Abstract
Although transcription activator-like effector nucleases (TALENs) can be designed to cleave chosen DNA sequences, TALENs have been shown to have activity against related off-target sequences. To better understand TALEN specificity and engineer TALENs with improved specificity, we profiled 30 unique TALENs with varying target sites, array length, and domain sequences for their ability to cleave any of 1012 potential off-target DNA sequences using in vitro selection and high-throughput sequencing. Computational analysis of the selection results predicted 76 off-target substrates in the human genome, 16 of which were accessible and modified by TALENs in human cells. The results collectively suggest that (i) TALE repeats bind DNA relatively independently; (ii) longer TALENs are more tolerant of mismatches, yet are more specific in a genomic context; and (iii) excessive DNA-binding energy can lead to reduced TALEN specificity in cells. Based on these findings, we engineered a TALEN variant, Q3, that exhibits equal on-target cleavage activity but 10-fold lower average off-target activity in human cells. Our results demonstrate that identifying and mutating residues that contribute to non-specific DNA-binding can yield genome editing reagents with improved DNA specificities.
Introduction
The ability to engineer site-specific changes in genomes is a powerful capability with significant research and therapeutic implications. Transcription activator-like effector nucleases (TALENs) are fusions of the FokI restriction endonuclease cleavage domain with a DNA-binding TALE repeat array (Fig. 1A). These arrays consist of multiple 34-amino acid TALE repeat sequences, each of which uses a repeat-variable di-residue (RVD), the amino acids at positions 12 and 13, to recognize a single DNA nucleotide.1, 2 Examples of RVDs that recognize each of the four DNA base pairs are known, enabling arrays of TALE repeats to be constructed that can bind virtually any DNA sequence. TALENs can be engineered to be active only as heterodimers through the use of obligate heterodimeric FokI variants.3, 4 In this configuration, two distinct TALEN monomers are each designed to bind one target half-site resulting in cleavage within the DNA spacer sequence between the two half-sites. In cells, TALEN-induced double-strand breaks can result in targeted gene knockout through non-homologous end joining (NHEJ)5 or precise targeted genomic sequence alteration through homology-directed repair (HDR) with an exogenous DNA template.6, 7 TALENs have been successfully used to manipulate genomes in a variety of organisms6, 8–11 and cell lines.5, 7, 12, 13
Figure 1. TALEN architecture and selection scheme.
(A) Architecture of a TALEN. A TALEN monomer contains an N-terminal domain (blue) followed by an array of TALE repeats (brown), a C-terminal domain (green), and a FokI nuclease cleavage domain (purple). The 12th and 13th amino acids (the RVD, red) of each TALE repeat recognize a specific DNA base pair. Two different TALENs bind their corresponding half-sites, allowing FokI dimerization and DNA cleavage. The C-terminal domain variants used in this study are shown in green. (B) A single-stranded library of DNA oligonucleotides containing partially randomized left half-site (L), spacer (S), right half-site (R) and constant region (thick black line) was circularized, then concatemerized by rolling circle amplification. The concatemerized double stranded DNA (double arrows) contained repeated target sites with L′ S′ R′ representing the reverse sequence complement of L S R. (C) The concatemerized DNA libraries of mutant target sites were incubated with an in vitro-translated TALEN of interest. Cleaved library members were blunted and ligated to adapter #1. The ligation products were amplified by PCR using one primer consisting of adapter #1 and the other primer consisting of adapter #2–constant sequence, which anneals to the constant regions. From the resulting ladder of amplicons containing a half-site with an integral number (n) of repeats of a target site (represented by brackets), amplicons corresponding to 1.5 target-sites in length were isolated by gel purification and subjected to high-throughput DNA sequencing and computational analysis (Supplementary Algorithms).
Although TALENs are sufficiently specific to show activity against their intended target sites without causing widespread and highly abundant genomic off-target modification,14–17 TALEN-mediated DNA cleavage at off-target sites can result in unintended mutations at genomic loci. While recent studies identify closely related off-target sites containing two or fewer mismatches in zebrafish18 and in human cell lines,13 more distantly related off-target sites are of particular interest since one would expect a typical 36-bp target site to be approximately eight or more mutations away from any sequence in the human genome. In previous studies, two distant genomic off-target sites were identified from 19 potential off-target sites predicted using SELEX,7 an in vitro method to identify binding sites of DNA-binding domains in isolation. Only a single heterodimeric off-target site was identified using an integrase-deficient lentiviral vector(IDLV)-based approach19, 20 to capture off-target double-strand break sites in cells. The limited number of off-target TALEN sites identified in previous studies suggest that further research is needed both to better understand the extent of TALEN-induced genomic off-target mutations and to improve TALEN specificity to minimize these unwanted effects.
The underlying principles that determine the specificities of TALEN proteins remain poorly characterized. While SELEX experiments and a high-throughput study of TALE activator specificity have described the DNA-binding specificities of monomeric TALE proteins5, 7,9 and a single TALE activator,21 respectively, the DNA cleavage specificities of active, dimeric nucleases can differ from the specificities of their component monomeric DNA-binding domains.22 For example, zinc finger nucleases (ZFNs), another type of engineered dimeric nuclease, demonstrate compensation effects between monomers.22 Cellular methods to study off-target genomic modification such as whole-genome sequencing or IDLV capture could be complicated by cellular factors such as DNA accessibility, which varies from site to site and between cell types,23 or DNA repair and integration pathways after cleavage that could obscure the determination of intrinsic TALEN protein specificity. Purely cellular studies are also inherently limited to the stochastic handful of off-target sites in a given genome that are similar to the target sequence and thus are unable to evaluate the ability of TALENs to cleave a very large number of off-target sites necessary for a broad and in-depth study of TALEN specificity.
Using a previously described in vitro selection method,22, 24 we interrogated TALENs for their abilities to each cleave 1012 potential off-target DNA substrates related to their intended target sequences. The resulting data provide the first comprehensive profiles of TALEN cleavage specificities in a manner that is not limited to the small number of typical target-related sites in a genome. The selection results suggest a model in which excess non-specific DNA-binding energy gives rise to greater off-target cleavage relative to on-target cleavage. Based on this model, we engineered TALENs with a modified architecture that show substantially improved DNA cleavage specificity in vitro. In human cells, these modified TALENs exhibit 24- to > 120-fold greater specificity for the most readily cleaved off-target site than currently used TALEN constructs.
