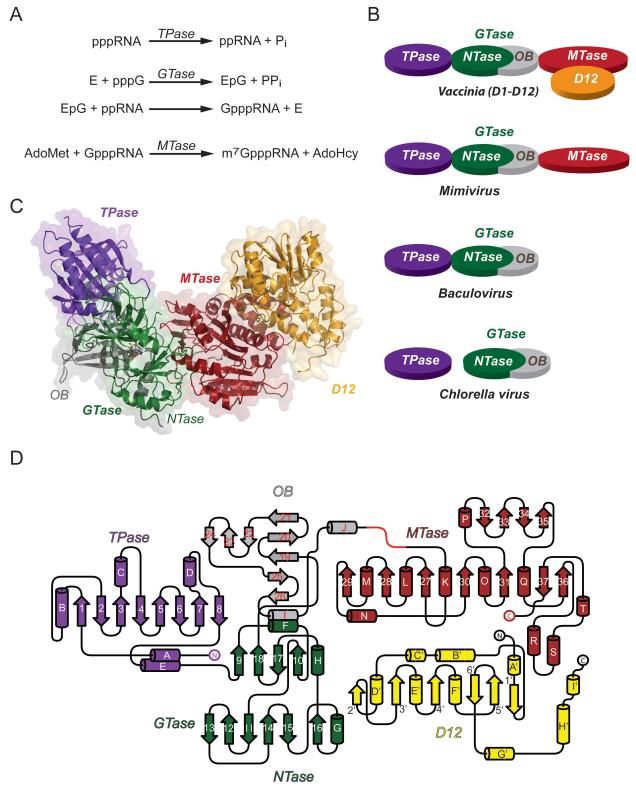
Summary
Vaccinia virus capping enzyme is a heterodimer of D1 (844-aa) and D12 (287-aa) polypeptides that executes all three steps in m7GpppRNA synthesis. The D1 subunit comprises an N-terminal RNA triphosphatase (TPase)–guanylyltransferase (GTase) module and a C-terminal guanine-N7-methyltransferase (MTase) module. The D12 subunit binds and allosterically stimulates the MTase module. Crystal structures of the complete D1•D12 heterodimer disclose the TPase and GTase as members of the triphosphate tunnel metalloenzyme and covalent nucleotidyltransferase superfamilies, respectively, albeit with distinctive active site features. An extensive TPase-GTase interface clamps the GTase nucleotidyltransferase and OB domains in a closed conformation around GTP. Mutagenesis confirms the importance of the TPase-GTase interface for GTase activity. The D1•D12 structure complements and rationalizes four decades of biochemical studies of this enzyme (the first capping enzyme to be purified and characterized) and provides new insights to the origins of the capping systems of other large DNA viruses.
Introduction
The 5′ cap is a distinctive feature of eukaryal cellular and viral messenger RNA that is required for mRNA stability and efficient translation. The cap consists of 7-methylguanosine linked via an inverted 5′-5′ triphosphate bridge to the initiating nucleoside of the transcript. The cap is formed by enzymatic modification of nascent pre-mRNAs synthesized by cellular or viral RNA polymerases. Capping entails three enzymatic reactions (Figure 1A). The 5′ triphosphate end of the pre-mRNA is first hydrolyzed to a diphosphate by RNA 5′ triphosphatase (TPase). The diphosphate RNA is then capped with GMP by RNA guanylyltransferase (GTase) via a two-step mechanism in which: (i) GTase reacts with GTP to form a covalent enzyme-(lysyl-Nζ)-GMP intermediate and PPi and (ii) GMP is transferred from GTase to the ppRNA end to form GpppRNA. Finally, the GpppRNA cap is converted to m7GpppRNA by AdoMet:RNA(guanine-N7)-methyltransferase (MTase). This pathway was elucidated via the analysis of the poxvirus capping enzyme purified from infectious vaccinia virions (Martin and Moss, 1975, 1976; Martin et al., 1975; Shuman and Hurwitz, 1981; Shuman et al., 1980; Venkatesan et al., 1980). The same pathway is conserved in all eukarya, several large DNA viruses, and reovirus (a dsRNA virus), though many other RNA viruses use alternative strategies to cap their RNAs (Decroly et al., 2012).
Figure 1. Overall structure and topology.
(A) The three reactions catalyzed by respectively the TPase, GTase and MTase to generate the cap 0 structure.
(B) Schematic diagram showing varying architecture of viral capping enzymes.
(C) Overall ribbon representation of the full-length capping enzyme showing separate domains. The TPase is colored in purple, the NTase in green, the OB-fold in grey, the MTase in red and the D12 subunit in yellow, as in (B). The bound SAH and GTP molecules are shown as sticks.
(D) Schematic topology diagram of the full-length capping enzyme with secondary structure elements labeled. The domains are colored as in (C).
Whereas the three capping reactions are universal in eukarya and DNA viruses, there is a remarkable diversity in the genetic and physical organization of the cap-forming enzymes in different taxa and in different viruses, ranging from separately encoded TPase, GTase, and MTase enzymes that have no physical interactions with each other (e.g., in fission yeast), to separately encoded enzymes that form complexes in trans (e.g. budding yeast), to fusions of two of the capping enzymes within a single polypeptide (metazoa, plants, baculovirus), to fusion of all three enzymes in a single polypeptide (poxviruses). There is also a complete divergence in the structure and catalytic mechanism of the TPase enzymes between taxa. The TPases fall into two classes: the metal-dependent RNA triphosphatases found in fungi, protozoa, and large DNA viruses (poxviruses, baculoviruses, Chlorella virus and mimivirus) versus the metal-independent cysteine-phosphatase-type RNA triphosphatases found in metazoans and plants (Shuman, 2002). In principle, these differences can be exploited to develop novel anti-infective agents that inhibit pathogen capping enzymes and they also provide instructive clues to eukaryal phylogeny and evolution of large DNA viruses (Shuman, 2002).
Vaccinia virus capping enzyme was the first capping enzyme to be purified and characterized (Martin et al., 1975). It is a heterodimer of 844-aa and 287-aa polypeptides, encoded by the D1 and D12 genes, respectively, that efficiently executes all three steps in m7GpppRNA synthesis. Consequently, vaccinia capping enzyme has been used widely as a reagent for capping and cap-labeling RNAs in vitro. The physical organization of vaccinia capping enzyme is unique (Figure 1B). D1-(1-545) constitutes an autonomous monomeric triphosphatase–guanylyltransferase domain (TPase-GTase) (Myette and Niles, 1996a, b; Yu and Shuman, 1996). The MTase domain consists of D1-(548-844) heterodimerized with the D12 subunit (Cong and Shuman, 1992; De la Pena et al., 2007; Higman et al., 1992; Higman et al., 1994; Mao and Shuman, 1994). The MTase active site is located entirely within D1-(548-844), which has a weak intrinsic methyltransferase activity that is stimulated allosterically by D12 (Higman et al., 1994; Mao and Shuman, 1994; Schwer et al., 2006).
The capping apparatus is organized differently in other large DNA viruses. Mimivirus, a parasite of amoeba, encodes an 1170-aa trifunctional capping enzyme (MimiCE) that resembles vaccinia D1, but it does not encode a homolog of vaccinia D12 (Figure 1B). Deletion analysis identified MimiCE-(1-668) as an autonomous TPase-GTase unit [analogous to vaccinia D1-(1-545)] and MimiCE-(1-237) as a minimal autonomous TPase domain (Benarroch et al., 2008). Baculoviruses, which replicate in arthropod hosts, encode a bifunctional TPase-GTase capping enzyme analogous to vaccinia D1-(1-545) (Gross and Shuman, 1998) (Figure 1B), but do not encode a cap guanine-N7 methyltransferase. The N-terminal 236-aa segment of baculovirus capping enzyme comprises an autonomous TPase catalytic domain (Martins and Shuman, 2001). Chlorella virus PBCV-1 encodes separate TPase and GTase enzymes (Häkansson et al., 1997; Gong and Shuman, 2002) (Figure 1B), but does not appear to encode a cap guanine-N7 methyltransferase. Physical separation of the mimivirus, baculovirus, and Chlorella virus TPase and GTase domains contrasts with the inability to isolate stand-alone TPase and GTase modules derived from vaccinia D1-(1-545) (Myette and Niles, 1996b; Yu et al., 1997).
The domains and three active sites of vaccinia capping enzyme have been charted in detail by mutagenesis with in vitro and in vivo readouts of TPase, GTase and MTase activities (Cong and Shuman, 1995; Gong and Shuman, 2003; Ho et al., 2000; Mao and Shuman, 1994, 1996; Saha and Shuman, 2001; Saha et al., 2003; Schwer et al., 2006; Yu et al., 1997; Yu and Shuman, 1996; Zheng and Shuman, 2008a, b). Analyses of the vaccinia MTase were guided initially by crystal structures of the homologous eukaryal cellular cap methyltransferase (Fabrega et al., 2004) and later by the crystal structure of the vaccinia D1(MTase)•D12 heterodimer (De la Pena et al., 2007). The combined collections of cellular and poxvirus cap MTase structures with bound substrates and products conferred great power in interpreting the extensive functional data, yielding important insights into: (i) the basis for cap methyl acceptor specificity; (ii) the mechanism of catalysis of methyl transfer; (iii) a unique methyl donor conformation and AdoMet binding pocket in the poxvirus MTase versus the cellular homolog (with strong implications for anti-poxviral inhibitor design); (iv) protein conformational steps accompanying substrate binding; and (v) the kinetic mechanism of D12’s stimulation of D1-MTase activity. Moving forward, we aspired to attain a comparably deep understanding of the TPase and GTase components of the vaccinia capping system.
