Abstract
Somatic evolution during cancer progression and therapy results in tumor cells that exhibit a wide range of phenotypes including rapid proliferation and quiescence. Evolutionary life history theory may help us understand the diversity of these phenotypes. Fast life history organisms reproduce rapidly while those with slow life histories show less fecundity and invest more resources in survival. Life history theory also provides an evolutionary framework for phenotypic plasticity with potential implications for understanding ‘cancer stem cells’. Life history theory suggests that different therapy dosing schedules could select for fast or slow life history cell phenotypes, with important clinical consequences.
Introduction
Cancer has been historically viewed as a disease of rapid proliferation and uncontrolled cell growth. However, cancer must also evolve survival or ‘hardiness’ strategies to persist in challenging environments that may include hypoxia, acidosis, and a predatory immune response. It is likely that these adaptations significantly contribute to the ability of cancers to metastasize to other organs and survive toxic therapies. Life history theory, a theoretical framework from organismal evolutionary biology1 suggests that cancer cells may be subject to tradeoffs between maximizing growth and maximizing survival (i.e., having maximal tolerance and flexibility to unfavorable conditions) – cellular equivalents of the metaphorical ‘tortoises’ and ‘hares.’ In cancer evolution both strategies can be successful depending on the environmental conditions and both strategies have important clinical implications for cancer patients.
In general, evolutionary life history theory proposes that a number of tradeoffs shape the evolution of phenotypes. They apply to all living things that are subject to natural selection and therefore should apply to neoplastic cells as well. The three most important tradeoffs that have been identified are: 1) reproduction versus survivorship, 2), offspring now versus offspring later and 3) offspring number versus offspring quality2. Life history theory developed from the observation that even though each living organism possesses a unique natural history, all organisms’ life histories seem to fall along the “axes” defined by the three major life history tradeoffs. In long-lived mammals such as elephants (Loxodonta africana) the adaptive strategy emphasizes: 1) survivorship over offspring, 2) delayed maturation and longevity, and 3) fewer but higher quality offspring. By contrast, small mammals such as meadow voles (Microtus pennsylvanicus) have low survivorship, rapid maturation, and frequent, large litters of young. In many species, frequent and prolific reproduction comes at the cost of longevity3. For example, the lifespan of some organisms, i.e. burying beetle (Nicrophorus orbicollis) and fruit fly (Drosophila melanogaster), is significantly shortened by reproduction4–6. In a dividing cell, there is no separation between number and timing of offspring. Cells can only divide rapidly or slowly. Therefore, the latter two tradeoffs collapse into a single: division rate versus offspring (competitive) quality. Life history tradeoffs have been already been observed in single celled organisms such as yeast, where slower growth rates (i.e. via mutations involved in ribosome biogenesis or RNA polymerase) are associated with increased survival in challenging environments.7 Here we explore the presence and ramifications of life history tradeoffs for understanding and treating cancer.
Evolutionary life history theory can provide a useful framework for understanding the evolutionary selection pressures and adaptive strategies that govern the tradeoff between maximizing proliferation versus survival in cancer phenotypes. Like the rest of nature, neoplastic cells face evolutionary tradeoffs with respect to resource allocation and growth constraints. In this evolutionary competition, the ‘proliferative hares’ may reproduce rapidly but at the cost of increased mortality in adverse environments, whilst ‘quiescent tortoises’ may proliferate more slowly but with the benefit of increased survival under stressful conditions including administration of therapy. We propose that these tradeoffs are critical but poorly recognized components of cancer biology that have important implications for neoplastic progression and treatment. It is currently unknown how these tradeoffs play out in normal and neoplastic cells and whether the nature of these tradeoffs changes during progression.