Results
Specificity Profiling of TALENs targeting the human CCR5 and ATM genes
We profiled the specificities of 30 unique heterodimeric TALEN pairs (hereafter referred to as TALENs) harboring different C-terminal, N-terminal and FokI domain variants and targeted to sites with half-sites of various lengths. Throughout this report, the number of base pairs recognized by each half-site is listed to include the 5′ T nucleotide recognized by the N-terminal domain. Most of the TALENs tested contained the obligate heterodimeric EL/KK FokI domain, although the more active heterodimeric ELD/KKR and homodimeric FokI nuclease domain were also used, as specified below.3, 26 TALENs were constructed as previously reported12 and designed to target one of three distinct sequences, which we refer to as CCR5A, CCR5B, or ATM, in two different human genes, CCR5 and ATM (Supplementary Fig. S1). The specificity profiles were generated using a previously described in vitro selection method.22, 24 Briefly, pre-selection libraries of > 1012 DNA sequences each were digested with 3 nM to 40 nM of an in vitro-translated TALEN (see Methods and Supplementary Results for more detail). The pre-selection DNA libraries were sufficiently large that they each contain, in theory, at least ten copies of all possible DNA sequences with six or fewer mutations relative to the on-target sequence. Cleaved library members harbored a free 5′ monophosphate that enabled them to be captured by adapter ligation (Fig. 1B). DNA fragments of length corresponding to 1.5 target sites (an intact target site and a repeated half-site up to the point of TALEN-induced DNA cleavage) were isolated by gel purification. High-throughput sequencing and computational analysis of TALEN-treated or control samples surviving this selection process revealed the abundance of all TALEN-cleaved sequences as well as the abundance of the corresponding sequences before selection. In the control sample, all members of the pre-selection library were cleaved by a restriction endonuclease at a constant sequence to enable them to be captured by adapter ligation and isolated by gel purification. The enrichment value for each library member surviving selection was calculated by dividing its post-selection sequence abundance by its pre-selection abundance.
For all TALEN variants and under all tested conditions, the DNA that survived the selection contained significantly fewer mean mutations in the targeted half-sites than were present in the preselection libraries (Fig. 2A and 2B; Supplementary Table S2 and S3). For all selections, the on-target sequences were enriched by 8- to 640-fold with an average enrichment value of 110-fold (Supplementary Table S4). To validate our selection results in vitro, we assayed the ability of the CCR5B TALEN targeting 13-bp left and right half-sites (L13+R13) to cleave each of 16 diverse off-target substrates (Fig. 2E and 2F). The efficiencies with which each of these 16 putative off-target substrates were cleaved by the TALEN in these discrete in vitro assays correlated well (r = 0.90) with the observed enrichment values from the selection (Fig. 2G).
Figure 2. In vitro selection results.
The fraction of sequences surviving selection (green) and before selection (black) are shown for CCR5A TALENs (A) and ATM TALENs (B) with EL/KK FokI domains as a function of the number of mutations in both half-sites (left and right half-sites combined excluding the spacer). (C) Specificity scores for the CCR5A TALENs at all positions in the target half-sites plus a single flanking position. The colors range from dark blue (maximum specificity score of 1.0) to white (no specificity, score of 0) to dark red (maximum negative score of −1.0); see the main text for details. Boxed bases represent the intended target base. Note for the right half-site, the R18 TALENs, the sense strand is shown. (D) Same as (C) for the ATM TALENs. For (A), (B), (C) and (D) sample statistics (sample sizes, means, standard deviations, and P-values) are given in Supplementary Table S2 and S3. (E) Enrichment values from the selection of L13+R13 CCR5B TALEN for 16 mutant DNA sequences (mutations in red) relative to on-target DNA (OnB). (F) Correspondence between discrete in vitro TALEN cleavage efficiency (cleaved DNA as a fraction of total DNA) for the sequences listed in (E) normalized to on-target cleavage (= 1) versus their enrichment values in the selection normalized to the on-target enrichment value (= 1). The Pearson’s r coefficient of correlation between normalized cleavage efficiency and normalized enrichment value is 0.90. (G) Discrete assays of on-target and off-target sequences used in (F) as analyzed by PAGE.
To quantify the DNA cleavage specificity at each position in the TALEN target site for all four possible base pairs, a specificity score was calculated as the difference between pre-selection and post-selection base pair frequencies, normalized to the maximum possible change of the pre-selection frequency from complete specificity (defined as 1.0) to complete anti-specificity (defined as −1.0). For all TALENs tested, the targeted base pair at every position in both half-sites is preferred, with the sole exception of the base pair closest to the spacer for some ATM TALENs at the right-half site (Fig. 2C, 2D and Supplementary Fig. S3 through S8). The 5′ T recognized by the N-terminal domain is highly specified, and the 3′ DNA end (targeted by the C-terminal TALEN end) generally tolerates more mutations than the 5′ DNA end; both of these observations are consistent with previous reports.27, 28 All 12 of the positions targeted by the NN RVDs in the ATM and CCR5A TALENs were enriched for G, confirming previous reports5, 7, 27, 29 that the NN RVD specifies G. Taken together, these results show that the selection data accurately predicts the efficiency of off-target TALEN cleavage in vitro, and that TALENs are overall quite specific across the entire target sequence.
TALEN Off-Target Cleavage in Cells
For TALENs targeting a total of 36 base pairs, potential off-target sites in the human genome are expected on average to contain approximately eight or more mutations relative to the on-target site (Supplementary Table S5), more mutations than theoretically are covered in the in vitro selection. Therefore, we used a machine-learning “classifier” algorithm25 trained on the tens of thousands of off-target sites revealed by the in vitro selection to identify rare TALEN candidate off-target sites in the human genome (see Supplementary Results for details). Using this algorithm, we identified the 36 best-scoring heterodimeric candidate off-target sites for the ATM TALENs and 48 of the best-scoring candidate off-target sites for the CCR5A TALENs (Supplementary Table S6). These sites differ from the on-target sequence at seven to fourteen positions.
