Abstract
The proteasome is the main proteolytic machine in the cytosol and nucleus of eukaryotic cells where it degrades hundreds of regulatory proteins, removes damaged proteins, and produces peptides that are presented by MHC complexes. New structures of the proteasome particle show how its subunits are arranged and provide insights into how the proteasome is regulated. Proteins are targeted to the proteasome by tags composed of several ubiquitin moieties. The structure of the tags tunes the order in which proteins are degraded. The proteasome itself edits the ubiquitin tags and drugs that interfere in this process can enhance the clearance of toxic proteins from cells. Finally, the proteasome initiates degradation at unstructured regions within its substrates and this step contributes to substrate selection.
Cellular protein concentrations are controlled through their rates of synthesis and degradation. In the cytosol and nucleus of eukaryotic cells, most of this degradation is by the ubiquitin proteasome system (UPS). At the center of the UPS is a single proteolytic machine, the proteasome, which controls the concentrations of hundreds of regulatory proteins and clears misfolded and damaged proteins from the cell. Thus, the proteasome has to be able to degrade any protein but do so while avoiding the accidental destruction of the rest of the cellular proteome. Here we review recent advances in our understanding of how the proteasome selects its substrates. Just as protein synthesis is regulated at many different levels, it is becoming increasingly clear how protein degradation is also.
The basic principle of proteasome substrate selection is well understood [1,2]. The proteasome is a large particle of ~33 different subunits that add up to a molecular weight of approximately 2.5 MDa. It combines three different proteolytic sites with broad and complementary sequence preferences to allow it to degrade many different amino acid sequences. The proteasome particle controls the activity of these sites by encapsulating them inside its structure and controlling access to them. Most proteins are targeted to the proteasome by the covalent attachment of ubiquitin molecules. The proteasome recognizes the ubiquitin signal and initiates degradation at an unstructured region in the protein. The substrate is then unfolded and translocated to the proteolytic sites in an ATP-dependent reaction. However, many questions remain. For example, the proteasome is able to extract individual subunits from complexes without degrading their binding partners, the proteasome degrades ubiquitinated proteins in a specific order and ubiquitin signals target proteins to processes that do not involve degradation. We do not know how the proteasome makes these distinctions. At the same time, some proteins that lack ubiquitin signals are degraded by the proteasome. Over the last few years, new proteasome structures and biochemical investigations have brought new insights into these questions.
1. Proteasome
The proteasome particle is functionally and structurally divided into two parts. Its core is formed by a cylindrical 20S particle composed of four heptameric rings that are stacked onto top of each other. The inner two rings each consist of seven related β-subunits that are arranged to form a large internal cavity and three of the subunits in each ring contain a proteolytic site that faces the internal cavity. A ring of seven related α-subunits on each side flanks the β-rings and substrates have to enter the proteolytic cavity formed by the β-rings through a pore at the top of the α-ring. The pore is too narrow to allow folded proteins to pass through it. In free core particle, access to the pores is further hindered by the N-termini of the α-subunits so that even unfolded peptides are degraded only poorly.
The core particle is activated by regulatory particles or caps that bind to the ends of the core particle and induce conformational changes that open the pores. Four different caps are known and the best understood of them is the19S regulatory particle. It consists of 19 subunits that add up to a molecular weight of ~900kD. The complex of one or two of these caps with the 20S core particle is called the 26S proteasome and this seems to be the most common form of the proteasome in cells. The subunits of the 19S cap recognize substrates, unfold and translocate them into the core particle for degradation into short peptides.
1.1 Structure of the 26S proteasome
The structure of the 26S proteasome proved difficult to determine, perhaps because a number of accessory factor associate with the particle non-stoichiometrically or because of the structure undergoes conformational changes. In a major breakthrough, a series of studies published over the last two years describe the structure of the 19S cap bound to the core particle at high resolution by combining cryo-electronmicroscopy, crystallography, biochemical data and computer modeling [3**-10**] (Figure 1).
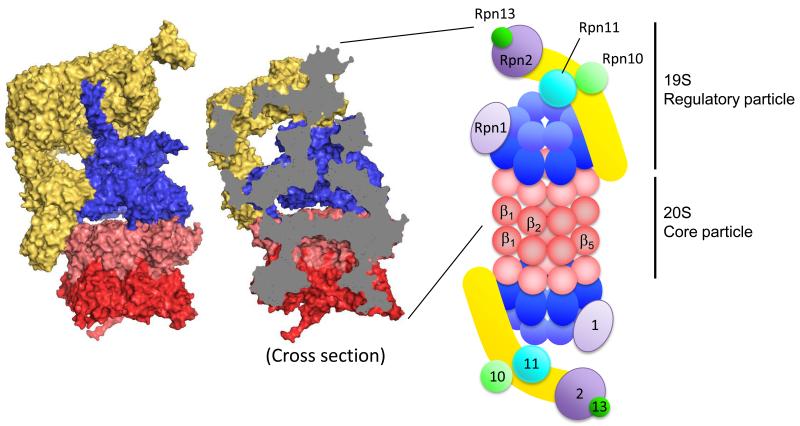
Figure 1.