A variety of mechanisms may underlie these life history tradeoffs, including those imposed by energy and time limitations as well as those constrained by cell state or configuration. One likely energetic tradeoff is that of ATP consumption by processes promoting proliferation versus those promoting survival. Drug resistant cell lines (i.e., those with enhanced survival in toxic conditions) have been shown to have approximately 50% less available ATP in a cell.8. Time limitations can also impose tradeoffs, such as between replicating DNA quickly (i.e., faster proliferation) versus replicating it accurately (indirectly enhancing survival). Finally, cells can be subject to tradeoffs among different states, as they cannot, for example, simultaneously be in an autophagous state (which enhances survival) and a proliferative state.
Mechanisms underlying fast life history strategies of cancer cells include abrogation of cell cycle checkpoints, shortening of the G1 phase of the cell cycle, increasing proliferation rate by suppression of DNA repair, use of fast (but error-prone) polymerases, activation of cell migration pathways, and even the switch to a glycolytic metabolism may facilitate increased proliferation9 while generating a toxic acidic environment10. Alternatively, mechanisms underlying slow life history strategies include suppression of apoptosis, up-regulation of efflux pumps, enhanced DNA repair, autophagy in starvation conditions, use of proof-reading polymerases, remodeling of the tumor microenvironment, greater ROS detoxification11, enhanced uptake and sequestering of resources along with their efficient utilization through oxidative phosphorylation at the expense of proliferation9.
The exact nature of tradeoffs between these mechanisms has yet to be determined in most cases. However, some of these mechanisms are likely to involve antagonistic pleiotropy12, in which a mutation that increases proliferation may have a negative effect on survival or vice versa. Antagonistic pleiotropy has been observed at the organismal level13, and appears to be instantiated by cell level characteristics including apoptosis and cell senescence14, 15. However, antagonistic pleiotropy is likely to apply to cancer cells as well, with tradeoffs between cell proliferation and survival emerging during somatic evolution. Because mutations early in cancer progression may enhance both survival and reproduction through inactivation of regulatory machinery (e.g., inactivation of TP53), we predict that antagonistic pleiotropy likely emerges later during progression (see ‘Life history tradeoffs may emerge late’).
Life history theory and tumor ecology
Ecologists have found that populations in stable environments with limited resources, evolve slow life histories16 (historically called “K-selection” after the carrying capacity term of the logistic model of population growth,17 though this terminology has lost favor). If resources are abundant and stable in the absence of predation (or other sources of high extrinsic mortality), the Malthusian law18 will eventually lead to the population growing until it is resource limited, and therefore slow life histories will eventually be selected. Organisms with slow life histories tend to evolve larger bodies, better somatic maintenance and repair, chemical defenses, mechanisms to survive environmental stresses, mechanisms to reduce the fitness of their competitors (direct competition), slower metabolism, efficient uptake and utilization of resources, little dispersal and longer lifespans16.
In contrast, environments with rapid and stochastic fluctuations in resource availability and/or high rates of extrinsic mortality (e.g., predation) select for fast life histories (historically called “r-selection“ after the maximum growth rate term of the logistic model17) that can exploit a temporary abundance of available resources and quickly repopulate following a disturbance or downturn. Organisms that evolve under fast life history selection tend to reproduce early and rapidly, overexploit their local environment, migrate (to escape competition, depleted local environments and to find new regions where they can proliferate19, 20), invest little in their own somatic maintenance, competitive capacity or their offspring, and die young21. There are exceptions to the general pattern of the tradeoff between survival and reproduction, but this is typically because of tradeoffs with other traits or other constraints (e.g., body size22, dispersal constraints23, age-dependent predation/mortality24, age dependent competition25 etc.).
The terms r- and K-selection have lost favor because they derive from a particular model of density dependent population growth that may not apply to many biological systems of interest and does not capture important selective forces such as competition, predation, etc. that may dominate the dynamics of many systems. In addition, the dynamics resulting from variation in extrinsic and age-specific mortality rates are often more important than the population growth rate (r)1, 21. We therefore use the terminology of fast and slow life history strategies rather than r- and K-selection.