These 84 predicted off-target sites for CCR5A and ATM TALENs were amplified from genomic DNA purified from human U2OS-EGFP cells expressing either CCR5A or ATM TALENs.12 Sequences containing insertions or deletions of three or more base pairs in the DNA spacer of the potential genomic off-target sites and present in significantly greater numbers in the TALEN-treated samples versus the untreated control sample were considered TALEN-induced modifications. Consistent with a previous report3, CCR5A or ATM TALENs containing ELD/KKR and homodimeric FokI domains demonstrated increased on-target activity compared to EL/KK FokI domains. Of the 45 CCR5A off-target sites that we successfully amplified, we identified nine off-target sites with TALEN-induced modifications; likewise, of the 31 ATM off-target sites that we successfully amplified, we observed seven off-target sites with TALEN-induced modifications (Fig. 3 and Supplementary Table S7 and S9). Therefore, in total, 16 out of 76 assayed off-target candidates were accessible and modified by TALENs in cells. The inspection of modified on-target and off-target sites yielded a prevalence of deletions ranging from three to dozens of base pairs (Supplementary Fig. S9), consistent with previously described characteristics of TALEN-induced genomic modification.31 These results collectively indicate in vitro selection data processed through a machine-learning algorithm, can predict bona fide off-target substrates that undergo TALEN-induced modification in human cells. We also directly compared our combined in vitro selection and machine learning method with TALENoffer, a recently described purely computational prediction method32 and found that our approach outperforms the purely computational approach for the identification of TALEN-induced off-target substrates in cells (Supplementary Tables S8 and S9 and Supplementary Results).
Figure 3. Cellular modification induced by TALENs at on-target and predicted off-target genomic sites.
(A) For cells treated with either no TALEN or CCR5A TALENs containing heterodimeric EL/KK, heterodimeric ELD/KKR, or the homodimeric (Homo) FokI cleavage domain variants, cellular modification rates are shown as the percentage of observed insertions or deletions (indels) consistent with TALEN cleavage relative to the total number of sequences for on-target (On) and predicted off-target sites (Off). See the main text for details. ND refers to no data collected since the cellular modification of off-target sites OffC-38, OffC-49, OffC-69 and OffC-76 was not assayed for CCR5A TALENs containing EL/KK and ELD/KKR FokI domains. (B) Same as (A) for ATM TALENs. For (A) and (B) sample sizes and P-values are given in Supplementary Tables S7 and S9.
TALE Repeat Binding Independence and Effects of TALEN Length on Specificity
The extensive number of quantitatively characterized off-target substrates in the selection data enabled us to address several key questions about TALEN specificity. First, we assessed whether mutations at one position in the target sequence affect the ability of TALEN repeats to productively bind other positions and concluded that TALE repeats bind their respective DNA base pairs independently beyond a slightly increased tolerance for adjacent mismatches (Supplementary Results).
The independent binding of TALE repeats simplistically predicts that TALEN specificity per base pair is independent of target-site length. To experimentally characterize the relationship between TALE array length and off-target cleavage, we constructed TALENs targeting 10, 13, or 16 bps (including the 5′ T) for both the left (L10, L13, L16) and right (R10, R13, R16) half-sites. TALENs representing all nine possible combinations of left and right CCR5B TALENs were subjected to in vitro selection. The results revealed that shorter TALENs have greater specificity per targeted base pair than longer TALENs (Supplementary Table S2). For example, sequences cleaved by the L10+R10 TALEN contained a mean of 0.032 mutations per recognized base pair, while those cleaved by the L16+R16 TALEN contained a mean of 0.067 mutations per recognized base pair.
For selections with the longest CCR5B TALENs targeting 16+16 base pairs or CCR5A and ATM TALENs targeting 18+18 bp, the mean selection enrichment values do not follow a simple exponential decrease as function of mutation number (Fig. 4 and Supplementary Table S10). It is possible these TALENs have greater affinity than is required to substantially bind and cleave the target site (referred to hereafter as “excess DNA-binding energy”). Thus, we hypothesize that excess DNA-binding energy from the larger number of TALE repeats in longer TALENs reduces specificity by enabling the cleavage of sequences with more mutations, without a corresponding increase in the cleavage of sequences with fewer mutations, because the latter are already nearly completely cleaved. Indeed, the in vitro cleavage efficiencies of discrete DNA sequences for these longer TALENs are independent of the presence of a small number of mutations in the target site (Fig. 5C–5F), suggesting there is nearly complete binding and cleavage of sequences containing few mutations. Likewise, higher TALEN concentrations also result in decreased enrichment values of sequences with few mutations while increasing the enrichment values of sequences with many mutations (Supplementary Table S4). These results together support a model in which excessive TALEN binding arising from either long TALE arrays or high TALEN concentrations decreases the observed TALEN DNA cleavage specificity for each recognized base pair. Despite being less specific per base pair, TALENs designed to cleave longer target sites are estimated to have higher overall specificity than those that target shorter sites when considering the number of potential off-target sites in the human genome (Supplementary Results).
Figure 4. In vitro specificity as a function of TALEN length.
The enrichment value of on-target (zero mutation) and off-target sequences containing one to six mutations are shown for CCR5B TALENs of varying TALE repeat array lengths with EL/KK FokI domains. The TALENs targeted DNA sites of 32 bp (L16+R16), 29 bp (L16+R13 or L13+R16), 26 bp (L16+R10 or L13+R13 or L10+R16), 23 bp (L13+R10 or L10+R13) or 20 bp (L10+R10) in length.
Figure 5. In vitro specificity and discrete cleavage efficiencies of TALENs containing canonical or engineered C-terminal domains.
(A and B) On-target enrichment values for selections of (A) CCR5A TALENs containing canonical, Q3, Q7, or 28-aa C-terminal domains with EL/KK FokI cleavage domains or (B) ATM TALENs containing canonical, Q3 or Q7 C-terminal domains with EL/KK FokI cleavage domains. (C) CCR5A on-target sequence (OnC) and double-mutant sequences with mutations in red. For CCR5A, sequences containing two mutations were assayed because one-mutation and zero-mutation sequences were similarly enriched (Supplementary Table S4A). (D) ATM on-target sequence (OnA), single-mutant sequences, and double-mutant sequences with mutations in red. (E) Discrete in vitro cleavage efficiency of DNA sequences listed in (C) with CCR5A TALENs containing either canonical or engineered Q7 C-terminal domains with EL/KK FokI domains. Error bars reflect s.d. from three biological replicates, except two replicates for C4. All pairwise P-values were calculated between the cleavage efficiencies of the on-target sequence (OnC) digested by CCR5A TALENs containing the canonical C-terminal vs. the cleavage efficiencies of a mutant sequence (C1, C2, …., or C8) digested by CCR5A TALENs containing the canonical C-terminal domain. All pairwise P-values were also calculated between the cleavage efficiencies of the on-target sequence (OnC) digested by CCR5A TALENs containing the Q7 C-terminal domain vs. the cleavage efficiencies of a mutant sequence (C1, C2, …., or C8) digested by CCR5A TALENs containing the Q7 C-terminal domain. The cleavage efficiencies of mutant sequences C1, C2, C6, C7 and C8 digested by CCR5A TALENs containing the canonical C-terminal domain demonstrate a P-value significantly different (P-value < 0.025) from the cleavage efficiencies of the on-target sequence (OnC) digested by CCR5A TALENs containing the canonical C-terminal domain. The cleavage efficiencies of mutant sequences C1, C3, C4, C6, C7 and C8 digested by CCR5A TALENs containing the Q7 C-terminal domain demonstrate a P-value significantly different (P-value < 0.025) from the cleavage efficiencies of the on-target sequence (OnC) digested by CCR5A TALENs containing the Q7 C-terminal domain. (F) Same as (E) for ATM TALENs. The cleavage efficiencies of mutant sequences A1, A2, A3, A5, A6, A7 and A8 digested by ATM TALENs containing the Q7 C-terminal domain demonstrate a P-value significantly different (P-value < 0.025) from the cleavage efficiencies of ATM TALENs containing the canonical C-terminal domain digestion of the on-target sequence (OnC).