Here we report the crystal structure of the full-length D1•D12 heterodimer with AdoHcy in the methyltransferase active site and with or without GTP in the guanylyltransferase active site at 2.8 and 2.9 Å resolution, respectively. The structures reveal that: (i) the poxvirus RNA triphosphatase domain is a bona fide “triphosphate tunnel metalloenzyme” (TTM) akin to the mimivirus and fungal TPases (Benarroch et al., 2008; Lima et al., 1999), and (ii) the guanylyltransferase domain consists of nucleotidyltransferase (NTase) and OB-fold (OB) modules as in previously characterized cellular/viral capping enzymes and DNA ligases (Shuman and Lima, 2004). Closely packed contacts between the TPase and GTase modules mutually stabilize their active conformations and account for the prior inability to isolate autonomous TPase and GTase catalytic domains. Furthermore, by introducing alanine mutations at the TPase-GTase interface, we identify lesions that decouple the TPase and GTase. The TPase-GTase contacts also clamp the NTase and OB domains of the GTase in a closed conformation, restricting inter-domain flexibility. In addition to structure-guided functional studies we discuss the insights into the evolution of the capping apparatus afforded by the structure of vaccinia D1•D12.
Results
Crystallization and structure determination of the full-length vaccinia D1•D12 heterodimer
The full-length vaccinia D1 and D12 subunits were co-expressed in bacteria and purified as a 1:1 heterodimer. The D1•D12 protein, supplemented variously with AdoHcy, AdoMet, m7GpppG, GTP and magnesium, crystallized in three different lattices, monoclinic (P21, forms 1 and 3) and triclinic (P1, form 2). The structure of the D1•D12 complex was solved by molecular replacement combined with cross-crystal averaging over four data sets, including native (2.9 Å resolution), platinum derivative (3.2 Å) and selenomethionine derivative (3.5 Å) crystals in space-group P21 (form 1) and a native crystal (2.9 Å) in space group P1 (see Materials and Methods and Table 1 for crystallographic statistics). Models of the triclinic and the monoclinic crystals were refined independently to R/Rfree values of 25.4/29.8 and 25.2/29.4, respectively (Table 1). The two refined structures are extremely similar except for minor differences in domain orientation. In all determined structures, AdoHcy is found in the MTase active site. No ligand was observed in the TPase site. Diffraction data collected for GTP co-crystals in the P1 form were used to derive the 2.8 Å structure of D1•D12 with GTP in the GTase active site, which was refined to R/Rfree values of 21.0/25.9. The final D1•D12 models included maximally 822/844 amino acids of the D1 subunit (lacking residues 28–33 in the TPase domain, 320-323 and usually 408-411 in the GTase, 531–543 from the linker between the GTase and MTase and, in the GTP bound structures, 554-561 in the MTase) and 283/287 amino acids of the D12 subunit (lacking residues 120–123). The structures have excellent stereochemistry, with >99% of residues in allowed regions of a Ramachandran plot (Table 1).
Table 1. Data collection and refinement statistics.
| Crystal |
Form 1 D1D12 + SAH SeMet-labelled |
Form1 D1D12 + SAH Pt soaked |
Form 1 D1D12 + SAH |
Form 2 D1D12 +SAH |
Form 2 D1D12 + SAH + GTP |
|---|---|---|---|---|---|
|
| |||||
| Data collection | |||||
| ESRF Beamline | BM14 | ID23-EH1 | BM14 | ID14-EH3 | ID14-EH4 |
| Wavelength | 0.979 | 0.921 | 0.979 | 0.931 | 0.976 |
| Space group | P21 | P21 | P21 | P1 | P1 |
| Cell dimensions: | |||||
| a, b, c (Å) | 61.5, 198.9, 128.3 | 61.7, 198.2, 116.4 | 61.7, 200.3, 128.1 | 61.1, 198.5, 62.0 | 60.3, 197.7, 61.1 |
| α, β, γ (deg) | 90.0, 102.2, 90.0 | 90.0, 101.2, 90.0 | 90.0, 102.2, 90.0 | 90.01, 108.31, 94.47 | 90.13, 109.27, 94.05 |
| Resolution (Å) a | 30.0-3.5 (3.8-3.5) | 30-3.2 (3.5-3.2) | 30-2.9 (3.0-2.9) | 30-2.9 (3.0 2.9) | 50-2.8 (2.9-2.8) |
| Rsyma | 0.204 (0.344) | 0.138 (0.301) | 0.143 (0.398) | 0.125 (0.447) | 0.078 (0.646) |
| I/σIa | 5.71 (3.52) | 4.57 (2.59) | 5.05 (2.13) | 5.42 (1.73) | 10.75 (1.43) |
| Completenessa | 98.4 (97.2) | 98.8 (97.1) | 92.0 (81.1) | 94.8 (95.4) | 96.1 (90.3) |
| Redundancya | 3.79 (3.51) | 1.93 (1.89) | 1.88 (1.83) | 1.90 (1.83) | 2.37 (2.31) |
|
| |||||
| Refinement | Not applicable | Not applicable | |||
| Resolutiona | 29.2 - 2.90 (2.97-2.90) | 41.8 – 2.90 (2.95-2.90) | 48.4-2.80 (2.88-2.80) | ||
| No. refls work (free) | 59651 (3174) | 55057 (2929) | 59782 (3184) | ||
| Rwork a | 25.5 (36.6) | 25.2 (36.1) | 20.9 (36.4) | ||
| Rfreea | 29.7 (37.8) | 29.4 (37.8) | 25.5 (42.7) | ||
| Total non-H atomsb | 18065 | 17813 | 17863 | ||
| Ligand (SAH, GTP) | 2 × 26 | 2 × 26 | 2 × 26, 1 × 32 | ||
| Solvent molecules | 82 | 76 | 25 | ||
| Average B-factor | 42.4 | 55.8 | 76.7 | ||
| RMS Deviations | |||||
| Bond lengths (Å) | 0.005 | 0.004 | 0.007 | ||
| Bond angles (deg) | 1.010 | 0.853 | 1.126 | ||
| Ramachandranc | |||||
| Allowed | 99.5 | 99.3 | 99.1 | ||
| Favoured | 93.7 | 91.3 | 94.5 | ||
|
| |||||
Highest resolution shell is shown in parentheses,
Two complexes per asymmetric unit,
From Molprobity: http://molprobity.biochem.duke.edu/
The overall structure of the D1•D12 complex resembles an elongated, U-shape with dimensions of 110 Å × 50 Å × 46 Å (Figure 1C). One lobe of this U-shape consists of the TPase tunnel and the N-terminal part of the NTase domain of the GTase and has a length of 56 Å. The other lobe is formed by the D12 subunit and is 60 Å long. These are connected by the distal half of the GTase NTase domain, the OB module, and the entire D1 MTase domain with a width of 110 Å. The relative arrangement of the three domains together with the D12 subunit opens a deep cleft with dimensions of 24 Å in width and 30 Å in depth (Figure 1C). The folding topology of the vaccinia capping enzyme is depicted in Figure 1D.
The TPase domain
The structure of the vaccinia TPase domain (residues 1-225) clearly indicates that it belongs to the triphosphate tunnel metalloenzyme (TTM) family of RNA triphosphatases, as shown by its structural homology to mimivirus TPase (Z=12.8 from DALI (Holm and Sander, 1993)) (Benarroch et al., 2008) and yeast Cet1 (Z=9.8) (Lima et al., 1999)(Figures 2 and S1). Similar to these other TPases, the central core of the vaccinia TPase consists of an anti-parallel eight-stranded β-barrel (β1 to β8; Figure 1D) that forms a topologically closed hydrophilic tunnel (Figure 2B-D). The vaccinia, mimivirus and yeast Cet1 TPases differ mainly in the position of the peripheral α helices that pack against the outer walls of the tunnel (Figure S1). Cet1 has four helices (αA-αD in Figure S1A) exclusively located below the bottom floor of the tunnel, three of which are conserved in the mimivirus and vaccinia TPases (as αA, αB and αE in Figure S1B and C). Vaccinia TPase has two additional helices located on the roof of the tunnel (αC and αD in Figure S1C, connecting strands β3 to β4 and β7 to β8, respectively). In contrast to the viral TPases, yeast Cet1 is an obligate homodimer (Hausmann et al., 2003; Lima et al., 1999) which explains the more complex structure of the Cet1 peripheral helices. An alignment of the viral and yeast TPase structures is shown in Figure S1D.
Figure 2. The triphosphatase domain. See also Figure S1.