Structure of the 26S proteasome. Molecular surface of the 19S activator particle bound to the 20S core particle (PDB 4C0V) (left). The 20S core particle is composed of central two β rings (dark red) and outer two α rings (light red) at either end. The 19S regulatory particle, which contains AAA ATPase subunits (blue) and non-ATPase subunits (yellow), caps either end of the 20S. Cross section reveals the degradation channel that connects the proteolytic chamber in the 20S core particle to the entrance of 19S activator (middle). Structures are produced by PyMOL. Schematic drawing of the 26S proteasome indicates the approximate locations of the enzymatic activities and binding platforms on the 19S activator cap (right). α (light red) and β (dark red) subunits of the 20S particle, ATPase domain (dark blue) and OB domain (light blue) of ATPase subunits, backbone of lid sub particle (yellow), docking subunits Rpn1 (light purple) and Rpn2 (dark purple), ubiquitin receptors Rpn10 (light green) and Rpn13 (dark green), and DUB metallo-protease subunit Rpn11 (sky blue).
The heart of the 19S cap is a ring of six ATPase subunits (Rpt1-Rpt6), which make up the motor that feeds substrates to the proteolytic sites. The subunits form a long channel at their center that runs through approximately two thirds of the 19S particle and ends in a ring of the AAA+ domains at the C-terminal end of the ATPase subunits. The very C-termini of the AAA+ domains dock into the 20S core particle and trigger pore opening. Two large subunits that serve as interaction platforms bind to the ATPase ring, Rpn1 to the outside of the ring, and Rpn2 to the top of the ring. Rpn1 provides the binding sites for a series of non-stoichiometric proteasome subunits called UbL-UBA proteins, which serve as additional ubiquitin receptors and we will discuss these briefly later, and Rpn2 organizes the two ubiquitin receptors Rpn10 and Rpn13 subunit near the outer end of the 19S cap. No single one of these receptors is essential in yeast [11**] so that it seems that the different receptors work together to form a versatile binding platform to capture proteasome substrates (Figure 3). The cap also contains a pair of JAMM or MPN domain metallo-protease subunits called Rpn11 and Rpn8. Only Rpn11 is enzymatically active and it cleaves entire ubiquitin chains of the substrates as these are degraded. Rpn11 is located near the entrance of the substrate channel formed by the ATPase subunits so that it is well place to interact with substrate protein feeding into the proteasome. Thus, the activities required for protein degradation are ordered sequentially along the long axis of the proteasome particle [2] (Figure 1).
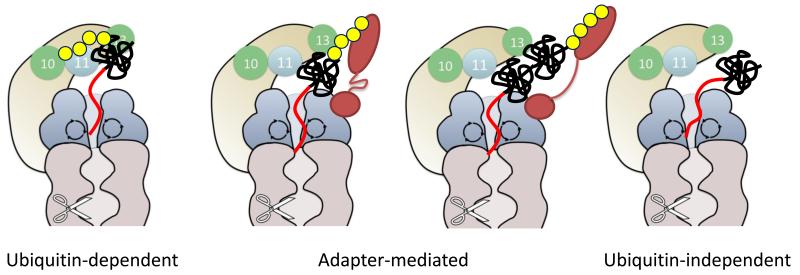
Figure 3.
The proteasome recognizes substrates in three different modes; ubiquitin-dependent (left), adapter-mediated (middle), and ubiquitin-independent (right) modes. In all three, an intrinsically disordered region in the substrate is recognized by the ATPase motor to allow the proteasome to initiate degradation. This aspect of proteasomal degradation resembles the targeting mechanisms predominant with the bacterial and archaeal analogues of the proteasome. Ubiquitin tags can be either recognized by the two intrinsic proteasome receptors Rpn10 and Rpn13 (left), or by non-stoichiometric proteasome subunits that serve as substrate adaptors such as UbL-UBA proteins (middle). The UbL-UBA proteins might bind substrates by themselves (second right) or together with the intrinsic substrate receptors (second left), which facilitate degradation of various substrates by positioning the disordered region properly. Finally, some substrates may be recognized only by their initiation sites.
The remainder of the cap is formed by seven scaffolding subunits that form a clamp that binds to the side of the cap reaching all the way from the end of the proteasome particle, where it interacts with Rpn2 and the ubiquitin receptor Rpn10, via the ATPase subunits, down to the α-ring of the core particle. The clamp subunits complete a network of interactions that seems to stabilize the proteasome particle and may allow allosteric regulation and coordination between the activities on the proteasome particle. Biochemical experiments have shown substrate and ATP binding can affect gating of the substrate channel and proteolytic activity and binding of polyubiquitinated substrate stimulates proteolysis [12-14]. Comparison of the proteasome structures in the presence of ATP but without substrate, in the presence of ATP and with substrate bound, and in the presence of a slowly hydrolysable ATP analogue reveal substantial conformational changes in the structure [9**,10**]. For example, substrate or ATP analog binding switches the cap from a presumably inactive conformation in the substrate channel is discontinuous to a conformation in which the channel is properly aligned through the entire proteasome particle and the active site of Rpn11 swings in and out of alignment with the channel entrance. The ATPase subunits switch between arrangements in which they form spiral or a planar ring but it is not clear to what extend these changes reflect motor action that drives substrate into the proteasome or switches between resting and active states.