Engineering TALENs with Improved Specificity
The findings above suggest that TALEN specificity could be improved by reducing non-specific DNA binding energy to only what is needed to support efficient on-target cleavage. The most widely used 63-aa C-terminal domain between the TALE repeat array and the FokI nuclease domain contains ten cationic residues.4, 5, 7, 9, 10, 12 A related C-terminal domain variant (89% homology), containing 11 cationic residues, has also been used in other studies.6, 20, 28 We hypothesized that reducing the cationic charge of the canonical 63-aa TALE C-terminal domain would decrease non-specific DNA binding33 and improve the specificity of TALENs.
We constructed two C-terminal domain variants in which three (“Q3”, consisting of K788Q, R792Q, and R801Q) or seven (“Q7”, consisting of K777Q, K778Q, K788Q, R789Q, R792Q, R793Q, and R801Q) cationic Arg or Lys residues in the canonical 63-aa C-terminal domain were mutated to Gln. We performed in vitro selections on CCR5A and ATM TALENs containing the canonical 63-aa, the engineered Q3, and the engineered Q7 C-terminal domains, as well as a previously reported 28-aa truncated C-terminal domain5 with a theoretical net charge (−1) identical to that of the Q7 C-terminal domain. The on-target sequence enrichment values for the CCR5A and ATM selections increased substantially as the net charge of the C-terminal domain decreased (Fig. 5A and 5B). For example, the ATM selections resulted in on-target enrichment values of 510, 50, and 20 for the Q7, Q3, and canonical 63-aa C-terminal variants, respectively. These results suggest that the TALEN variants in which cationic residues in the C-terminal domain have been partially replaced by neutral residues or completely removed are substantially more specific in vitro than the TALENs that contain the canonical, commonly used 63-aa C-terminal domain. Similarly, mutating one, two, or three cationic residues in the TALEN N-terminus to Gln also increased cleavage specificity (Supplementary Table S4, and Supplementary Fig. S3–S6, and see Supplementary Results).
In order to confirm the greater DNA cleavage specificity of Q7 over canonical 63-aa C-terminal domains in vitro, a representative set of 16 off-target DNA substrates was digested in vitro with TALENs containing either canonical 63-aa or engineered Q7 C-terminal domains. ATM TALENs with the canonical 63-aa C-terminal domain demonstrated comparable in vitro cleavage activity on target sites with zero, one, or two mutations (Fig. 5C–5F). CCR5A TALENs with the canonical 63-aa C-terminal domain TALEN demonstrate comparable in vitro cleavage activity on target sites with zero or two mutations. In contrast, for 11 of the 16 off-target substrates tested, the engineered Q7 TALEN variants showed substantially higher (~4-fold or greater) discrimination against off-target DNA substrates with one or two mutations than the canonical 63-aa C-terminal domain TALENs, even though the Q7 TALENs cleaved their respective on-target sequences in vitro with comparable or greater efficiency than TALENs with the canonical 63-aa C-terminal domains (Fig. 5C–5F). For both the ATM and CCR5A Q7 C-terminal TALENs, some sequences are cleaved with greater specificity than others. Sequence-dependent specificity is expected based on the variable specificity at each position (Fig. 2C and D). Overall, the discrete cleavage assays are consistent with the selection results and indicate that TALENs with engineered Q3 or Q7 C-terminal domains can be substantially more specific than TALENs with canonical 63-aa C-terminal domains in vitro.
Improved Specificity of Engineered TALENs in Human Cells
To determine if the increased specificity of the engineered TALENs observed in vitro also occurs in human cells, TALEN-induced modification rates of the on-target and top 36 predicted off-target sites were measured for CCR5A and ATM TALENs containing all six possible combinations of the canonical 63-aa, Q3, or Q7 C-terminal domains and the EL/KK or ELD/KKR FokI domains (12 TALENs total). We did not analyze TALENs containing a 28-aa C-terminal domain in these experiments because both the ATM and CCR5A on-target sites have DNA spacer lengths of 18 bp, which lies outside the 28-aa C-terminal domain’s preferred DNA spacer length range (Supplementary Fig. S14). For both FokI variants, the TALENs with Q3 C-terminal domains demonstrate significant on-target activities ranging from 8% to 24% modification, comparable to the activity of TALENs with the canonical 63-aa C-terminal domains. TALENs with canonical 63-aa or Q3 C-terminal domains and the ELD/KKR FokI domain are both more active in modifying the CCR5A and ATM on-target site in cells than the corresponding TALENs with the Q7 C-terminal domain by 5- to 9-fold (Fig. 6A–C and Supplementary Table S7).
Figure 6. Specificity of engineered TALENs in human cells.