(A) Two orthogonal views of the vaccinia TPase in ribbon representation with secondary structure elements labeled as in Figure 1D and the structure based alignment of Figure S1D. (B-D) Comparison of TPase tunnel interiors of the vaccinia (B), Mimivirus (C) (PDB entry 2QZE) and yeast Cet1 (D) (PDB entry 1D8I) enzymes. The structural similarity of vaccinia TPase as given by DALI to that of Mimivirus is RMSD of 3.8 Å over 183 aligned Cα positions, 12% sequence identity, Z-score 12.8 and to Cet1 is RMSD of 3.5 Å for 163 aligned Cα positions, 15% sequence identity, Z-score 9.8. Waters are depicted as red spheres. The manganese ion in the active site of Cet1 is shown as a purple sphere. The bound acetate (Mimivirus) and sulphate (Cet1) ions are shown as sticks. Selected amino acid side chains in the vaccinia, mimivirus and Cet1 tunnels are depicted as stick models with blue, gray and red carbons, respectively. Residues of TPase motifs A, B and C are colored in black, green and brown, respectively, in B, C and D (See also Figure S2).
The TPase active site
The TPase tunnel accommodates the 5′ triphosphate end of the substrate RNA together with at least one divalent metal ion, which promotes hydrolysis of the γ-phosphate (Lima et al., 1999). The tunnel is lined with charged residues that are partitioned such that one half of the internal surface (the ‘roof’) is populated almost exclusively by basic residues (Lys65, Lys75, Arg77, Lys79, Lys107, Lys161, Lys163, exceptionally Glu144) and the other half (the ‘floor’) by acidic residues (Glu37, Glu39, Glu126, Asp159, Glu192, Glu194, exceptionally Arg122) (Figure 2B). These include two conserved signature motifs of tunnel-forming TPases (motifs A and C, Figure S1D), each containing two metal-coordinating glutamates: Glu37 and Glu39 in β1 and Glu192 and Glu194 in β8 for Vaccinia TPase, which correspond to Glu307, Glu309, Glu494 and Glu496 in Cet1 and Glu37, Glu39, Glu212 and Glu214 in mimivirus, respectively (Figure 2B-D). Previous mutational analysis of the vaccinia TPase showed that six acidic and three basic residues are critical for enzymatic activity: Glu37, Glu39, Arg77, Lys107, Glu126, Asp159, Lys161, Glu192 and Glu194 (Gong and Shuman, 2003; Yu et al., 1997; Yu and Shuman, 1996).
Cet1 and mimivirus TPases contain a third motif (motif B), which comprises three basic side chains (Arg454, Lys456 and Arg458 in Cet1) projecting from strand β6 on the side wall of the tunnel. Based on the Cet1 complex with sulfate anion, these are predicted to coordinate the negatively charged RNA 5′ triphosphate (Arg454 to the β-phosphate, Arg458 to the γ-phosphate), either by direct hydrogen bonding or by water-mediated hydrogen bonds (Lima et al., 1999). Motif B is critical for the TPase activity of fungal and Chlorella virus TPase (Bisaillon and Shuman, 2001; Gong and Shuman, 2002; Pei et al., 1999; Pei et al., 2000). Vaccinia TPase lacks a basic motif B, the equivalent positions being substituted by Glu144, Val146 and Leu148 (Figure S1D). Instead, two basic side chains on strand β7, Lys161 and Lys163 are positioned to functionally replace respectively Arg458 and Arg454 in Cet1. Indeed Lys161 is essential for vaccinia TPase function; its mutation to alanine or glutamine elicits a 50-fold decrement in TPase activity with no impact on GTase activity (Gong and Shuman, 2003). However Lys163 has been shown to be dispensable for vaccinia TPase activity (Gong and Shuman, 2003).
Vaccinia TPase Arg77 (β3) and Lys107 (β4) are the counterparts of Arg393 and Lys409 in Cet1 and are essential for activity (Gong and Shuman, 2003; Yu et al., 1997) probably by coordinating the γ-phosphate. The active site of the vaccinia TPase is completed by two acidic side chains: Glu126 (β5) and Asp159 (β7). Glu126 (Glu433 in Cet1) is proposed to serve as a general base catalyst that coordinates and abstracts a proton from the water nucleophile that attacks the γ-phosphorus. Consistent with this, mutating Glu126 effaces vaccinia TPase activity (Gong and Shuman, 2003). An acidic functional group is apparently important at position 159, insofar as D159E retains full TPase activity, whereas D159A and D159N are 11% and 2% as active as wild-type TPase (Gong and Shuman, 2003). The equivalent aspartate in Cet1 (Asp471) forms a salt bridge with Cet1 Arg469, which in turn is thought to make a hydrogen bond with one of the waters in the octahedral manganese coordination complex. In vaccinia TPase, the arginine position is occupied by Thr157 (β7) which is unlikely to fulfill the same role.
Lys75 and Lys79 (in strand β3) are constituents of the triphosphate tunnel of vaccinia TPase that are conserved in the capping enzymes of other Poxviridae (Figure S1D) but not as basic residues in Cet1 or mimivirus TPase. To test whether Lys75 and Lys79 are relevant for vaccinia TPase activity, we changed them to alanine in the context of D1(1-545) and assayed the recombinant proteins (Figure S1E) for TPase activity in parallel with wild-type D1(1-545). The K75A and K79A mutations had no deleterious effect on TPase function (Figure S1E). We also mutated 12 other charged or hydrophilic residues that either project into the TPase tunnel from the β-strands (Arg65, Glu122, Ser124, Glu144, Gln150, Lys152, Thr157, Ser190 and Thr196) or out to solvent from the tunnel rim (Arg128, Lys152 on one end of the tunnel; Arg142, Glu112 on the other end). Most of the positions chosen for this alanine scan are conserved among poxvirus TPases except (Arg65, Thr157 and Thr196), but not in Cet1 or mimivirus TPase (Figure S1D). Our criterion for judging an amino acid important for capping enzyme function is that its replacement by alanine causes at least a four-fold decrement in activity. By this test, none of the mutated residues are important for NTP phosphohydrolase activity.
The guanylyltransferase
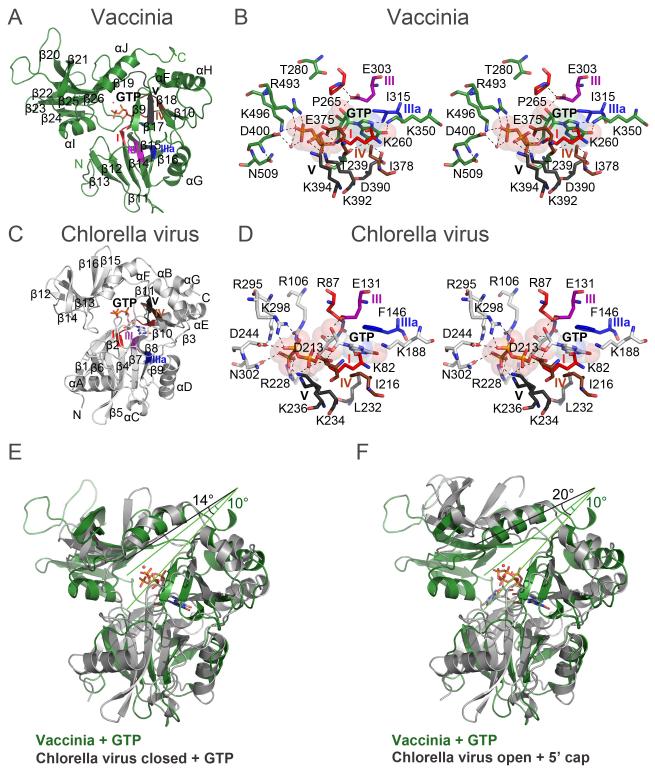
Capping enzyme guanylyltransferases (GTases), with the exception of those of some dsRNA viruses, share a common fold with ATP-dependent DNA ligases (Shuman and Lima, 2004) consisting of two domains: an N-terminal nucleotidyltransferase (NTase) and a C-terminal OB-fold (OB). Structural homology searches with vaccinia GTase (residues 226-530) using DALI identified four capping enzyme GTases. In order of similarity they are: Chlorella virus GTase (DALI Z=14.7) (Hakansson et al., 1997), the GTase domain of human capping enzyme (Z=14.1) (Chu et al., 2011), Candida albicans Cgt1 (Z=11.5) (Fabrega et al., 2003) and Saccharomyces cerevisiae Ceg1 (Z=10.3) (Gu et al., 2010). The vaccinia NTase fold consists of: (i) a central six-stranded antiparallel β-sheet (β13,12,11,14,15,16), (ii) a four-stranded mixed β-sheet (β9,18,17,10), and (iii) three α-helices (αF,G,H) (Figure 1D, 3AC). A structure-based amino acid sequence alignment of the viral and human GTases is shown in Figure S2. We will focus our discussion on a comparison of the vaccinia and Chlorella virus GTases.
Figure 3. The guanylyltransferase domain. See also Figure S2.
(A) Ribbon representation of the vaccinia virus GTase in green with bound GTP and secondary structure elements labelled. Residues of the defining structural motifs I, III, IIIa, IV and V of the covalent nucleotidyl-transferase family are highlighted and labelled in red, purple, blue, black and brown, respectively. The same color coding and labelling is used in 3B, 3C and 3D.