1-2. Alternative proteasome activators
Recently, Barthelme and Sauer found that the chaperone Cdc48 can also form a complex with the 20S core particle and support the degradation of substrate proteins [15**,16**]. Cdc48, called p97 or VCP in animals, is a cytosolic chaperone distantly related to ATPase subunits in the 19S regulatory particle and involved in the degradation of a subset of proteasome substrates by a poorly defined mechanism. For example, it is part of the quality control process for endoplasmic reticulum proteins (ERAD) where it is required for the translocation of misfolded proteins from the ER to cytosolic proteasomes [17]. It now appears that Cdc48/p97/VCP may be directly involved in degradation by serving as an alternative proteasome cap, perhaps to unfold a different subsets of proteins than the 19S cap. Proteasome with Cdc48 caps would resemble the archaeal proteasome and the analogous bacterial AAA+ proteases. These proteases fulfill similar functions as the eukaryotic proteasome and share the same overall architecture [18].
Two other further types of proteasome caps are known, called the 11S particle and the PA200 activator. These caps neither recognize ubiquitin nor hydrolyze ATP and their role seems to be to degrade a specific subset of substrate and some unstructured proteins [1] [19].
2. Ubiquitination
2-1. Ubiquitination system
Most proteins are targeted to the proteasome by ubiquitin tags or degrons. Ubiquitin is attached to the target proteins through the sequential action of a ubiquitin activating enzyme (E1), a ubiquitin conjugating enzyme (E2), and a ubiquitin ligase (E3). In most cases, ubiquitin forms an isopeptide bond through its C-terminal carboxy group (Gly76 of ubiquitin) with the ε-amino group of lysine residues in the substrate, and more rarely with the N-terminus of the polypeptide chain or the side chain of a cysteine residue in the substrate protein [20-22]. Typically ubiquitin is attached to more than one residue in the target proteins and in many cases, a second ubiquitin is then attached to a lysine residue in the first ubiquitin and so on to create polyubiquitin chains. In addition, cells contains are large number of deubiquitinating enzymes (DUBs) that remove ubiquitin chains again [23].
2-2. Ubiquitin signals
Thousands of proteins are ubiquitinated in yeasts cell, but almost half of ubiquitinated proteins are not targeted to the proteasomal degradation [24] and it is not clear how the cell differentiates between the different ubiquitin signals. The canonical view is that ubiquitin chain linked through Lys48 of ubiquitin target to the proteasome and biochemical experiments show that chains of at least four ubiquitin moieties are required for proper recognition [20,25]. Modification with a single ubiquitin molecule or through polyubiquitin chains linked through other Lys residues such as Lys63 and even linear ubiquitin chains play roles in cellular processes that do not involve the proteasome such as the regulation of chromatin structure, membrane trafficking and a signal transduction. However, the distinctions are not strict and Lys63-linked polyubiquitin chains [26,27] and even monoubiquitin tags [28-30] can target some substrates to the proteasome for degradation. Purified proteasome binds the Lys63-linked polyubiquitin chain with almost the same affinity as the Lys48-linked polyubiquitin chain [31*] and so specificity may come from accessory proteins. For example the ESCRT0 protein involved in membrane trafficking binds Lys63-linked polyubiquitin chains better than Lys48-linked chains whereas the UbL-UBA proteins that can serve as non-stoichiometric ubiquitin receptors for the proteasome have the opposite preference [31*]. Therefore, a Lys48-linked polyubiquitin chain has a greater chance to be delivered to the proteasome than the Lys63-linked polyubiquitin chain. A different possibility is that physical properties of the substrate proteins themselves, such as their stability against unfolding [32] and the presence of initiation sites for the proteasome [33**, 34] contribute to specificity as processes such as membrane trafficking or the formation of signaling complexes do not require protein unfolding and do not involve initiation.
2-3 Dynamic regulation of ubiquitination
Ubiquitination is not a simple switch that turns degradation on and off, but rather an adjustable signal that fine-tunes degradation and can determine the order in which proteins in a regulatory pathway are degraded. For example, the progression of cells through the cell division cycle requires the degradation of regulatory proteins in the correct sequence. Degradation can be ordered by timing the ubiquitination event and many E3s recognize their substrates only when their interaction site is first phosphorylated by a kinase [22]. Degradation order is also controlled by the nature of the ubiquitin modification and during the cell cycle, regulators that acquire long ubiquitination chains are degraded before the regulators that are ubiquitinated with multiple shorter chains [35*,36*]. The regulators are ubiquitinated by the same E3 but for the early substrates ubiquitination is more processive than for the late substrates probably because the substrates have different dissociation rates from the E3 [35*,36*].