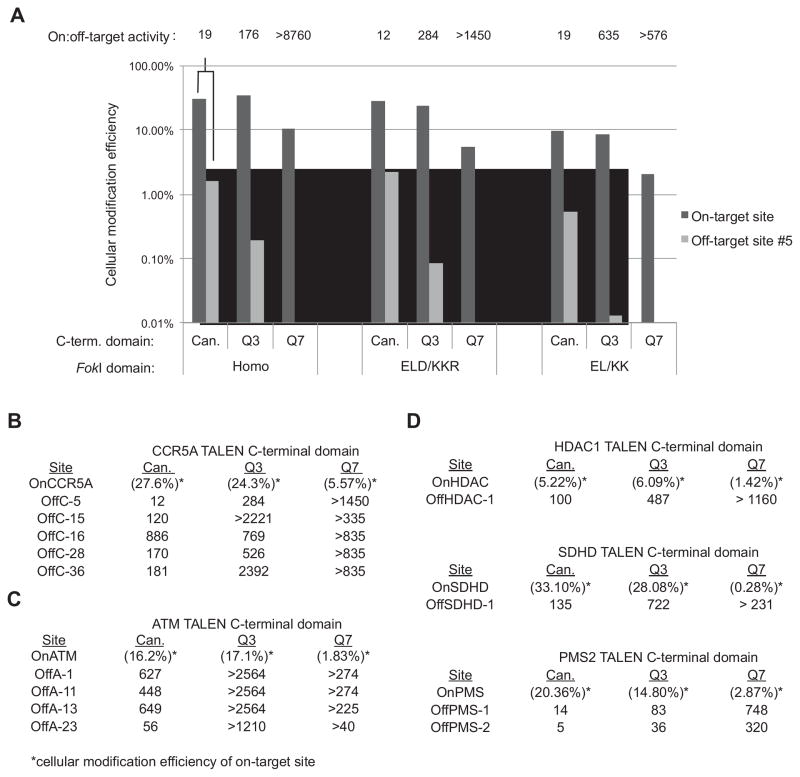
(A) The cellular modification efficiency of canonical and engineered TALENs expressed as a percentage of indels consistent with TALEN-induced modification out of total sequences is shown for the on-target CCR5A site (OnCCR5A) and for CCR5A off-target site #5 (OffC5), the most highly cleaved off-target substrate tested. All pairwise P-values comparing the number of observed sequences containing insertions or deletions consistent with TALEN-induced cleavage vs. the total number of sequences were calculated with a Fischer exact test between samples (see Supplementary Table S7). P-values are < 0.005 for samples of canonical vs. Q3 vs. Q7 TALENs in the same FokI background for both on-target and off-target sites with the exception of off-target site #5 modified with Q3 vs. Q7 TALENs in the EL/KK FokI background (P-value < 0.087). On:off target activity, defined as the ratio of on-target to off-target modification, is shown above each pair of bars. (B) The on:off target activity of the canonical, Q3, and Q7 TALENs for each detected genomic off-target substrate of the CCR5A TALEN with the ELD/KKR FokI domain are shown. The absolute genomic modification frequency for the on-target site is in parentheses. (C) Same as (B) for the ATM TALENs and off-target sites. (D) The on:off target activities of the canonical, Q3, and Q7 TALENs for each detected genomic off-target substrate of the PMS2, SDHD, and HDAC1 TALENs with the ELD/KKR FokI domain are shown. The absolute genome modification frequency for the on-target site is in parentheses.
Compared to the canonical 63-aa C-terminal domains, TALENs with Q3 C-terminal domains demonstrate a mean increase in on-target:off-target activity ratio of more than 12-fold and more than 9-fold for CCR5A and ATM sites, respectively, with the ELD/KKR FokI domain (Fig. 6A–C & Supplementary Table S7). These mean improvements can only be expressed as lower limits due to the absence or near-absence of observed cleavage events by the engineered TALENs for many off-target sequences. For the ATM TALENs containing Q7 C-terminal domains, the cleavage efficiency of both the on-target and off-target sites is so low that their specificity cannot be determined (Fig. 6C). For the most abundantly cleaved off-target site (CCR5A off-target site #5), the Q3 C-terminal domain is 24-fold more specific, and the Q7 C-terminal domain is > 120-fold more specific (Fig. 6A), than the canonical 63-aa C-terminal domain. Consistent with the improved on-target:off-target activity ratio observed in vitro, the engineered Q7 TALENs are more specific than the Q3 variants, which in turn are more specific than the canonical 63-aa C-terminal domain TALENs.
To determine if the increased specificity of the engineered TALENs observed for CCR5A and ATM TALENs applies more generally, three new TALENs targeting sequences in the PMS2, SDHD, and HDAC1 genes12 were constructed using the canonical 63-aa, Q3, or Q7 C-terminal domains and ELD/KKR FokI domains. Of the 64 TALENs reported previously in Reyon et al, these three TALENs had target sequences with closely homologous genomic off-target sites containing one to five mutations. For each of these TALENs, modification rates were measured for genomic on-target and off-target sites. PMS2, SDHD, and HDAC-1 TALENs with Q3 C-terminal domains demonstrate on-target activities ranging from 6% to 28% modification, comparable to the activity of TALENs with the canonical 63-aa C-terminal domains (Fig. 6D and Supplementary Table S8). While demonstrating similar on-target activities to TALENs with canonical domains, PMS2, SDHD, and HDAC1 TALENs with Q3 C-terminal domains demonstrated a 5- to 7-fold increase in on-target:off-target activity ratio. For the PMS2 TALENs, the Q7 C-terminal domains demonstrated a 53- and 64-fold increase in on-target:off-target activity ratio in cells, although as observed above the Q7 TALENs were less active on the target site than TALENs containing the canonical and Q3 C-terminal domains (Fig. 6D and Supplementary Table S8).
Together, these results reveal that for five families of TALENs targeting the CCR5, ATM, PMS2, SDHD, and HDAC1 genes, replacing the canonical 63-aa C-terminal domain with the engineered Q3 C-terminal domain results in comparable activity for the on-target site in cells, and an average 10-fold increase in specificity for all assayed off-target sites. The engineered Q7 C-terminal domain can offer additional gains in specificity beyond that of the Q3 TALENs, but with reduced on-target activity. Collectively, these results validate a method to evaluate the specificity of TALEN variants that also revealed underlying principles resulting in engineered TALENs with improved DNA cleavage specificity in cells.
Discussion
The in vitro selection results for 30 unique TALENs each challenged with 1012 closed related off-target sequences and subsequent analysis inform our understanding of TALEN specificity through four key findings: (i) TALENs are highly specific for their intended target base pair at 103 of the 104 positions profiled with specificity increasing near the N-terminal TALEN end of each TALE repeat array (corresponding to the 5′ end of the bound DNA); (ii) longer TALENs are more specific in a genomic context while shorter TALENs have higher specificity per nucleotide; (iii) TALE repeats each bind their respective base pairs relatively independently; and (iv) excess DNA-binding affinity leads to increased TALEN activity against off-target sites and therefore decreased specificity.
The 16 confirmed TALEN off-target sites containing eight to 12 mutations identified from the 76 predicted sites assayed in this study represent more bona fide genomic off-target sites in the human genome than have been revealed collectively to date by other methods. These 16 sites were modified at efficiencies ranging from 2.3% to 0.03% in human cells, demonstrating that TALENs can have appreciable off-target activities in human cells even at sites that are eight or more mutations away from the on-target sequence. Site accessibility in cells, mediated by histone proteins, transcription factors, and DNA modification,23 likely account for at least some of the difference between our in vitro, computational, and cell-based results. For a comparison of our method with others for characterizing TALEN specificity and identifying genomic TALEN off-target sites, see the Supplementary Discussion.