(B) Stereo view of the active site of the vaccinia GTase showing bound GTP and key interacting residues with hydrogen bonds indicated by dotted black lines.
(C) Ribbon representation of the Chlorella virus GTase with bound GTP in the closed conformation (PDB entry 1CKN chain B, light grey). The structural similarity of vaccinia GTase as given by DALI to that of Chlorella virus is an RMSD of 3.2 Å over 216 aligned Cα positions, 17% sequence identity, Z-score 14.7.
(D) Active site of the Chlorella virus GTase in the closed conformation with bound GTP.
(E) Superposition of vaccinia (green) and the closed form of Chlorella virus (grey) GTases (grey), both with bound GTP. The angles between the NTase and OB-fold domains are indicated.
(F) Superposition of vaccinia GTase with bound GTP (green) and the open form of the Chlorella virus GTase with bound GpppG (grey). The angles between the NTase and OB-fold domains are indicated. The angle between the vaccinia domains does not change, whereas that of Chlorella virus opens with GpppG present.
The vaccinia and Chlorella NTase domains differ most strongly at their N-termini. Chlorella virus GTase has an additional module – composed of a two-stranded β-sheet (β1-β2) and an α-helix (αA) – at its N-terminus that packs against the central β-sheet (Figure 3C). In the vaccinia NTase domain, this helix is replaced by a long loop that runs across the central β-sheet, and by the β9 strand, which is absent in the Chlorella virus enzyme.
The C-terminal OB module comprises a five-stranded anti-parallel β-sheet (β21,20,19,25,26) with an inserted three-stranded antiparallel β-sheet (β22,23,24) located beneath the five-stranded β-sheet (Figure 1D, 3A, S3). The inserted β-sheet is a unique feature of the vaccinia GTase. Towards the C-terminus there are two α helices (αI and αJ), flanking the three- and the five-stranded β-sheet, respectively (Figure 1D). The OB domain of Chlorella virus GTase lacks helix αI and has instead a C-terminal helix (αG) that folds back on the NTase domain (Figure 3C). In vaccinia GTase, the equivalent region interacts weakly with αH of the NTase domain (Figure 3A) and leads into a long, disordered loop (530-544) connecting the GTase with the MTase; this loop is protease-sensitive in the native vaccinia capping enzyme (De la Pena et al., 2007; Shuman, 1989).
The NTase and the OB-fold domains of the GTase form a deep substrate binding cleft. Chlorella virus GTase was crystallized in two different conformations: an unreactive open conformation with bound GTP, and a catalytically active closed conformation with bound GTP that could be converted in situ into the covalent enzyme-GMP adduct upon addition of manganese (Figure 3C)(Hakansson et al., 1997). Even in the absence of nucleotide, the vaccinia GTase resembles the closed conformation of the GTP-bound Chlorella virus GTase (Figure 3E), whereas for human GTase (with sulphate in the active site) a range of conformations from closed to half-open were observed (Chu et al., 2011). In vaccinia GTase, the residues 467EVG468 at the tip of the surface loop between strands β23 and β24 of the OB module interacts with two surface loops of the NTase module, thereby reinforcing the closed conformation of the GTase. Specifically, the Glu467 main chain carbonyl accepts a hydrogen bond from the Lys306 main chain amide; the Val468 carbonyl makes van der Waals contacts with the Trp310-Cζ2,CH2 indole atoms; and Val468-Cγ1 makes van der Waals contacts to His281-Cδ,Nε. The tip of this vaccinia OB-fold loop is physically at the same place as the turn of the extra N-terminal β-hairpin of the Chlorella virus NTase domain.
The guanylyltransferase active site
Co-crystals of vaccinia capping enzyme with GTP revealed density for the nucleotide in one of the two D1•D12 complexes in the asymmetric unit (Figure S5C). GTP binds between the six-stranded and four-stranded β-sheets of the NTase module and induces no large scale changes in the enzyme, only affecting the conformations of a few side chains in direct contact with the ligand. The defining features of the vaccinia GTase active site are six conserved motifs that interact with the guanine base (motifs IIIa and IV), the ribose hydroxyls (motifs I and III) and the triphosphate moiety (motifs I, V and VI) of GTP (Shuman and Lima, 2004; Wang et al., 1997). Vaccinia GTase Motif I (260KTDGIP265) is located in the loop between β10 and β11 and contains Lys260, the active site lysine nucleophile to which GMP becomes covalently attached (Niles and Christen, 1993).
The guanosine nucleoside adopts a syn conformation in the vaccinia GTase. The base stacks between Ile315 (β15, motif IIIa) and Ile378 (β17, motif IV), which correspond to Phe146 and Ile216 in Chlorella virus GTase. Guanine specificity is conferred by hydrogen bonding between amino acid side chains and the polar atoms of the nucleobase (Lys350 to O6 and N7, Thr239 and Asp390 to N2). The triphosphate conformation in the vaccinia complex differs from that in the closed form of the Chlorella virus GTP complex and is not optimal for catalysis. In the active Chlorella GTase, the Lys82-Nζ nucleophile is 3.2 Å from the α-phosphorus and is apical to the bridging α-β O3A oxygen of the PPi leaving group. In the vaccinia GTase, Lys260 contacts the GTP α and β phosphate oxygens, but the α-phosphate is not oriented properly for a nucleophilic attack by Lys260 to form the enzyme-GMP adduct. Lys260-Nζ is too distant (3.8 Å) from the α-phosphorus and makes an acute angle with respect to the PPi leaving group. We envision that this unfavorable geometry is rectified when the essential Mg2+ ion is bound in the active site. This ion is likely to be bound by the essential motif IV acidic residue (Glu375 in Vaccinia and Asp213 in Chlorella virus GTases) that is conserved in all ATP-dependent DNA and RNA ligases, NAD+-dependent DNA ligases, and GTP-dependent mRNA capping enzymes (Cong and Shuman, 1995; Sawaya and Shuman, 2003; Wang et al., 1997). The metal is postulated to promote catalysis by: (i) coordinating the α-phosphate and stabilizing the negative charge developed on a phosphorane transition state (Hakansson et al., 1997; Nandakumar et al., 2006), and (ii) coordinating the Lys-Nζ and favoring its deprotonation for nucleophilic attack on the α-phosphorus.
The GTP triphosphate moiety in the vaccinia GTase is coordinated by basic residues in motifs V (390DFKIKKENTID400) and VI (493RIDK496). Motif V comprises strands β18 and β19 and their connecting loop, which joins the NTase module to the OB module. Motif V Lys392, which is essential for RNA capping activity (Cong and Shuman, 1995), makes a bifurcated contact via Nζ with the α-β bridging O3A and the β-phosphate O2B. Lys394 points towards the β-phosphate, but is not within hydrogen bonding distance. We suspect that the vaccinia motif V lysine contacts with the phosphates may be reorganized in the context of a Michaelis complex with Mg2+.
Motif VI (RxDK) in the OB module is essential for the activity of cellular GTases (Sawaya and Shuman, 2003; Wang et al., 1997). Motif VI of Chlorella virus GTase is located in a loop (295RSDKNQAN302) connecting a β strand and an α helix of the OB domain and is similarly situated in the S. cerevisiae, C. albicans, and human GTases. In the catalytically poised closed conformation of Chlorella virus GTase, Arg295 coordinates the GTP β phosphate while Lys298 and Asn302 engage the γ phosphate, thereby orienting the PPi leaving group apical to the lysine nucleophile. However, in the vaccinia GTase, motif VI resides entirely within helix αI (493RIDKTMKYN502). Despite the distinctive secondary structure of motif VI in the poxvirus enzyme, the Arg493 and Lys496 side chains engage the GTP γ-phosphate oxygens, signifying a functional correspondence to motif VI of the cellular and Chlorella virus GTases. Lys496 also makes a salt bridge to the putative metal-binding Asp400 carboxylate. Vaccinia residue Asn502 occupies a position similar to that of Asn302 in Chlorella virus GTase, but it is 5 Å away from the GTP γ-phosphate, again suggestive that the Vaccinia active site configuration is non-active.
In sum, the structure of the GTase teaches us that the poxvirus capping enzyme shares features of NTP recognition and catalysis with other viral and cellular capping enzymes, and with polynucleotide ligases, while highlighting multiple distinctive components of the poxvirus active site.
The interface between TPase and GTase
The TPase (Cet1) and GTase (Ceg1) components of the S. cerevisiae capping apparatus are encoded as separate polypeptides that associate in vivo and in vitro to form a stable Cet1•Ceg1 complex. The Cet1•Ceg1 crystal structure revealed a hetero-tetramer composed of one Cet1 homodimer and two molecules of Ceg1 (Gu et al., 2010). The main Cet1•Ceg1 interaction surface (1900 Å2) is between the OB domain of Ceg1 and the extremity of the N-terminal extension of the distal Cet1, after the latter has crossed over the proximal subunit of the Cet1 homodimer. There is minimal interaction of the NTase module of Ceg1 with Cet1, thereby allowing flexibility between the OB and NT domains.