Ubiquitin tags on proteins can grow and shrink even while bound to the proteasome through the action of E3 and DUB enzymes associated with the proteasome. In yeast, the DUB Ubp6 and in mammalian cells the Ubp6 homologue Usp14 and the DUB Uch37 bind to the 19S proteasome cap [1]. These DUBs trim ubiquitin chains from the distal end of the chain in steps of one or a few ubiquitin moieties at a time and thus limit the time that a substrate remains associated with the proteasome [37,38**]. Hence, proteins that are difficult to degrade because they cannot be unfolded or because they lack good initiations sites would dissociate from the proteasome after it tried to degrade them for a limited time, freeing up the proteasome for a different substrate and preventing it from clogging up. On the flipside, inhibitors of proteasome DUB Usp14 show promise as drug for the treatment of neurodegenerative diseases by increasing the proteasome’s ability to degrade resistant substrates, presumably by increasing their interaction with the proteasome [38**]. Small molecular inhibitors of proteasomal DUBs are also tested in cancer therapy but here the drugs affect degradation differently and lead to the accumulation of ubiquitinated proteins [39] so that the biological effect may be similar to that of the proteasome inhibitors already used to treat multiple myeloma [40].
E3s also bind the proteasome [41]. In particular, the E3 Hul5 is associated with Ubp6 on the 19S activator of the proteasome where it counteracts the activity of Ubp6 by increasing the length of polyubiquitin chains [42**]. Ubiquitin chain editing may serve to fine-tune degradation rates or to make protein targeting more robust by buffering fluctuations in ubiquitin chains and substrate stability. Another possibility is that ubiquitin ligation on the proteasome makes degradation more processive to avoid the formation of partially degraded protein fragments [43] by re-ubiquitinating long proteins as the proteasome runs along their polypeptide chain [44].
3. A second component to the proteasome targeting code?
3-1. Initiation of degradation
The proteasome recognizes and binds its substrates through their polyubiquitin tag but initiates degradation at a disordered region in the substrate [33**,45] (Figure 2). Once the substrate is engaged at the initiation site, the proteasome proceeds along the polypeptide chain from there to unfold and degrade the entire protein sequentially [32]. The initiation region is reminiscent of the linear targeting signals found in substrates of the archaeal and bacterial analogues of the proteasome [18]. Bacterial AAA+ proteases recognize their linear degrons through loops that line the pore at the center of the ring of ATPase subunits and it seems likely that the proteasome recognizes its initiation sites similarly [46]. In the proteasome, the equivalent loops line the degradation channel at a position some 30 - 60 Å in from the entry pore. The diameter of the pore is too narrow to allow folded proteins to pass through it so that a disordered polypeptide tail would have to be at least 20-30 amino acids long to be able to reach the ATPase loops. This length requirement agrees roughly with the results of in vitro degradation experiments with model proteasome substrates, where proteins become degraded rapidly by purified yeast proteasome once they contain an unstructured tail of approximately 30 amino acids in length [33**,45,47].
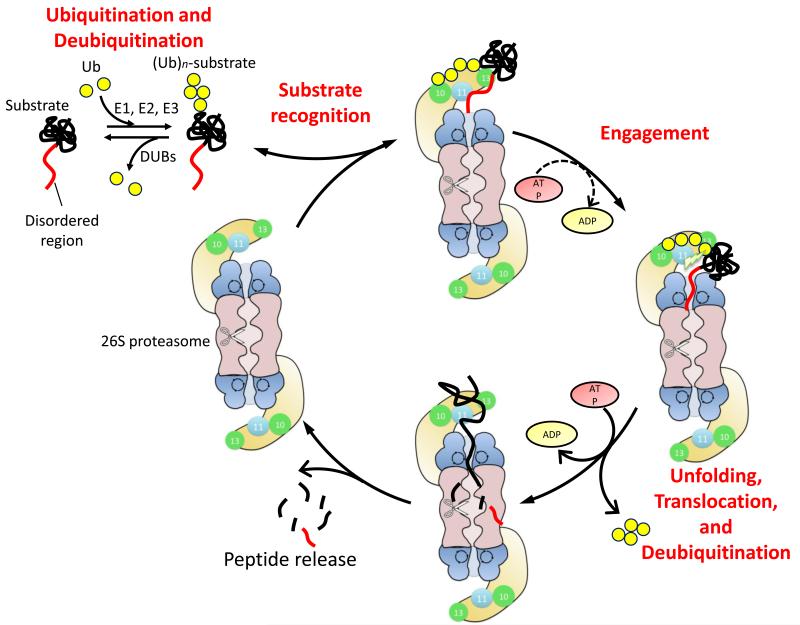
Figure 2.
Schematic representation of the degradation cycle of the ubiquitin proteasome system. Proteins are targeted to the proteasome by a two-part degradation signal or degron. It consists of an intrinsically disordered region within the substrate and a reversibly attached polyubiquitin tag (Ubn). Polyubiquitin tag is attached by E1-E2-E3 ubiquitination cascade and this process can be reversed by DUBs (top left). The proteasome recognizes its substrates at the ubiquitin tag through ubiquitin receptors (Rpn10 and Rpn13; green) (top) and initiates degradation at the unstructured region (right). Once the proteasome has engaged its substrate, it unravels the protein by translocating it into a central cavity in the core particle, where the protein is proteolysed sequentially (bottom). Polyubiquitin tag is cleaved off by the intrinsic DUB Rpn11 (skyblue) immediately before the degradation.