The observed decrease in specificity for TALENs with more TALE repeats or more cationic residues in the C-terminal domain or N-terminus are consistent with a model in which excess TALEN binding affinity leads to increased promiscuity. This excess binding energy model may explain reports that NN RVDs bind either A or G.2,28,35 These studies used TALE arrays of more than 14 RVDs, which could have created a scenario in which excess DNA-binding energy permits a suboptimal NN RVD interaction with A compared to G. We observed NN RVDs can discriminate between A and G, consistent with reports using shorter TALE arrays of 13 RVDs29 and by direct biochemical interrogation.27 Excess DNA-binding energy could also explain the previously reported promiscuity at the 5′ terminal T of TALENs with longer C-terminal domains36 and is consistent with a report of higher TALEN protein concentrations resulting in more off-target site cleavage in vivo.9 While decreasing TALEN protein expression in cells in theory could reduce off-target cleavage, TALE arrays are reported with on-target DNA binding affinities as high as Kd = 2.8 nM,27 which is sufficient to theoretically saturate target sites even when expressed at modest, mid-nM concentrations in the cell. The difficulty of improving the specificity of such TALENs by lowering their expression levels, coupled with the need to maintain sufficient TALEN concentrations to effect desired levels of on-target cleavage, highlight the value of engineering TALENs with higher intrinsic specificity such as those described in this work.
Our findings suggest that mutant C-terminal domains with reduced non-specific DNA binding may be used to alter the DNA-binding affinity of TALENs such that on-target sequences are cleaved efficiently but with minimal excess DNA-binding energy, resulting in better discrimination between on-target and off-target sites. Since TALENs targeting up to 46 total base pairs have been shown to be active in cells,14 it may be possible to further improve specificity by engineering TALENs with a combination of mutant N-terminal and C-terminal domains that impart reduced non-specific DNA-binding, a greater number of TALE repeats to contribute additional on-target DNA binding, and lower-affinity RVDs such as the NK RVD to recognize G.28, 29 It is tempting to speculate that the strategy of mutating residues that contribute to non-specific DNA binding to improve DNA specificity may also apply to other genome engineering proteins including Cas9 and ZFNs.
Our model and the resulting improved TALENs would have been difficult to derive or validate using purely cellular off-target cleavage methods. The ability of our profiling method to reveal the broad, unobscured DNA cleavage specificity of TALENs in the absence of cellular complications enabled the elucidation of the inherent DNA-cleavage specificity of TALENs. Studies of cellular off-target cleavage are also intrinsically limited by the small number of sequences present in a genome that may be closely related to a target sequence of interest. In contrast, each active, dimeric TALEN in this study was evaluated for its ability to cleave any of 1012 close variants of its on-target sequence, a library size several orders of magnitude greater than the number of different sequences in a mammalian genome. This dense coverage of off-target sequence space enabled the elucidation of detailed relationships between DNA-cleavage specificity and target base pair position, TALE repeat length, TALEN concentration, mismatch location, and engineered TALEN composition. These results collectively reveal principles for characterizing and improving TALENs with greater specificity that may enable a wider range of genome engineering applications.
ONLINE METHODS
Oligonucleotides, PCR and DNA Purification
All oligonucleotides were purchased from Integrated DNA Technologies (IDT). Oligonucleotide sequences are listed in Supplementary Table S12. PCR was performed with 0.4 μL of 2 U/μL Phusion Hot Start II DNA polymerase (Thermo-Fisher) in 50 μL with 1x HF Buffer, 0.2 mM dNTP mix (0.2 mM dATP, 0.2 mM dCTP, 0.2 mM dGTP, 0.2 mM dTTP) (NEB), 0.5 μM to 1 μM of each primer and a program of: 98 °C, 1 min; 35 cycles of [98 °C, 15 s; 62 °C, 15 s; 72 °C, 1 min] unless otherwise noted. Many DNA reactions were purified with a QIAquick PCR Purification Kit (Qiagen) referred to below as Q-column purification or MinElute PCR Purification Kit (Qiagen) referred to below as M-column purification.
TALEN Construction
The canonical TALEN plasmids were constructed by the FLASH method12 with each TALEN targeting 10–18 base pairs. N-terminal mutations were cloned by PCR with Q5 Hot Start Master Mix (NEB) [98 °C, 22 s; 62 °C, 15 s; 72 °C, 7 min]) using phosphorylated TAL-N1fwd (for N1), phosphorylated TAL-N2fwd (for N2), or phosphorylated TAL-N3fwd (for N3) and phosphorylated TAL-Nrev as primers. 1 μL DpnI (NEB) was added and the reaction was incubated at 37 °C for 30 min then M-column purified. ~25 ng of eluted DNA was blunt-end ligated intramolecularly in 10 μL 2x Quick Ligase Buffer, 1 μL of Quick Ligase (NEB) in a total volume of 20 μL at room temperature (~21 °C) for 15 min. 1 μL of this ligation reaction was transformed into Top10 chemically competent cells (Invitrogen). C-terminal domain mutations were cloned by PCR using TAL-Cifwd and TAL-Cirev primers, then Q-column purified. ~1 ng of this eluted DNA was used as the template for PCR with TAL-Cifwd and either TAL-Q3 (for Q3) or TAL-Q7 (for Q7) for primers, then Q-column purified. ~1 ng of this eluted DNA was used as the template for PCR with TAL-Cifwd and TAL-Ciirev for primers, then Q-column purified. ~1 μg of this DNA fragment was digested with HpaI and BamHI in 1x NEBuffer 4 and cloned22 into ~2 μg of desired TALEN plasmid pre-digested with HpaI and BamHI. TALENs containing the N-terminal mutant domains, the Q3 C-terminal domains and the Q7 C-terminal will be available from Addgene.
In Vitro TALEN Expression
TALEN proteins, all containing a 3xFLAG tag, were expressed by in vitro transcription/translation. 800 ng of TALEN-encoding plasmid or no plasmid (“empty lysate” control) was added to an in vitro transcription/translation reaction using the TNT® Quick Coupled Transcription/Translation System, T7 Variant (Promega) in a final volume of 20 μL at 30 °C for 1.5 h. Western blots were used to visualize protein using the anti-FLAG M2 monoclonal antibody (Sigma-Aldrich). TALEN concentrations were calculated by comparison to standard curve of 1 ng to 16 ng N-terminally FLAG-tagged bacterial alkaline phosphatase (Sigma-Aldrich).