The vaccinia TPase-GTase interface differs significantly from that of Cet1 and Ceg1, although both bury comparable surface accessible areas (2155 Å2 in the vaccinia interface versus 2400 Å2 for the Cet1-Ceg1 interface). The main difference is that the vaccinia TPase makes substantial interactions with the NTase (807 Å2) and OB (1348 Å2) modules of the GTase, holding them in a closed conformation (Figure 4A). In comparison, Ceg1 GTase is in a wide open conformation (Gu et al., 2010). The vaccinia TPase contacts the GTase mainly via the helices packed against the TPase β-barrel and the loops connecting these helices to the tunnel. This results in a more rigid connection between the two enzymes, in contrast to the flexible connection between Cet1 and Ceg1. Finally, the relative orientation of the TPase tunnel to the GTase differs markedly. In the vaccinia enzyme the tunnel entrance points directly towards the GTase active site (Figure 6), whereas in the Cet1-Ceg1 structure the tunnel is rotated almost 45° around the axis perpendicular to the longitudinal tunnel with the entrance pointing towards the OB-fold domain of Ceg1.
Figure 4. The TPase/GTase interface. See also Figure S3.
(A) Overall ribbon representation of the vaccinia TPase /GTase interface, with coloring as in Figure 1C and secondary structural elements involved in the TPase/GTase interface labelled.
(B) and (C) Close-up stereo views of the corresponding areas marked in (A). For clarity the view in (B) is shown 180° rotated around the Y-axis, allowing a view from the rear of the interface. Side chains involved in interface formation are shown as stick models, hydrophobic interactions and hydrogen bonds are presented as dotted lines in magenta and black, respectively.
(D) Effects of alanine mutations at the TPase/GTase interface on TPase and GTase activity. Guanylyltransferase reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 8.0), 5 mM DTT, 5 mM MgCl2, 5 μM [α32 P]GTP, and 200 ng wild-type or mutant His10Smt3-D1-(1-545) were incubated for 5 min at 37°C. The reactions were quenched with SDS and the products were analyzed by SDS-PAGE. The covalent enzyme-[32P]GMP complex was visualized by autoradiography of the dried gel and quantified by scanning the gel with a phosphorimager. Triphosphatase reaction mixtures (10 μl) containing 50 mM Tris-HCl (pH 8.0), 2 mM DTT, 10 mM MgCl2, 1 mM [α32P]ATP, and 150 ng wild-type or mutant His10Smt3-D1-(1-545) were incubated for 30 min at 37°C. The reactions were quenched with 20 mM EDTA. An aliquot (6 μl) of the reaction mixture was spotted on a polyethyleneimine-cellulose TLC plate, which was developed in 0.45 M (NH4)2SO4. The release of [α32P]ADP was quantified by scanning the TLC plate with a phosphorimager. The GTase and TPase activities of the mutants were normalized to the wild-type activity (defined as 100%).
Figure 6. mRNA cap synthesis pathway between active sites.
Two orthogonal views of the complete trifunctional enzyme, in ribbon and surface representation, showing the spatial relationship between the three active sites. The colour scheme is as in Figure 1C and ligands GTP and SAH are shown as space-filling models. Dotted black arrows and brackets indicate ligands that are obscured in a particular view. Possible paths of the mRNA 5′ end to enter successively the TPAse, GTase and MTase are shown in red (see text).
The interface of the vaccinia TPase and GTase comprises three separate sub-interfaces between: (a) the TPase N-terminal peptide and residues of the GTase β-strand β11 of the central β-sheet and the long N-terminal loop located on top of the central β-sheet; (b) the TPase helix αB and GTase residues in helix αI and β24 of the OB domain and (c) the TPase residues in αD and the loop connecting αD with β8 and GTase OB residues from β21, β24, β25, their connecting loops and helix αI. These interfaces are illustrated in Figure 4 and described in more detail in the supplementary information.
Mutational analysis of the TPase-GTase interface
To address the importance of the inter-domain contacts for capping enzyme function, we introduced interfacial mutations T10A, L47A-L50A-T51A, N181A, and R186A in the TPase domain and K478A in the GTase domain of D1-(1-545) and assayed the affinity-purified wild-type and mutant recombinant proteins (Figure S3A) for TPase and GTase activities. We purified and tested in parallel a TPase-defective mutant E192A that eliminates one of the metal-binding side chains in the TPase active site tunnel. Velocity sedimentation analysis of the wild-type D1-(1-545) protein verified that it sedimented as a discrete monomeric component and that the GTase and TPase activity profiles coincided with that of the D1-(1-545) polypeptide (Figure S3B,C). The relative activities of the D1-(1-545)-Ala mutants vis à vis the wild-type enzyme are shown in Figure 4D. As noted above, our criterion for deeming an amino acid important for capping enzyme function is that its replacement by alanine elicits at least a four-fold decrement in activity. There was a clear hierarchy of mutational effects, the most instructive of which were the lesions that resulted in a complete separation of function. For example, the E192A enzyme with a TPase active site mutation was fully active as a guanylyltransferase, but was crippled as a triphosphatase (4% of wild-type activity) (Figure 4D). Two of the interfacial mutations, L47A-L50A-T51A in the TPase domain and K478A in the GTase domain, reduced GTase activity to <1% of the wild-type level while sparing the TPase (42-43% of wild-type activity). Lys478 is at the center of a rich network of cross-domain contacts between the δD helix and flanking loop of the TPase domain and the OB domain of the GTase, especially the αI helix that includes motif VI side chains needed for GTase activity (Fig. 4C). We surmise that the K478A mutation destabilizes the active conformation of the GTase. Note that other mutations in this portion of the interface (N181A and R186A) did not exert a significant or selective impact on GTase versus TPase activity (Figure 4D).
The L47A-L50A-T51A triple-mutation, which eliminates all of the atomic contacts between the TPase αB helix and the OB domain of the GTase (including the direct contacts of Leu47 to the αI helix), also abolished GTase activity while preserving substantial TPase function (Figure 4D). These results highlight the key role of the TPase interface with the OB domain as a determinant of guanylyltransferase activity. TPase interactions with the NTase domain are also relevant, insofar as the TPase T10A mutation, which severs the contacts to the 224PEVV227 peptide at the proximal margin of the NTase domain, results in a 5-fold decrement in GTase activity with no effect on the TPase (Figure 4D).
These findings explain the long-standing conundrum in analysis of the domain organization of vaccinia capping enzyme, whereby it has not been possible to define by N-terminal deletion an autonomous GTase domain.
The interface between the GTase and MTase domains
A comparison with the previously described MTase•D12 structure (De la Pena et al., 2007) reveals no significant structural differences of the sub-complex in the full length capping enzyme. The vaccinia MTase is connected to the GTase via a flexible linker (residues 529-543) that is disordered in all crystal structures. This is followed by a region containing several residues required for the coordination of the methyl-donor AdoMet and for catalysis (De la Pena et al., 2007; Zheng and Shuman, 2008a). In the full length capping enzyme this peptide is well defined in the P21 crystal form 1 but residues 553-561 are disordered in the P1 crystal form.
The crystal structure of full length D1•D12 provides the first visualization of an interface between an RNA (guanine-N7)-methyltransferase and an RNA guanylyltransferase. This interface buries a surface area of around 917 Å2 (432 Å2 with NT and 485 Å2 with OB), which is significantly less than the TPase–GTase interface. The secondary structure elements of the MTase contributing to the interface include the N-terminal peptide, helix αO, the β-strands β33, β34, β35 (inserted between β31 and αQ) and their connecting loops. Their GTase components at the interface are β9, αF, β17 and β18 from the NTase module and β20, β21 and αJ from the OB module. This interface is illustrated in Figure 5A and described in more detail in the supplementary information.
Figure 5. MTase interfaces with GTase and D12. See also Figure S4.
(A) Left: Overall ribbon representation of the vaccinia GTase/MTase interface. The colour scheme is as in Figure 1C. Secondary structure elements contributing to the interface are labeled. Right: Stereo view of the marked area in left hand diagram. Side chains involved in the interface and SAH are shown as stick models, hydrophobic interactions and hydrogen bonds are presented as dotted lines in magenta and black, respectively.
(B) Same as (A) but for the MTase/D12 interface.