The requirement of unstructured initiation regions may also be reflected in the global stability profiles of proteins. At least 30% of eukaryotic proteins contain intrinsically disordered regions (IDRs) and these are involved in various cellular activities [48,49]. There is some evidence from bioinformatics studies that proteins that contain intrinsically disordered regions have on average shorter half-lives than proteins lacking these regions [50,51] but so far the evidence for this relationship is not consistent. Other studies do not find these correlations [52-54] and there is some evidence that ubiquitination sites of proteasome substrates are preferentially located in unstructured regions [55,56]. Even when the unstructured region is not found in a substrate protein, ubiquitination itself may induce the local unfolding near the ubiquitinated residue, which, in turn, could create an initiation site for the proteasome [57].
3-2. Degradation of protein complexes
Ubiquitin tag and initiation site do not have to be located on the same polypeptide chain but can work together in trans so that a ubiquitinated subunit in a complex can target a binding partner for degradation [34]. The ubiquitinated subunit serves as an adaptor that binds to the proteasome and presents the bound protein for proteolysis. Presumably, UbL-UBA proteins function in this manner to serve non-stoichiometric ubiquitin receptors for the proteasome [1,58]. These proteins bind to the proteasome through their UbL (ubiquitin-like) domains and to ubiquitinated proteins through their UBA (ubiquitin associated) domains and stimulate degradation of the ubiquitinated protein while the UbL-UBA proteins themselves escape degradation. The mechanism behind this unexpected stability of UbL proteins has been investigated for yeast Rad23 [59-61]. These experiments showed that Rad23 escapes degradation because it lacks an effective proteasome initiation site [60,61].
The flipside of this mechanism is also observed and the proteasome is able to remodel protein complexes by degrading only the ubiquitinated subunit and leaving other proteins in the complex intact [62,63]. This remodeling activity is important in many regulatory processes in the cell. For example, during cell cycle regulation in yeast, the proteasome extracts the cyclin-dependent kinase inhibitor Sic1 from its complex with cyclin and cyclin-dependent kinase to degrade solely Sic1 [64]. Shortly afterwards, the cyclin is ubiquitinated and then degraded to release intact but inactive kinase [65]. Since the proteasome is able to degrade proteins that are bound to the proteasome indirectly it is unlikely that ubiquitination by itself specifies target selection. Presumably, the proteasome instead determines which subunit is degraded by where it initiates degradation. Once the polypeptide chain of a subunit is fed into the degradation channel, the proteasome will proceed along that chain and hydrolyze the protein sequentially [32]. The most likely initiation site for the proteasome is probably the unstructured region closest to the entrance to the degradation channel. Indeed, biochemical experiments show that initiation regions must be placed at the appropriate distance from the ubiquitin tag for a protein to be degraded, presumably so that the proteasome can bind the ubiquitin tag and engage the initiation region simultaneously [47]. Thus, under some circumstances, the proteasome may select substrates at the initiation step.
3-4. Ubiquitin-independent substrates
A range of proteins is degraded by the proteasome without being ubiquitinated [66] and the best understood example is ornithine decarboxylase (ODC) [67,68]. Degradation of ODC requires ATP and an accessory protein called antizyme and begins a 37 amino acid long unstructured region at the C terminus of ODC [68]. To some extent, this ODC tail can function as a transferable degradation signal and attaching it to certain other proteins causes their degradation. One plausible explanation for the ubiquitin-independent degradation is that the unstructured regions themselves have bind sufficiently tightly to the to the ATPase ring loops so that ubiquitin is not required for proteasome association (Figure 3). Thus, this targeting mechanism can be taken as a variation of the conventional proteasome degron in which the ubiquitin tag component is missing and which resembles the degrons observed in the archaea and bacteria [18].
Several other proteasome substrates including p21/Cip1, c-Jun, c-Fos, p53, p73 IκBα, T-cell antigen receptor chain α, Fra-1, and Hif-1α, can also be degraded in an ubiquitin-independent manner [69-71]. The mechanisms of these processes are not well understood and it is possible that these proteins are degraded by isolated 20S core particle in the absence of ATP [69], and in vivo perhaps more likely by 20S core particle activated by alternative caps [70] or even by 26S proteasome [71]. The proteins in this group of ubiquitin-independent proteasome substrates are largely unstructured, but their degradation can still be regulated. The best understood example of this regulation is given by NQO1 [72,73*] NQO1 is largely unstructured and can be degraded by 20S proteasome in vitro. Binding of NQO1’s cofactor FAD stabilizes the protein’s structure and inhibits its proteasomal degradation. Quite interestingly, FAD binding to NQO1 also stabilizes other ubiquitin-independent proteasome substrates, setting up a regulatory circuit controlled by the availability of FAD and thus the metabolic state of the cell.