In Vitro Selection for DNA Cleavage
Pre-selection libraries were prepared with 10 pmol of oligo libraries containing partially randomized target half-site sequences (CCR5A, ATM, or CCR5B) and fully randomized 10- to 24-bp spacer sequences (Supplementary Table S12). Oligonucleotide libraries were separately circularized by incubation with 100 units of CircLigase II ssDNA Ligase (Epicentre) in 1x CircLigase II Reaction Buffer (33 mM Tris-acetate, 66 mM potassium acetate, 0.5 mM dithiothreitol, pH 7.5) supplemented with 2.5 mM MnCl2 in 20 μL total for 16 h at 60 °C then incubated at 80 °C for 10 min. 2.5 μL of each circularization reaction was used as a substrate for rolling-circle amplification at 30 °C for 16 h in a 50-μL reaction using the Illustra TempliPhi 100 Amplification Kit (GE Healthcare). The resulting concatemerized libraries were quantified with Quant-iT™ PicoGreen ® dsDNA Kit (Invitrogen) and libraries with different spacer lengths were combined in an equimolar ratio.
For selections on the CCR5B sequence libraries, 500 ng of pre-selection library was digested for 2 h at 37 °C in 1x NEBuffer 3 with in vitro transcribed/translated TALEN plus empty lysate (30 μL total). For all CCR5B TALENs, in vitro transcribed/translated TALEN concentrations were quantified by Western blot (during the blot, TALENs were stored for 16 h at 4 °C) and then TALEN was added to 40 nM final concentration per monomer. For selections on CCR5A and ATM sequence libraries, the combined pre-selection library was further purified in a 300,000 MWCO spin column (Sartorius) with three 500-μL washes in 1x NEBuffer 3. 125 ng pre-selection library was digested for 30 min at 37 °C in 1x NEBuffer 3 with a total 24 μL of fresh in vitro transcribed/translated TALENs and empty lysate. For all CCR5A and ATM TALENs, 6 μL of in vitro transcription/translation left TALEN and 6 μL of right TALEN were used, corresponding to a final concentration in a cleavage reaction of 16 nM ± 2 nM or 12 nM ± 1.5 nM for CC5A or ATM TALENs, respectively. These TALEN concentrations were quantified by Western blot performed in parallel with digestion.
For all selections, the TALEN-digested library was incubated with 1 μL of 100 μg/μL RNase A (Qiagen) for 2 min and then Q-column purified. 50 μL of purified DNA was incubated with 3 μL of 10 mM dNTP mix (10 mM dATP, 10 mM dCTP, 10 mM dGTP, 10 mM dTTP) (NEB), 6 μL of 10x NEBuffer 2, and 1 μL of 5 U/μL Klenow Fragment DNA Polymerase (NEB) for 30 min at room temperature and Q-column purified. 50 μL of the eluted DNA was ligated with 2 pmol of heated and cooled #1 adapters containing barcodes corresponding to each sample (selections with different TALEN concentrations or constructs) (Supplementary Table S12A). Ligation was performed in 1x T4 DNA Ligase Buffer (50 mM Tris-HCl, 10 mM MgCl2, 1 mM ATP, 10 mM DTT, pH 7.5) with 1 μL of 400 U/μL T4 DNA ligase (NEB) in 60 μL total volume for 16 h at room temperature, then Q-column purified.
6 μL of the eluted DNA was amplified by PCR in 150 μL total reaction volume (divided into 3x 50 μL reactions) for 14 to 22 cycles using the #2A adapter primers in Supplementary Table S12A. The PCR products were purified by Q-column. Each DNA sample was quantified with Quant-iT™ PicoGreen® dsDNA Kit (Invitrogen) and then pooled into an equimolar mixture. 500 ng of pooled DNA was run a 5% TBE 18-well Criterion PAGE gel (BioRad) for 30 min at 200 V and DNAs of length ~230 bp (corresponding to 1.5 target site repeats plus adapter sequences) were isolated and purified by Q-column. ~2 ng of eluted DNA was amplified by PCR for 5 to 8 cycles with #2B adapter primers (Supplementary Table S12A) and purified by M-column.
10 μL of eluted DNA was purified using 12 μL of AMPure XP beads (Agencourt) and quantified with an Illumina/Universal Library Quantification Kit (Kapa Biosystems). DNA was prepared for high-throughput DNA sequencing according to Illumina instructions and sequenced using a MiSeq DNA Sequencer (Illumina) using a 12 pM final solution and 156-bp paired-end reads. To prepare the preselection library for sequencing, the pre-selection library was digested with 1 μL to 4 μL of appropriate restriction enzyme (CCR5A = Tsp45I, ATM = Acc65I, CCR5B = AvaI (NEB)) for 1 h at 37 °C then ligated as described above with 2 pmol of heated and cooled #1 library adapters (Supplementary Table S9A). Pre-selection library DNA was prepared as described above using #2A library adapter primers and #2B library adapter primers in place of #2A adapter primers and #2B adapter primers, respectively (Supplementary Table S12A). The resulting pre-selection library DNA was sequenced together with the TALEN-digested samples.
Discrete In Vitro TALEN Cleavage Assays
Discrete DNA substrates for TALEN digestion were constructed by combining pairs of oligonucleotides as specified in Supplementary Table 12B with restriction cloning22 into pUC19 (NEB). Corresponding cloned plasmids were amplified by PCR (59 °C annealing for 15 s) for 24 cycles with pUC19Ofwd and pUC19Orev primers (Supplementary Table S12B) and Q-column purified. 50 ng of amplified DNAs were digested in 1x NEBuffer 3 with 3 μL each of in vitro transcribed/translated TALEN left and right monomers (corresponding to a ~16 nM to ~12 nM final TALEN concentration), and 6 μL of empty lysate in a total reaction volume of 120 μL. The digestion reaction was incubated for 30 min at 37 °C, then incubated with 1 μL of 100 μg/μL RNase A (Qiagen) for 2 min and purified by M-column. The entire 10 μL of eluted DNA with glycerol added to 15% was analyzed on a 5% TBE 18-well Criterion PAGE gel (Bio-Rad) for 45 min at 200 V, then stained with 1x SYBR Gold (Invitrogen) for 10 min. Bands were visualized and quantified on an AlphaImager HP (Alpha Innotech).