The interface between the MTase catalytic domain and D12
We previously described the interface between the vaccinia MTase with the stimulatory D12 subunit and proposed a mechanism for the allosteric effect of D12 on the MTase activity, whereby the D12 helix αB’ packs against the C-terminal part of the MTase helix αK mainly via hydrophobic interactions. This interaction stabilizes this helix, which contains Asn570 and Lys573, two conserved residues coordinating the methyl acceptor GTP and the methyl donor AdoMet, respectively (De la Pena et al., 2007). To further investigate this interface we introduced alanine mutations for MTase residues Tyr571, Leu575, Leu576, Met579, Phe585, Leu586, Asp587, Asp784, Asn785, Arg794, Phe798, Met805 and Glu806 at the interface with the D12 subunit. The His-tagged wild-type and mutant MTase catalytic domains D1-(540-844) were co-expressed in E. coli with untagged D12. SDS-PAGE analysis of the affinity-purified proteins revealed that D12 co-purified with D1-(540-844) in every case (Figure S4A), signifying that none of the interfacial mutations affected the heterodimerization of the MTase catalytic domain with D12 during protein production in vivo in bacteria. The recombinant proteins were assayed for methyltransferase activity by label transfer from [3H-CH3]AdoMet to GpppA cap dinucleotide. None of the alanine changes elicited a significant decrement in MTase activity (Figure S4B). In addition we tested the effects of the alanine mutations on cap methylation activity in vivo in yeast, taking advantage of the fact that co-expression of vaccinia D1 MTase catalytic domain and the D12 stimulatory subunit in S. cerevisiae complements the lethality of a yeast abd1Δ mutant that lacks the endogenous yeast cap guanine-N7 methyltransferase enzyme Abd1 (Saha et al., 2003). Co-expression of both vaccinia subunits is required for abd1Δ complementation and the genetic readout is sensitive to D1 mutations that affect methyltransferase catalytic activity (Saha et al., 2003; Zheng and Shuman, 2008a, b) and to certain D12 mutations (Saha et al., 2003; Schwer and Shuman, 2006). Here we found that all of the interfacial alanine mutants in the D1 MTase were active in abd1Δ complementation (Figure S4B). We conclude that none of the residues probed by alanine scanning at the MTase-•D12 interface is critical per se for biochemical or biological activity.
Discussion
The vaccinia virus mRNA capping enzyme combines all three enzymatic activities for cap 0 synthesis within one polypeptide chain. Our current and previous (De la Pena et al., 2007) structural studies reveal that the folds of the single components are similar to capping enzyme components already structurally characterized: the TPase belongs to the triphosphate tunnel metalloenzyme (TTM) family that includes yeast and mimivirus RNA triphosphatases, the GTase is a member of the covalent nucleotidyltransferase superfamily that comprises capping enzymes and polynucleotide ligases, and the MTase belongs to the class I family of AdoMet-dependent MTases.
The unification of the vaccinia RNA triphosphatase with the TTM family resolves a longstanding conundrum about the structure and origins of this enzyme. Early studies of the vaccinia TPase revealed what would emerge as the signature biochemical feature of the TTM-type RNA triphosphatases: the ability to hydrolyze NTP to NDP and Pi in the presence of Mn2+ or Co2+ (Shuman et al., 1980). The yeast Cet1 structure unveiled a then novel tunnel tertiary structure (Lima et al., 1999) that has since been recognized as the founder of a TTM superfamily that extends well beyond capping enzyme TPases (Gong et al., 2006; Jain and Shuman, 2008). Amino acid sequence comparisons and mutational analyses of metal-dependent RNA triphosphatases of fungi, microsporidia, protozoa and Chlorella virus highlighted conservation of the β strands and essential amino acids that compose the active site tunnel of yeast Cet1, implying a common structure and evolutionary origin for these enzymes (Shuman, 2002), a view later fortified by the structure of mimivirus TPase (Benarroch et al., 2008). However the vaccinia TPase had no recognizable primary structure similarity to the fungal, protozoan, Chlorella virus or mimivirus TPases, except for the metal-binding motifs A and C. Whereas mutational studies had identified constituents of the vaccinia TPase active site, and it was predicted that some of these would reside in β strands (e.g., 74VKIRTKI80 and 158IDFKLKY164), it was inferred that the poxvirus TPases lack a topologically closed tunnel architecture (Gong and Shuman, 2003). The present structure vitiates that notion, while at the same time showing that several of the active site constituents of vaccinia TPase are indeed deployed in a distinctive manner (i.e. in different β strands of the tunnel) vis à vis other TTM RNA triphosphatases.
The structure of the full-length D1 protein adds new information on the disposition of the catalytic domains and their interfaces. The N-terminal decapeptide and tunnel-surrounding helices of the TPase domain interact, respectively, with the NTase and OB modules of the GTase domain. In this way, the vaccinia TPase clamps the GTase in a conformation similar to the closed conformation of Chlorella virus capping enzyme, but unlike the Cet1-Ceg1 complex or the isolated C. albicans and human GTases, in which the relative orientation of the NTase and OB modules is flexible. It is therefore unlikely that the vaccinia GTase undergoes the same degree of inter-NTase–OB domain movement as observed in Chlorella virus and cellular GTases and ATP-dependent DNA ligases. Consistent with this idea is our observation that residues in the vaccinia TPase/GTase interface have profound effects on GTase activity. In particular, the mutations L47A-L50A-T51A, K478A and T10A resulted in separations of function, whereby the mutant enzymes were inactive or partially defective in TPase activity, but spared with respect to GTase function.
The salient characteristic of the open conformations of GTases and DNA ligases is that motif VI is remote from the NTP β and γ phosphates. In the vaccinia GTase, motif VI engages the GTP phosphates, but the geometry of the triphosphate moiety observed in our structure is inimical to catalysis. We presume that the triphosphate conformation and certain atomic contacts of the active site functional groups will be remodeled in the Michaelis complex of the GTase reaction, especially when the catalytic Mg2+ engages the GTP α-phosphate and the non-catalytic Mg2+ bridges the β- and γ-phosphates.
The orientation of the TPase tunnel relative to the GTase active site in the vaccinia capping enzyme is well suited for sequential action. Although bound nucleoside triphosphate substrate has not been observed in any available RNA triphosphatase structure, it is hypothesized from positions of a bound divalent cation and a sulphate (mimicking the γ-phosphate) in the yeast Cet1 TPase structure that the 5′ pppRNA end substrate enters the tunnel from the direction depicted in Figure 6. In the vaccinia system, the TPase tunnel is positioned so that its entrance points directly towards the GTase active site. Based on the vaccinia enzyme structure, we can imagine that the mRNA 5′ triphosphate end enters the deep groove formed by the TPase, OB-fold and MTase, giving it access to the TPase and then, after conversion to a 5′ diphosphate RNA, to the GTase active site (Figure 6 left). By contrast, the transition from the GTase to the MTase active site is less clear, insofar as the MTase site is on the opposite side of the D1•D12 heterodimer (Figure 6 right). It is possible that the GpppRNA continues to track around the protein surface in the same direction in the groove between the NTase, MTase and D12 subunits (path A), taking advantage of the U-shaped form of the complex, but this is a long way round. Alternatively the GpppRNA could dissociate from the GTase and rebind to the MTase active site (path B). In any case, it should be borne in mind that the capping enzyme is directly associated with the vaccinia RNA polymerase (Hagler and Shuman, 1992) so that the 5′ end of the nascent mRNA will stay in proximity to the capping enzyme even if dissociated from a particular capping enzyme active site. Furthermore, mRNA elongation by the polymerase could be a driving force to move the 5′ end from one active site to the next. Although our structures provide new insights to the architecture of the complete vaccinia capping enzyme, we must await a next generation of structures of the enzymes in complexes with pppRNA, ppRNA and GpppRNA ligands to illuminate how the mRNA 5′ end proceeds from one site to the next and whether significant conformational changes, not so far observed in any crystal form, play a role in these transitions.
The D1•D12 structure, the first of a complete trifunctional DNA virus capping enzyme, raises interesting questions about the evolution of the mRNA capping apparatus. The poxvirus D1 subunit comprises TPase, GTase and MTase catalytic domains, arrayed in the same linear order in the D1 polypeptide as the sequential capping reactions they perform. The structural similarities of the poxvirus TPase, GTase and MTase modules to the separately encoded TPase, GTase, and MTase enzymes of fungi and microsporidia (Fabrega et al., 2004; Fabrega et al., 2003; Lima et al., 1999) argues that the poxvirus capping enzyme evolved by gene fusion events. The fact that other large eukaryal DNA viruses, such as African swine fever virus and mimivirus, also encode D1-like TPase-GTase-MTase polypeptides (Benarroch et al., 2008; Pena et al., 1993), as do virus-like cytoplasmic plasmids of fungi (Tiggemann et al., 2001), suggests a common ancestry for the DNA virus and virus-like trifunctional capping enzymes (Shuman, 2002).
New clues to this ancestry can be found in the growing collection of giant DNA viruses that have been isolated from marine protozoa and eukaryal phytoplankton in the wake of the discovery of mimivirus. The giant DNA viruses that encode poxvirus/mimivirus-like TPase-GTae-MTase polypeptides are as follows (with viral genome size and capping enzyme polypeptide accession number and size indicated in parentheses): Megavirus chiliensis (1.26 Mb; YP_004894563: 1164 aa); Acanthamoeba polyphaga moumouvirus (1.02 Mb; YP_007354410: 1162 aa); Cafeteria roenbergensis virus (0.73 Mb; YP_003969844: 1015 aa); Phaeocystis globosa virus (0.46 Mb; YP_008052553: 1191 aa); Marseillevirus (0.37 Mb; YP_003407048: 819 aa); and Organic Lake phycodnavirus 2 (~0.3 Mb; ADX06468: 942 aa). By contrast, the largest known eukaryal DNA virus, Pandoravirus salinus (2.5 Mb genome) encodes three separate capping enzymes: a TTM-type TPase (YP_008436986: 333 aa), a GTase (YP_008437195: 482 aa), and a MTase (YP_008436891: 452 aa).