4. System-wide studies of the UPS
The mechanisms described above are largely derived from investigations of the behavior specific proteins in vitro or in the cell. Over the last five years, high-through studies have begun to provide a system-level picture of how the UPS regulates protein concentrations. Improvements in mass spectroscopy technology and in the strategies for sample preparation are making it possible to define the set of proteins that are ubiquitinated in the cells and the nature of their ubiquitin modifications [74-76]. So far, the sets of ubiquitinated proteins identified overlap only partially suggesting that current the experiments do not yet capture all ubiquitinated proteins [74]. The studies still provide valuable insights, for example by describing the wide range of polyubiquitin chains made in cells [27] and the fraction of nascent proteins that are ubiquitinated as part of protein quality control surveillance [77,78].
Other approaches measure the stability and turnover rates of a large fraction of the proteins in cells. The first experiments used the tagged protein collection in yeast and followed their degradation by cycloheximide shut-off and Western blotting [79] and later measurements in mammalian cells use SILAC [80] or fluorescent protein fusions [52,81]. These studies show that protein halftimes in eukaryotic cells range over at least two orders of magnitudes and thus that proteins concentrations are indeed adjusted by the balance of synthesis and degradation. Combining protein stability measurements with the ubiquitination databases, or with chemical inhibition of protein ubiquitin ligases provides increasing depth to our understanding of the regulation of cellular protein stability [50-53,82].
Summary
As we begin to understand the mechanism of the UPS in increasing biochemical detail it is becoming increasing clear that the regulation of degradation is far richer than the binary decision of degradation or no degradation. Just like protein synthesis is tuned by a myriad of processes, we are discovering new ways in which their degradation is tuned. Recent structural and biochemical discoveries have provided a range of novel paradigms that govern proteasome action and new experimental strategies make it possible to observe protein ubiquitination and degradation system-wide. It will be interesting to see whether and how they are used in the cell.
The UPS adjusts intracellular protein concentrations through degradation.
High-resolution structures of entire proteasome particle provide clues on mechanism
Ubiquitin tags can be tuned to adjust the order and rate of degradation
Ubiquitin tags are edited on the proteasome and this process can be targeted by drugs
Degradation signal has a second component in the proteasome initiation region
System-wide studies of ubiquitin tags and degradation rates are emerging
Acknowledgements
This work was supported by Welch Foundation grant F-1817, Gates Foundation grant OPP1061182, and NIH grant U54GM105816.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomko RJ, Jr., Hochstrasser M. Molecular Architecture and Assembly of the Eukaryotic Proteasome. Annu Rev Biochem. 2013;82:415–445. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3**.Bohn S, Beck F, Sakata E, Walzthoeni T, Beck M, Aebersold R, Förster F, Baumeister W, Nickell S. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc Natl Acad Sci USA. 2010;107:20992–20997. doi: 10.1073/pnas.1015530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4**.Sakata E, Bohn S, Mihalache O, Kiss P, Beck F, Nagy I, Nickell S, Tanaka K, Saeki Y, Förster F, et al. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc Natl Acad Sci USA. 2012;109:1479–1484. doi: 10.1073/pnas.1119394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6**.Lasker K, Förster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci USA. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7**.Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci USA. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8**.da Fonseca PCA, He J, Morris EP. Molecular model of the human 26S proteasome. Mol Cell. 2012;46:54–66. doi: 10.1016/j.molcel.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 9**.Matyskiela ME, Lander GC, Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol. 2013;20:781–788. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10**.Śledź P, Unverdorben P, Beck F, Pfeifer G, Schweitzer A, Förster F, Baumeister W. Structure of the 26S proteasome with ATP-γS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc Natl Acad Sci USA. 2013;110:7264–7269. doi: 10.1073/pnas.1305782110. This set of papers [3-10] describes the structure of the 26S proteasome in almost atomic resolution by combining a range of experimental approaches to complement cryo-electron microscopy structures. The papers reveal the location of the major ubiquitin receptors, the location of the main deubiquitinating activity, ATPase motors, as well as the network of interactions between them and some of the conformational changes they undergo.
- 11**.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. This paper describes the discovery that Rpn13 serves as a second intrinsic ubiquitin receptor and describes that the different ubiquitin receptors have overlapping functions.
- 12.Bech-Otschir D, Helfrich A, Enenkel C, Consiglieri G, Seeger M, Holzhütter H-G, Dahlmann B, Kloetzel P-M. Polyubiquitin substrates allosterically activate their own degradation by the 26S proteasome. Nat Struct Mol Biol. 2009;16:219–225. doi: 10.1038/nsmb.1547. [DOI] [PubMed] [Google Scholar]
- 13.Peth A, Besche HC, Goldberg AL. Ubiquitinated Proteins Activate the Proteasome by Binding to Usp14/Ubp6, which Causes 20S Gate Opening. Mol Cell. 2009;36:794–804. doi: 10.1016/j.molcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Demartino GN. Variably modulated gating of the 26S proteasome by ATP and polyubiquitin. Biochem J. 2009;2009;421:397–404. doi: 10.1042/BJ20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.Barthelme D, Sauer RT. Identification of the Cdc48*20S proteasome as an ancient AAA+ proteolytic machine. Science. 2012;337:843–846. doi: 10.1126/science.1224352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Barthelme D, Sauer RT. Bipartite determinants mediate an evolutionarily conserved interaction between Cdc48 and the 20S peptidase. Proc Natl Acad Sci USA. 2013;110:3327–3332. doi: 10.1073/pnas.1300408110. In [15] and [16], the authors find that the Cdc48 AAA ATPase can form a complex with the 20S core particle to degrade proteins. Eukaryotic Cdc48 may serve as an alternative proteasome activator than the 19S particle.