Cellular TALEN Cleavage Assays
TALENs were cloned into mammalian expression vectors12 and the resulting TALEN vectors transfected into U2OS-EGFP cells as previously described.12 Genomic DNA was isolated after 2 days as previously described.12 For each assay, 50 ng of isolated genomic DNA was amplified by PCR [98 °C, 15 s 67.5 °C, 15 s; 72 °C, 22s] for 35 cycles with pairs of primers with or without 4% DMSO as specified in Supplementary Table S12C. Two PCR reactions were performed for OffC-5 to improve the limit of detection. The relative dsDNA content of the PCR reaction for each genomic site was quantified with Quant-iT™ PicoGreen ® dsDNA Kit (Invitrogen) and then pooled into an equimolar mixture, keeping no-TALEN and all TALEN-treated samples separate. DNA corresponding to 150 to 350 bp was purified by PAGE as described above.
44 μL of eluted DNA was incubated with 5 μL of 1x T4 DNA Ligase Buffer and 1 μL of 10 U/μL Polynucleotide kinase (NEB) for 30 min at 37 °C and Q-column purified. 43 μL of eluted DNA was incubated with 1 μL of 10 mM dATP (NEB), 5 μL of 10x NEBuffer 2, and 1 μL of 5 U/μL DNA Klenow Fragment (3′ → 5′ exo−) (NEB) for 30 min at 37 °C and purified by M-column. 10μL of eluted DNA was ligated as above with 10 pmol of heated and cooled G (genomic) adapters (Supplementary Table S12A) and purified by Q-column. 8 μL of eluted DNA was amplified by PCR for 6 to 8 cycles with G-B primers containing barcodes corresponding to each sample. Each sample DNA was quantified with Quant-iT™ PicoGreen ® dsDNA Kit (Invitrogen) and then pooled into an equimolar mixture. The combined DNA was subjected to high-throughput sequencing using a MiSeq as described above.
Data Analysis
Illumina sequencing reads were filtered and parsed with scripts written in Unix Bash as outlined in the Supplementary Algorithms. DNA sequences and source code are available upon request. Specificity scores were calculated as previously described.22 Sample sizes for sequencing experiments were maximized (within practical experimental considerations) to ensure greatest power to detect effects. Statistical analysis on the distribution of number of mutations in various TALEN selections in Supplementary Table S2 was performed as previously described22. Statistical analysis of TALEN modified genomic sites in Supplementary Tables S7, S8 and S9 was performed as previously described25 with multiple comparison correction using the Benjamini-Hochberg method.37, 38
To determine extrapolated mean enrichment curves mutation enrichment value as function of mutation number were fit to an exponential function, a*eb, with R2 reported utilizing the non-linear least squares method. The a, b and R2 values and the mutation range for these fits are reported in Supplementary Table S11. These exponential decrease, b, were used to extrapolate all mean enrichment values beyond five mutations to determine the extrapolated mean enrichment.
Supplementary Material
Acknowledgments
J.P.G., V.P. and D.R.L. were supported by DARPA HR0011-11-2-0003, DARPA N66001-12-C-4207, NIH/NIGMS R01 GM095501 (DRL), and the Howard Hughes Medical Institute. V.P. was supported by award Number T32GM007753 from the National Institute of General Medical Sciences. D.R.L. is a HHMI Investigator. D.R., S.Q.T., J.D.S., and J.K.J. were supported by a National Institutes of Health (NIH) Director Pioneer Award (DP1 GM105378). J.K.J. was supported by the Jim and Ann Orr MGH Research Scholar Award. We thank Morgan L. Maeder for preforming transfections and isolating genomic DNA. We are grateful to Cyd Khayter and Mathew Goodwin for technical assistance.
Footnotes
Accession Codes
SRP035232
AUTHOR CONTRIBUTIONS
J.P.G., V.P., D.R., J.D.S., and S.Q.T. performed the experiments, designed the research, analyzed the data, and wrote the manuscript. J. K. J. and D.R.L. designed the research, analyzed the data, and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
D.R.L and J.K.J. have filed a provisional patent related to this work and are consultants for Editas Medicine, a company that applies genome engineering technologies. J.K.J. has financial interests in Editas Medicine and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
References Cited
- 1.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 2.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 3.Doyon Y, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nature methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 4.Cade L, et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 6.Bedell VM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tesson L, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 10.Moore FE, et al. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs) PLoS One. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood AJ, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mussolino C, et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding Q, et al. A TALEN Genome-Editing System for Generating Human Stem Cell-Based Disease Models. Cell Stem Cell. 2012 doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei Y, et al. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs) Proc Natl Acad Sci U S A. 2012;109:17484–17489. doi: 10.1073/pnas.1215421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y, et al. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol. 2013;31:251–258. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- 18.Dahlem TJ, et al. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 20.Osborn MJ, et al. TALEN-based Gene Correction for Epidermolysis Bullosa. Molecular Therapy. 2013 doi: 10.1038/mt.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature biotechnology. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeder ML, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nature biotechnology. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sander JD, et al. In silico abstraction of zinc finger nuclease cleavage profiles reveals an expanded landscape of off-target sites. Nucleic Acid Research. 2013;41 doi: 10.1093/nar/gkt716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 27.Meckler JF, et al. Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christian ML, et al. Targeting G with TAL effectors: a comparison of activities of TALENs constructed with NN and NK repeat variable di-residues. PloS one. 2012;7:e45383. doi: 10.1371/journal.pone.0045383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Witten IH, Frank E. Data mining: practical machine learning tools and techniques. 2. Morgan Kaufman; San Francisco: 2005. [Google Scholar]
- 31.Kim Y, Kweon J, Kim JS. TALENs and ZFNs are associated with different mutation signatures. Nat Methods. 2013;10:185. doi: 10.1038/nmeth.2364. [DOI] [PubMed] [Google Scholar]
- 32.Grau J, Boch J, Posch S. TALENoffer: genome-wide TALEN off-target prediction. Bioinformatics. 2013;29:2931–2932. doi: 10.1093/bioinformatics/btt501. [DOI] [PubMed] [Google Scholar]
- 33.McNaughton BR, Cronican JJ, Thompson DB, Liu DR. Mammalian cell penetration, siRNA transfection, and DNA transfection by supercharged proteins. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6111–6116. doi: 10.1073/pnas.0807883106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nature biotechnology. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat Biotechnol. 2012;30:593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 36.Sun N, Liang J, Abil Z, Zhao H. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol Biosyst. 2012;8:1255–1263. doi: 10.1039/c2mb05461b. [DOI] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 38.Noble WS. How does multiple testing correction work? Nature Biotechnology. 2009;27:1135–1137. doi: 10.1038/nbt1209-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.