The D12 subunit is a distinctive feature of the poxvirus capping enzyme. As noted previously, the D12 structure resembles a degenerate cap 2′-O-MTase domain, insofar as it is structurally similar to the cap 2′-O-MTases of reovirus λ and flavivirus RNA polymerase, whilst lacking the AdoMet binding site (De la Pena et al., 2007). We surmise that poxvirus D1•D12 descended from a multi-functional viral enzyme that was capable of synthesizing a cap 1 structure, m7GpppNm. Erosion of the erstwhile MTase function of vaccinia D12 was presumably compensated by the acquisition of a separately encoded cap 2′-O-MTase enzyme, VP39 (Schnierle et al., 1992). There is apparent selective pressure to maintain D12 as a subunit of the vaccinia capping enzyme, by virtue of: (i) its necessity for optimal MTase activity of the D1 subunit; and (ii) its requirement for the transcription termination factor activity of vaccinia capping enzyme during synthesis of poxvirus early mRNAs by the viral RNA polymerase (Luo et al., 1995).
PDB deposition
Co-ordinates and structure factors have been deposited in the wwPDB for the P21 (code 4CKC) and P1 (code 4CKE) forms of the SAH bound trifunctional enzyme and the SAH and GTP bound P1 form (code 4CKB).
Material and Methods
Full methods are given in the supplementary information.
Expression and crystallisation of vaccinia capping enzyme
D1, with an N-terminal tobacco etch virus (TEV) protease cleavable His10-tag, was co-expressed with D12 in E. coli strain BL21Star(DE3)pLysS and the complex purified by two steps of Ni-NTA affinity chromatography (before and after His-tag cleavage by His-tagged TEV), followed by heparin and then Superdex 200 columns (Pharmacia). Purified complex at 4 mg/ml was used with 5 mM AdoMet, 1 mM magnesium chloride, and 2 mM m7GpppG for crystal screening. After optimization, plate-like crystals, often in stacks, were grown from 0.1 M MES, pH 6.0, 1 M LiCl, 10% PEG-6000 at 20°C using the sitting drop vapor-diffusion technique. These crystals belong to space group P1 with two molecules per asymmetric unit (form 2). Crystals with similar morphology were grown from 0.1 M Tris-HCl, pH 7.5, 0.2 M LiCl, 0.2M (NH4)2SO4, 15% PEG-4000. These crystals belong to the space group P21 with two molecules per asymmetric unit (form 1). Form 1 crystals containing GTP were grown in the presence of 100 mM GTP, 5 mM manganese chloride, and 5 mM AdoMet from 0.1 M citric acid, pH 6.5, 1.2 M LiCl, 6% PEG-6000.
Data collection, phase determination and structure refinement
Diffraction data from cryo-protected crystals were collected at the European Synchrotron Radiation Facility (ESRF) beamlines and processed with XDS (Kabsch, 1993). The D1•D12 structure was solved by molecular replacement, using the MTase•D12 subcomplex (pdb 2VDW, 51% of the total), in combination with cross-crystal averaging over four data sets and the two D1•D12 complexes in each asymmetric unit: native datasets in P21 and P1, a selenomethionine labelled derivative and a non-isomorphous platinum derivative, with resolutions of 2.9 Å, 2.9 Å 3.5 Å and 3.2 Å, respectively. The RESOLVE-build script, which performs automatic, iterative, density modification, NCS averaging and model building, was very useful model construction (Terwilliger, 2003). Structures were refined with standard procedures using NCS restraints and TLS parameters.
Supplementary Material
Highlights.
2.8 Å resolution crystal structure of the complete vaccinia capping enzyme
The TPase has a triphosphate tunnel metalloenzyme fold
The extensive TPase-GTase interface clamps the GTase in a closed conformation.
TPase-GTase interfacial mutations selectively cripple the GTase activity.
Acknowledgements
We are grateful for access to platforms of the Grenoble Partnership for Structural Biology especially the EMBL HTX lab for robotic crystallization. We thank the staff of the ESRF-EMBL Joint Structural Biology Group for help with data collection on beamlines BM14, ID14-4, ID14-3 and ID23-1. We acknowledge the help of Dr. Heinz Gut in setting up the cross-crystal averaging. This work was supported by NIH grant GM42498 (to S.S.). S.S. is an American Cancer Society Research Professor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Benarroch D, Smith P, Shuman S. Characterization of a trifunctional mimivirus mRNA capping enzyme and crystal structure of the RNA triphosphatase domain. Structure. 2008;16:501–512. doi: 10.1016/j.str.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Bisaillon M, Shuman S. Structure-function analysis of the active site tunnel of yeast RNA triphosphatase. J Biol Chem. 2001;276:17261–17266. doi: 10.1074/jbc.M100980200. [DOI] [PubMed] [Google Scholar]
- Chu C, Das K, Tyminski JR, Bauman JD, Guan R, Qiu W, Montelione GT, Arnold E, Shatkin AJ. Structure of the guanylyltransferase domain of human mRNA capping enzyme. Proc Natl Acad Sci U S A. 2011;108:10104–10108. doi: 10.1073/pnas.1106610108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong P, Shuman S. Methyltransferase and subunit association domains of vaccinia virus mRNA capping enzyme. J Biol Chem. 1992;267:16424–16429. [PubMed] [Google Scholar]
- Cong PJ, Shuman S. Mutational Analysis of messenger-RNA capping enzyme identifies amino-acids involved in GTP-binding, enzyme-guanylate formation, and GMP transfer to RNA. Mol Cell Biol. 1995;15:6222–6231. doi: 10.1128/mcb.15.11.6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Pena M, Kyrieleis OJP, Cusack S. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. Embo Journal. 2007;26:4913–4925. doi: 10.1038/sj.emboj.7601912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E, Ferron F, Lescar J, Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol. 2012;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Schnaufer A, Salavati R, Stuart KD, Hol WG. High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. Journal of molecular biology. 2004;343:601–613. doi: 10.1016/j.jmb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- Fabrega C, Hausmann S, Shen V, Shuman S, Lima CD. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol Cell. 2004;13:77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- Fabrega C, Shen V, Shuman S, Lima CD. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell. 2003;11:1549–1561. doi: 10.1016/s1097-2765(03)00187-4. [DOI] [PubMed] [Google Scholar]
- Gong C, Shuman S. Chlorella virus RNA triphosphatase. Mutational analysis and mechanism of inhibition by tripolyphosphate. J Biol Chem. 2002;277:15317–15324. doi: 10.1074/jbc.M200532200. [DOI] [PubMed] [Google Scholar]
- Gong C, Smith P, Shuman S. Structure-function analysis of Plasmodium RNA triphosphatase and description of a triphosphate tunnel metalloenzyme superfamily that includes Cet1-like RNA triphosphatases and CYTH proteins. Rna. 2006;12:1468–1474. doi: 10.1261/rna.119806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong CL, Shuman S. Mapping the active site of vaccinia virus RNA triphosphatase. Virology. 2003;309:125–134. doi: 10.1016/s0042-6822(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Gross CH, Shuman S. RNA 5′-triphosphatase, nucleoside triphosphatase, and guanylyltransferase activities of baculovirus LEF-4 protein. J Virol. 1998;72:10020–10028. doi: 10.1128/jvi.72.12.10020-10028.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Rajashankar KR, Lima CD. Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA Capping Apparatus. Structure. 2010;18:216–227. doi: 10.1016/j.str.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagler J, Shuman S. A freeze-frame view of eukaryotic transcription during elongation and capping of nascent mRNA. Science. 1992;255:983–986. doi: 10.1126/science.1546295. [DOI] [PubMed] [Google Scholar]
- Hakansson K, Doherty AJ, Shuman S, Wigley, D X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- Hausmann S, Pei Y, Shuman S. Homodimeric quaternary structure is required for the in vivo function and thermal stability of Saccharomyces cerevisiae and Schizosaccharomyces pombe RNA triphosphatases. J Biol Chem. 2003;278:30487–30496. doi: 10.1074/jbc.M303060200. [DOI] [PubMed] [Google Scholar]
- Higman MA, Bourgeois N, Niles EG. The vaccinia virus mRNA (guanine-N7-)-methyltransferase requires both subunits of the mRNA capping enzyme for activity. J Biol Chem. 1992;267:16430–16437. [PubMed] [Google Scholar]
- Higman MA, Christen LA, Niles EG. The mRNA (guanine-7-)methyltransferase domain of the vaccinia virus mRNA capping enzyme. Expression in Escherichia coli and structural and kinetic comparison to the intact capping enzyme. J Biol Chem. 1994;269:14974–14981. [PubMed] [Google Scholar]
