Mannosamine analogues are classic bioorthogonal metabolic labeling agents, as pioneered by the Bertozzi group.[1a,10] It has been well demonstrated that the sialic acid biosynthetic pathway is tolerant of unnatural mannosamines bearing small unnatural N-acyl substituents.[11] Mannosamines bearing short azide-modified acyl chains (ManNAz) are readily accepted in the pathway and have been elegantly labeled by fluorescent cyclooctynes and Staudinger ligation probes after incorporation into glycans.[1a,10a] Several studies have established that lengthy or branched substituents are not well tolerated and extending the N-acyl substituent by more than five carbon atoms severely decreases uptake.[11b,12] This work has demonstrated that cellular metabolism is likely limited by phosphorylation of the unnatural analogues by ManNAc 6-kinase.[12] Thus, large norbornene- and trans-cyclooctene-N-acylmannosamine derivatives are not expected to be taken up by the biosynthetic machinery. Based on this prior work, we decided to test if cyclopropene-mannosamine analogues could be incorporated by live human cancer cells. Successful incorporation would provide evidence that methylcyclopropene tags can substitute for azides in stringent metabolic imaging or engineering applications.
We synthesized unnatural peracetylated mannosamine analogue bearing an N-acyl cyclopropene (Ac4ManNCyc, 3) by coupling peracetylated mannosamine to a highly reactive methylcyclopropene acid chloride that we previously described (Scheme 1 B).[5] Initially, we approached the synthesis of 3 by reacting unprotected mannosamine with highly reactive electrophilic methylcyclopropene precursors. This yielded multiple products and was not practical for isolating the desired compound. Instead, we first protected the amine functional group of mannosamine (to afford intermediate 1) followed by acetylation of the remaining alcohols, using acetic anhydride, and chromatographic separation to afford compound 2. Deprotection of the amine using trifluoroacetic acid yielded peracetylated mannosamine 3, which readily reacted with a methylcyclopropene acid chloride to yield the desired peracetyled cyclopropene-sugar 3. Alternatively, we could also react the primary amine with a methylcyclopropene-NHS derivative followed by peracetylation (see the Supporting information). We chose to utilize the peracetylated derivatives given the well-known ability of such lipophilic precursors to enter cells and accumulate in glycans, dramatically lowering the concentration of probe the cells are required to be exposed to.[13]
Methylcyclopropene-amides are known to react slower with tetrazine handles compared to faster methylcyclopropenyl carbamates.[5] However, the reported reaction rates for the methylcyclopropene-amides are comparable to alternative bioorthogonal reactions used for live-cell imaging. Additionally, the molecular weight of the methylcyclopropene-amide handle is similar to the azide handles that have been previously used for imaging glycans, making it an attractive derivative (Scheme 1 A).[1a] We and others have demonstrated that one of the benefits of tetrazine-based cycloadditions is the ability to utilize fluorogenic probes that increase in emission intensity after cycloaddition.[3b] Indeed, reaction of 3 elicits fluorogenic responses from quenched tetrazine probes such as tetrazine-Alexa Fluor 488 (Figure S1 A). Such fluorogenic reactions are valuable for live-cell imaging and can improve the signal to background by diminishing signal from nonspecifically bound or trapped fluorescent probes.