- 17.Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–1167. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 19.Stadtmueller BM, Hill CP. Proteasome activators. Mol Cell. 2011;41:8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 21.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deshaies RJ, Joazeiro CAP. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 23.Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 24.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh-E A, Tanaka K. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009;28:359–371. doi: 10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boutet SC, Disatnik M-H, Chan LS, Iori K, Rando TA. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130:349–362. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 29.Shabek N, Herman-Bachinsky Y, Buchsbaum S, Lewinson O, Haj-Yahya M, Hejjaoui M, Lashuel HA, Sommer T, Brik A, Ciechanover A. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol Cell. 2012;48:87–97. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Dimova NV, Hathaway NA, Lee B-H, Kirkpatrick DS, Berkowitz ML, Gygi SP, Finley D, King RW. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat Cell Biol. 2012;14:168–176. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31*.Nathan JA, Kim HT, Ting L, Gygi SP, Goldberg AL. Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 2013;32:552–565. doi: 10.1038/emboj.2012.354. The authors provide insights into how the cell interprets poly ubiquitin chains with different linkages by showing that Lys63-linked polyubiquitin chain binds accessory proteins that compete for proteasome association, resulting in inefficient targeting of Lys63-linked chain for degradation.
- 32.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 33**.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. This paper suggests that the proteasome degron has a second component in the form of a unstructured region at which the proteasome initiates degradation
- 34.Prakash S, Inobe T, Hatch AJ, Matouschek A. Substrate selection by the proteasome during degradation of protein complexes. Nat Chem Biol. 2009;5:29–36. doi: 10.1038/nchembio.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35*.Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. The authors reveal that the E3 anaphase-promoting complex (APC) coordinates the order of substrate degradation during the cell cycle. They show that the timing by which substrates are degraded depends on the processivity of their ubiquitination by APC.
- 36*.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 37.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 38**.Lee B-H, Lee MJ, Park S, Oh D-C, Elsasser S, Chen P-C, Gartner C, Dimova N, Hanna J, Gygi SP, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. Inhibition of Usp14 by its specific inhibitor enhances the proteasomal degradation, probably because the inhibitor suspends deubiquitination and increases the chance to be recognized by the proteasome. It introduces DUB inhibitors as a therapeutic strategy for neurodegenerative diseases.
- 39.D’Arcy P, Brnjic S, Olofsson MH, Fryknäs M, Lindsten K, De Cesare M, Perego P, Sadeghi B, Hassan M, Larsson R, et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17:1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 40.Kisselev AF, van der Linden WA, Overkleeft HS. Proteasome inhibitors: an expanding army attacking a unique target. Chem Biol. 2012;19:99–115. doi: 10.1016/j.chembiol.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- 42**.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. The paper shows that proteasome associates with ubiquitin ligases and deubiquitinating enzymes and proposes that the balance between these two opposing activities may regulate the proteasome’s substrate specificity.