- Ho CK, Martins A, Shuman S. A yeast-based genetic system for functional analysis of viral mRNA capping enzymes. J Virol. 2000;74:5486–5494. doi: 10.1128/jvi.74.12.5486-5494.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein-Structure comparison by alignment of distance matrices. Journal of molecular biology. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Jain R, Shuman S. Polyphosphatase activity of CthTTM, a bacterial triphosphate tunnel metalloenzyme. J Biol Chem. 2008;283:31047–31057. doi: 10.1074/jbc.M805392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of roatation diffraction data from crystals of initially unknown symmetry and cell constants. Journal of applied crystallography. 1993;26:795–800. [Google Scholar]
- Lima CD, Wang LK, Shuman S. Structure and mechanism of yeast RNA triphosphatase: An essential component of the mRNA capping apparatus. Cell. 1999;99:533–543. doi: 10.1016/s0092-8674(00)81541-x. [DOI] [PubMed] [Google Scholar]
- Luo Y, Mao X, Deng L, Cong P, Shuman S. The D1 and D12 subunits are both essential for the transcription termination factor activity of vaccinia virus capping enzyme. J Virol. 1995;69:3852–3856. doi: 10.1128/jvi.69.6.3852-3856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Shuman S. Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit. Identification of amino acid residues in the D1 protein required for subunit association and methyl group transfer. J Biol Chem. 1994;269:24472–24479. [PubMed] [Google Scholar]
- Mao X, Shuman S. Vaccinia virus mRNA (guanine-7-)methyltransferase: mutational effects on cap methylation and AdoHcy-dependent photo-cross-linking of the cap to the methyl acceptor site. Biochemistry. 1996;35:6900–6910. doi: 10.1021/bi960221a. [DOI] [PubMed] [Google Scholar]
- Martin SA, Moss B. Modification of RNA by mRNA guanylyltransferase and mRNA (guanine-7-)methyltransferase from vaccinia virions. J Biol Chem. 1975;250:9330–9335. [PubMed] [Google Scholar]
- Martin SA, Moss B. mRNA guanylyltransferase and mRNA (guanine-7-)-methyltransferase from vaccinia virions. Donor and acceptor substrate specificites. J Biol Chem. 1976;251:7313–7321. [PubMed] [Google Scholar]
- Martin SA, Paoletti E, Moss B. Purification of mRNA guanylyltransferase and mRNA (guanine-7-) methyltransferase from vaccinia virions. J Biol Chem. 1975;250:9322–9329. [PubMed] [Google Scholar]
- Martins A, Shuman S. Mutational analysis of baculovirus capping enzyme Lef4 delineates an autonomous triphosphatase domain and structural determinants of divalent cation specificity. J Biol Chem. 2001;276:45522–45529. doi: 10.1074/jbc.M107615200. [DOI] [PubMed] [Google Scholar]
- Myette JR, Niles EG. Characterization of the vaccinia virus RNA 5′-triphosphatase and nucleoside triphosphate phosphohydrolase activities - Demonstration that both activities are carried out at the same active site. Journal of Biological Chemistry. 1996a;271:11945–11952. doi: 10.1074/jbc.271.20.11945. [DOI] [PubMed] [Google Scholar]
- Myette JR, Niles EG. Domain structure of the vaccinia virus mRNA capping enzyme - Expression in Escherichia coli of a subdomain possessing the RNA 5′-triphosphatase and guanylyltransferase activities and a kinetic comparison to the full-size enzyme. Journal of Biological Chemistry. 1996b;271:11936–11944. doi: 10.1074/jbc.271.20.11936. [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Shuman S, Lima CD. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell. 2006;127:71–84. doi: 10.1016/j.cell.2006.08.038. [DOI] [PubMed] [Google Scholar]
- Niles EG, Christen L. Identification of the Vaccinia virus messenger-RNA guanyltransferase active-site lysine. Journal of Biological Chemistry. 1993;268:24986–24989. [PubMed] [Google Scholar]
- Pei Y, Ho CK, Schwer B, Shuman S. Mutational analyses of yeast RNA triphosphatases highlight a common mechanism of metal-dependent NTP hydrolysis and a means of targeting enzymes to pre-mRNAs in vivo by fusion to the guanylyltransferase component of the capping apparatus. J Biol Chem. 1999;274:28865–28874. doi: 10.1074/jbc.274.41.28865. [DOI] [PubMed] [Google Scholar]
- Pei Y, Lehman K, Tian L, Shuman S. Characterization of Candida albicans RNA triphosphatase and mutational analysis of its active site. Nucleic Acids Res. 2000;28:1885–1892. doi: 10.1093/nar/28.9.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena L, Yanez RJ, Revilla Y, Vinuela E, Salas ML. African swine fever virus guanylyltransferase. Virology. 1993;193:319–328. doi: 10.1006/viro.1993.1128. [DOI] [PubMed] [Google Scholar]
- Saha N, Shuman S. Effects of alanine cluster mutations in the D12 subunit of vaccinia virus mRNA (guanine-N7) methyltransferase. Virology. 2001;287:40–48. doi: 10.1006/viro.2001.1006. [DOI] [PubMed] [Google Scholar]
- Saha N, Shuman S, Schwer B. Yeast-based genetic system for functional analysis of poxvirus mRNA cap methyltransferase. J Virol. 2003;77:7300–7307. doi: 10.1128/JVI.77.13.7300-7307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya R, Shuman S. Mutational analysis of the guanylyltransferase component of Mammalian mRNA capping enzyme. Biochemistry. 2003;42:8240–8249. doi: 10.1021/bi034396d. [DOI] [PubMed] [Google Scholar]
- Schnierle BS, Gershon PD, Moss B. Cap-specific mRNA (nucleoside-O2′-)-methyltransferase and poly(A) polymerase stimulatory activities of vaccinia virus are mediated by a single protein. Proc Natl Acad Sci U S A. 1992;89:2897–2901. doi: 10.1073/pnas.89.7.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Hausmann S, Schneider S, Shuman S. Poxvirus mRNA cap methyltransferase. Bypass of the requirement for the stimulatory subunit by mutations in the catalytic subunit and evidence for intersubunit allostery. J Biol Chem. 2006;281:18953–18960. doi: 10.1074/jbc.M602867200. [DOI] [PubMed] [Google Scholar]
- Schwer B, Shuman S. Genetic analysis of poxvirus mRNA cap methyltransferase: suppression of conditional mutations in the stimulatory D12 subunit by second-site mutations in the catalytic D1 subunit. Virology. 2006;352:145–156. doi: 10.1016/j.virol.2006.03.050. [DOI] [PubMed] [Google Scholar]
- Shuman S. Functional domains of Vaccinia virus messenger-RNA capping enzyme - Analysis by limited tryptic digestion. Journal of Biological Chemistry. 1989;264:9690–9695. [PubMed] [Google Scholar]
- Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nature Reviews Molecular Cell Biology. 2002;3:619–625. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- Shuman S, Hurwitz J. Mechanism of mRNA capping by vaccinia virus guanylyltransferase: characterization of an enzyme--guanylate intermediate. Proc Natl Acad Sci U S A. 1981;78:187–191. doi: 10.1073/pnas.78.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S, Lima CD. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Current Opinion in Structural Biology. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Shuman S, Surks M, Furneaux H, Hurwitz J. Purification and characterization of GTP-pyrophosphate exchange activity from Vaccinis virions - Association of the GTP-pyrophosphate exchange activity with Vaccinia messenger-RNA guanyltransferase RNA(Guanine-7-)methyltransferase complex (capping enzyme) Journal of Biological Chemistry. 1980;255:1588–1598. [PubMed] [Google Scholar]
- Terwilliger TC. Improving macromolecular atomic models at moderate resolution by automated iterative model building, statistical density modification and refinement. Acta crystallographica. Section D, Biological crystallography. 2003;59:1174–1182. doi: 10.1107/S0907444903009922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiggemann M, Jeske S, Larsen M, Meinhardt F. Kluyveromyces lactis cytoplasmic plasmid pGKL2: heterologous expression of Orf3p and proof of guanylyltransferase and mRNA-triphosphatase activities. Yeast. 2001;18:815–825. doi: 10.1002/yea.728. [DOI] [PubMed] [Google Scholar]
- Venkatesan S, Gershowitz A, Moss B. Purification and characterization of mRNA guanylyltransferase from HeLa cell nuclei. J Biol Chem. 1980;255:2829–2834. [PubMed] [Google Scholar]
- Wang SP, Deng L, Ho CK, Shuman S. Phylogeny of mRNA capping enzymes. Proc Natl Acad Sci U S A. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Martins A, Deng L, Shuman S. Structure-function analysis of the triphosphatase component of vaccinia virus mRNA capping enzyme. J Virol. 1997;71:9837–9843. doi: 10.1128/jvi.71.12.9837-9843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Shuman S. Mutational analysis of the RNA triphosphatase component of vaccinia virus mRNA capping enzyme. J Virol. 1996;70:6162–6168. doi: 10.1128/jvi.70.9.6162-6168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Shuman S. Mutational analysis of vaccinia virus mRNA cap (guanine-N7) methyltransferase reveals essential contributions of the N-terminal peptide that closes over the active site. Rna. 2008a;14:2297–2304. doi: 10.1261/rna.1201308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Shuman S. Structure-function analysis of vaccinia virus mRNA cap (guanine-N7) methyltransferase. Rna. 2008b;14:696–705. doi: 10.1261/rna.928208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.