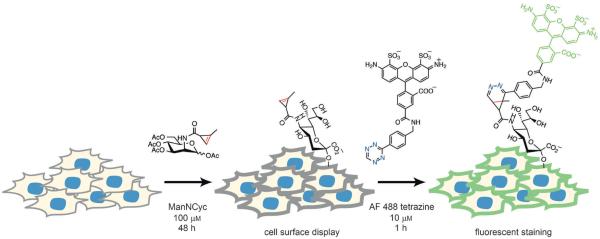
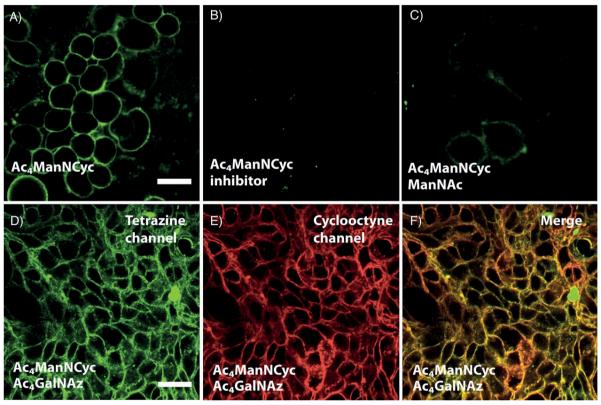
In order to determine if Ac4ManNCyc was incorporated effectively by cells, we incubated adherent human breast cancer (SKBR3) and colon cancer (LS174T) cell lines with 3 (100 μm) for 48 h in cell medium and serum. After incubation, cells were thoroughly washed and then reacted for one hour with fluorogenic Alexa Fluor 488 tetrazine (10 μm; Scheme 2). After a second wash, cells were imaged by confocal microscopy (Olympus FV1000). Cells that were exposed to 3 showed bright surface staining (Figure 1A) while control cells that were not treated with unnatural sugar had a complete absence of staining (Figure S2). In order to ensure that the staining was due to glycan uptake of the probe, we also performed controls by exposing cells to 3 (100 μm) and inhibiting uptake by using glycosylation inhibitors (tunicamycin or αBnGalNAc) or ManNAc (20 mm) as a competitive substrate.[14] Inhibitors severely diminished staining to levels that were similar to controls (Figure 1 B). The use of ManNAc as competitor also lowered the fluorescent signal, with faint surface staining visible (Figure 1 C). These experiments provide evidence that the staining patterns are reporting on glycosylation and uptake of the unnatural cyclopropene-mannosamine. We also performed additional studies using flow cytometry to quantitate the relative uptake of sugars, which corroborated the imaging data (Figure S1 B). Cells exposed to 3 followed by the tetrazine imaging probe showed increased fluorescence intensity compared to controls which only received the tetrazine imaging probe.
Scheme 2.
Cartoon outlining proposed fluorescent staining of glycans using tetrazine–cyclopropene chemistry. Ac4ManNCyc (3) is incubated with live cells for 48 h. If the unnatural mannosamine derivative is processed by the cell, methylcyclopropenes will be displayed on the surface and tagged by fluorogenic tetrazine-AlexaFluor 488. This staining pattern will be visible by confocal microscopy.
Figure 1.
Imaging glycans using cyclopropene-mannosamine derivatives. A) SKBR3 cell surfaces were stained by incubation with Ac4ManNCyc (3; 100 μm) followed by reaction with tetrazine-Alexa Fluor 488 (10 μm). B) Staining was significantly lower when SKBR3 cells were incubated with 3 as well as tunicamycin (1.2 μm), a glycosylation inhibitor. C) Staining was also reduced when SKBR3 cells were incubated with 3 as well as of ManNAc (20 mm) as a competitor. Simultaneous imaging of 3 and Ac4GalNAz incorporation. LS174T cells were incubated with of 3 (100 μm) and Ac4GalNAz (100 μm) for 48 h. After incubation, cells were reacted sequentially with tetrazine-Alexa Fluor 488 (10 μm) and DIBO 647 (15 μm). Confocal microscopy revealed extensive surface staining in the D) 488 nm channel and E) 647 nm channel with good colocalization of surface staining (F). Controls revealed minimal cross staining of tetrazine-Alexa Fluor 488 with cells incubated only with Ac4GalNAz and DIBO 647 with cells incubated only with 3 (see the Supporting information). Scale bars: 20 μm.