- 43.Tian L, Holmgren RA, Matouschek A. A conserved processing mechanism regulates the activity of transcription factors Cubitus interruptus and NF-kappaB. Nat Struct Mol Biol. 2005;12:1045–1053. doi: 10.1038/nsmb1018. [DOI] [PubMed] [Google Scholar]
- 44.Aviram S, Kornitzer D. The ubiquitin ligase Hul5 promotes proteasomal processivity. Mol Cell Biol. 2010;30:985–994. doi: 10.1128/MCB.00909-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeuchi J, Chen H, Coffino P. Proteasome substrate degradation requires association plus extended peptide. EMBO J. 2007;26:123–131. doi: 10.1038/sj.emboj.7601476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beckwith R, Estrin E, Worden EJ, Martin A. Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nat Struct Mol Biol. 2013;20:1164–1172. doi: 10.1038/nsmb.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inobe T, Fishbain S, Prakash S, Matouschek A. Defining the geometry of the two-component proteasome degron. Nat Chem Biol. 2011;7:161–167. doi: 10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 49.Babu MM, van der Lee R, de Groot NS, Gsponer J. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol. 2011;21:432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 50.Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tompa P, Prilusky J, Silman I, Sussman JL. Structural disorder serves as a weak signal for intracellular protein degradation. Proteins. 2008;71:903–909. doi: 10.1002/prot.21773. [DOI] [PubMed] [Google Scholar]
- 52.Yen H-CS, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 53.Yen H-CS, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923–929. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- 54.Suskiewicz MJ, Sussman JL, Silman I, Shaul Y. Context-dependent resistance to proteolysis of intrinsically disordered proteins. Protein Sci. 2011;20:1285–1297. doi: 10.1002/pro.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, Goebl MG, Iakoucheva LM. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 2010;78:365–380. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hagai T, Azia A, Tóth-Petróczy Á , Levy Y. Intrinsic disorder in ubiquitination substrates. J Mol Biol. 2011;412:319–324. doi: 10.1016/j.jmb.2011.07.024. [DOI] [PubMed] [Google Scholar]
- 57.Hagai T, Levy Y. Ubiquitin not only serves as a tag but also assists degradation by inducing protein unfolding. Proc Natl Acad Sci USA. 2010;107:2001–2006. doi: 10.1073/pnas.0912335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 59.Heessen S, Masucci M, Dantuma N. The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol Cell. 2005;18:225–235. doi: 10.1016/j.molcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 60.Fishbain S, Prakash S, Herrig A, Elsasser S, Matouschek A. Rad23 escapes degradation because it lacks a proteasome initiation region. Nat Commun. 2011;2:192. doi: 10.1038/ncomms1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heinen C, Acs K, Hoogstraten D, Dantuma NP. C-terminal UBA domains protect ubiquitin receptors by preventing initiation of protein degradation. Nat Commun. 2011;2:191. doi: 10.1038/ncomms1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson E, Gonda D, Varshavsky A. cis-trans recognition and subunit-specific degradation of short-lived proteins. Nature. 1990;346:287–291. doi: 10.1038/346287a0. [DOI] [PubMed] [Google Scholar]
- 63.Hochstrasser M, Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast alpha 2 repressor. Cell. 1990;61:697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- 64.Verma R, McDonald H, Yates JR, Deshaies RJ. Selective degradation of ubiquitinated Sic1 by purified 26S proteasome yields active S phase cyclin-Cdk. Mol Cell. 2001;8:439–448. doi: 10.1016/s1097-2765(01)00308-2. [DOI] [PubMed] [Google Scholar]
- 65.Nasmyth K, Shirayama M, Tóth A, Gálová M. APC: Cdc20: promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5: Article: Nature. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- 66.Baugh JM, Viktorova EG, Pilipenko EV. Proteasomes Can Degrade a Significant Proportion of Cellular Proteins Independent of Ubiquitination [Internet] Journal of Molecular Biology. 2009;386:814–827. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 68.Zhang M, Pickart CM, Coffino P. Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin-independent substrate. EMBO J. 2003;22:1488–1496. doi: 10.1093/emboj/cdg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Orlowski M, Wilk S. Catalytic activities of the 20 S proteasome, a multicatalytic proteinase complex. Arch Biochem Biophys. 2000;383:1–16. doi: 10.1006/abbi.2000.2036. [DOI] [PubMed] [Google Scholar]
- 70.Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol Cell. 2007;26:843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Erales J, Coffino P. Ubiquitin-independent proteasomal degradation. 2014;1843:216–221. doi: 10.1016/j.bbamcr.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Asher G, Bercovich Z, Tsvetkov P, Shaul Y, Kahana C. 20S proteasomal degradation of ornithine decarboxylase is regulated by NQO1. Mol Cell. 2005;17:645–655. doi: 10.1016/j.molcel.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 73*.Moscovitz O, Tsvetkov P, Hazan N, Michaelevski I, Keisar H, Ben-Nissan G, Shaul Y, Sharon M. A Mutually Inhibitory Feedback Loop between the 20S Proteasome and Its Regulator, NQO1. Mol Cell. 2012;47:76–86. doi: 10.1016/j.molcel.2012.05.049. Paper describes a novel mechanism by which ubiquitin independent degradation by the 20S proteasome may be regulated and linked to the metabolic state of the cell.
- 74.Sylvestersen KB, Young C, Nielsen ML. Advances in characterizing ubiquitylation sites by mass spectrometry. Curr Opin Chem Biol. 2013;17:49–58. doi: 10.1016/j.cbpa.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 75.Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wagner SA, Beli P, Weinert BT, Schölz C, Kelstrup CD, Young C, Nielsen ML, Olsen JV, Brakebusch C, Choudhary C. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol Cell Proteomics. 2012;11:1578–1585. doi: 10.1074/mcp.M112.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang F, Durfee LA, Huibregtse JM. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol Cell. 2013;50:368–378. doi: 10.1016/j.molcel.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duttler S, Pechmann S, Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol Cell. 2013;50:379–393. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci U S A. 2006;103:13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 81.Eden E, Geva-Zatorsky N, Issaeva I, Cohen A, Dekel E, Danon T, Cohen L, Mayo A, Alon U. Proteome half-life dynamics in living human cells. Science. 2011;331:764–768. doi: 10.1126/science.1199784. [DOI] [PubMed] [Google Scholar]
- 82.Emanuele MJ, Elia AEH, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen Y-N, Rush J, Hsu PW-C, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]