Finally, we tested whether or not we could simultaneously image two different metabolically incorporated unnatural sugar derivatives, 3 and peracetylated N-azidoacetylgalactosamine (Ac4GalNAz). The latter unnatural galactosamine azide has been shown to incorporate into O-linked mucins and can be tagged using commercially available fluorescent cyclooctynes.[15,16] Recent experimental work has shown that alkenes and azides can be mutually orthogonal to each other, even when using highly strained trans-cyclooctenes.[6f] Additionally, the Houk group has calculated that methylcyclopropene handles, such as those we have previously developed, should be mutually orthogonal to azide in reactions with cyclooctynes.[17] We incubated LS174T human colon cancer cells with 100 μm 3 and Ac4GalNAz for two days. After incubation, we sequentially stained the cells with tetrazine-AlexaFluor 488 (10 μm) and dibenzocyclooctyne (DIBO) 647 (15 μm). Fluorescent microscopy demonstrated cell-surface staining in both the fluorescent channels. Similar results were obtained with the SKBR3 cell line. Cells that were incubated with only 3 and DIBO 647 or 3 and tetrazine-Alexa Fluor 488 showed minimal surface staining (Figure S3). The ability to simultaneously label two different metabolically incorporated molecules using live-cell compatible inverse-electron-demand Diels–Alder and Huisgen reactions expands the capabilities of bioorthogonal metabolic imaging.
In conclusion, we have demonstrated that recently developed methylcyclopropene bioorthogonal handles can be used for metabolic imaging of unnatural mannosamine derivatives on live-cell surfaces.[9] The ability of methylcyclopropenes to substitute for azides should significantly expand the use of bioorthogonal reactions for metabolic imaging applications. This technology also enables multicolor imaging of two different metabolically incorporated mini-tags. These results highlight the potential utility of cyclopropenes as reactive mini-tags for a myriad of applications in the profiling and imaging of small molecules.
Supplementary Material
Acknowledgements
The authors acknowledge helpful discussions with Prof. Simpson Joseph. This work was funded in part by NIH grant K01EB010078, the University of California, San Diego, by a SDCSB Seed grant, by the NIH Center of Excellence grant P50 GM085763, and NIH grant P30 NS047101 supported by Jennifer Santini.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.201200719.
References
- [1].a) Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Proc. Natl. Acad. Sci. USA. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lang K, Davis L, Torres-Kolbus J, Chou C, Deiters A, Chin J. Nat. Chem. 2012;4:298–304. doi: 10.1038/nchem.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ngo J, Tirrell D. Acc. Chem. Res. 2011;44:677–685. doi: 10.1021/ar200144y. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Rangan K, Yang Y, Charron G, Hang H. J. Am. Chem. Soc. 2010;132:10628–10629. doi: 10.1021/ja101387b. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Salic A, Mitchison T. Proc. Natl. Acad. Sci. USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Devaraj N, Weissleder R. Acc. Chem. Res. 2011;44:816–827. doi: 10.1021/ar200037t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Devaraj N, Upadhyay R, Haun J, Hilderbrand S, Weissleder R. Angew. Chem. 2009;121:7147–7150. doi: 10.1002/anie.200903233. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2009;48:7013–7016. doi: 10.1002/anie.200903233. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Devaraj N, Hilderbrand S, Upadhyay R, Mazitschek R, Weissleder R. Angew. Chem. 2010;122:2931–2934. doi: 10.1002/anie.200906120. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2010;49:2869–2872. doi: 10.1002/anie.200906120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Plass T, Milles S, Koehler C, Szymanski J, Mueller R, Wiessler M, Schultz C, Lemke E. Angew. Chem. 2012;124:4242–4246. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2012;51:4166–4170. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]; b) Budin G, Yang K, Reiner T, Weissleder R. Angew. Chem. 2011;123:9550–9553. doi: 10.1002/anie.201103273. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2011;50:9378–9381. doi: 10.1002/anie.201103273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yang J, Šečkutė J, Cole C, Devaraj N. Angew. Chem. 2012;124:7594–7597. doi: 10.1002/anie.201202122. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2012;51:7476–7479. doi: 10.1002/anie.201202122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].a) Blackman M, Royzen M, Fox J. J. Am. Chem. Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Devaraj N, Weissleder R, Hilderbrand S. Bioconjugate Chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wiessler M, Waldeck W, Kliem C, Pipkorn R, Braun K. Int. J. Med. Sci. 2010;7:19–28. doi: 10.7150/ijms.7.19. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Devaraj N, Thurber G, Keliher E, Marinelli B, Weissleder R. Proc. Natl. Acad. Sci. USA. 2012;109:4762–4767. doi: 10.1073/pnas.1113466109. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Rossin R, Verkerk P, van den Bosch S, Vulders R, Verel I, Lub J, Robillard M. Angew. Chem. 2010;122:3447–3450. doi: 10.1002/anie.200906294. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2010;49:3375–3378. doi: 10.1002/anie.200906294. [DOI] [PubMed] [Google Scholar]; f) Karver M, Weissleder R, Hilderbrand S. Angew. Chem. 2012;124:944–946. doi: 10.1002/anie.201104389. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2012;51:920–922. doi: 10.1002/anie.201104389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Yu Z, Pan Y, Wang Z, Wang J, Lin Q. Angew. Chem. 2012;124:10752–10756. [Google Scholar]; Angew. Chem. Int. Ed. 2012;51:10600–10604. doi: 10.1002/anie.201205352. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Patterson D, Nazarova L, Xie B, Kamber D, Prescher J. J. Am. Chem. Soc. 2012;134:18638–18643. doi: 10.1021/ja3060436. [DOI] [PubMed] [Google Scholar]
- [8].a) Luchansky S, Goon S, Bertozzi C. ChemBioChem. 2004;5:371–374. doi: 10.1002/cbic.200300789. [DOI] [PubMed] [Google Scholar]; b) Oetke C, Brossmer R, Mantey L, Hinderlich S, Isecke R, Reutter W, Keppler O, Pawlita M. J. Biol. Chem. 2002;277:6688–6695. doi: 10.1074/jbc.M109973200. [DOI] [PubMed] [Google Scholar]
- [9].Han S, Collins B, Bengtson P. J. Paulson, Nat. Chem. Biol. 2005;1:93–97. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- [10].a) Saxon E, Bertozzi C. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]; b) Sletten E, Bertozzi C. Acc. Chem. Res. 2011;44:666–676. doi: 10.1021/ar200148z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Kayser H, Zeitler R, Kannicht C, Grunow D, Nuck R, Reutter W. J. Biol. Chem. 1992;267:16934–16938. [PubMed] [Google Scholar]; b) Keppler O, Horstkorte R, Pawlita M, Schmidt C, Reutter W. Glycobiology. 2001;11:11R–18R. doi: 10.1093/glycob/11.2.11r. [DOI] [PubMed] [Google Scholar]
- [12].Jacobs C, Goon S, Yarema K, Hinderlich S, Hang H, Chai D, Bertozzi C. Biochemistry. 2001;40:12864–12874. doi: 10.1021/bi010862s. [DOI] [PubMed] [Google Scholar]
- [13].Jacobs C, Yarema K, Mahal L, Nauman D, Charters N, Bertozzi C. Methods Enzymol. 2000;327:260–275. doi: 10.1016/s0076-6879(00)27282-0. [DOI] [PubMed] [Google Scholar]
- [14].Saxon E, Luchansky S, Hang H, Yu C, Lee S, Bertozzi C. J. Am. Chem. Soc. 2002;124:14893–14902. doi: 10.1021/ja027748x. [DOI] [PubMed] [Google Scholar]
- [15].Hang H, Yu C, Kato D, Bertozzi C. Proc. Natl. Acad. Sci. USA. 2003;100:14846–14851. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ning X, Guo J, Wolfert MA, Boons G-J. Angew. Chem. 2008;120:2285–2287. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2008;47:2253–2255. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liang Y, Mackey J, Lopez S, Liu F, Houk K. J. Am. Chem. Soc. 2012;134:17904–17907. doi: 10.1021/ja309241e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.