Seed sizes in plants illustrate several aspects of life history tradeoffs: larger seeds represent greater investment and generally have a competitive advantage over smaller seeds. At low planting densities there is little correlation between the success of a species and its seed size. However, at high planting densities, larger seeded species tend to be highly over-represented relative to small seeded plants26, demonstrating the advantage deriving from investment in quality over quantity in competitive, resource limited environments.
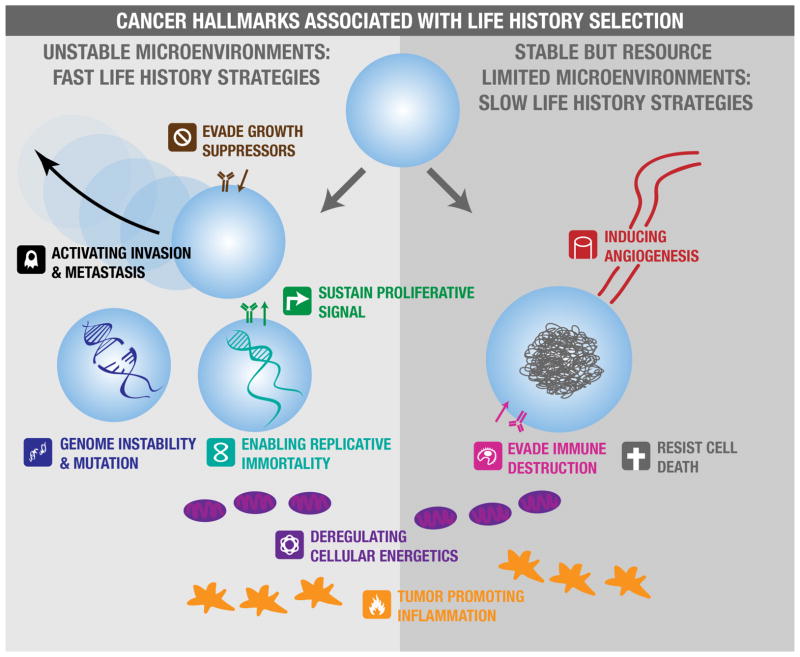
Neoplastic cells have phenotypes that reflect many characteristics of fast and slow life history strategies. In fact, all of the cancer “hallmarks”27, 28 fall under the categories of proliferation promoting phenotypes, survival promoting phenotypes, or both (FIG. 1). Sustaining proliferative signals, evading growth suppressors and enabling replicative immortality are clear fast life history hallmarks, enhancing the capacity of cells to proliferate quickly to vast numbers. Invasion and metastasis are other fast life history phenotypes, analogous to dispersal in organismal fast life history strategies. On the other hand, evading immune destruction and resisting cell death are clearly slow life history hallmarks, enhancing the ability of the cell to survive in challenging conditions. In addition, resource limited environments would be expected to select for angiogenesis, which is analogous to ‘niche construction’ behavior in slow life history organisms. Other hallmarks and enabling characteristics, such as deregulating cellular energetics and promoting tumor inflammation, may arise under fast or slow life history selection depending on the details of how these processes are affecting the cells. Early mutations may confer both proliferation and survival benefit though disrupting regulatory machinery, so tradeoffs between fast and slow life history characteristics of cells may not emerge until later in progression.
Figure 1. Hallmarks of cancer associated with life history selection.
Many of the hallmarks and enabling characteristics of cancer evolve under fast or slow life history selection. Environments that are unstable with regard to available resources and threats to cell survival, such as those characterized by wounding, variable blood flow, or rapid changes in the availability of growth factors, will select for fast life history hallmarks (left panel). Environments characterized by less disruption but limited availability of resources, or other population limitations such as immune predation, will select for slow life history hallmarks that increase cell survival or acquisition of resources (right panel). Some cancer hallmarks and enabling characteristics (positioned between the two panels) are associated with both fast and slow life history regimes.
An evolutionary life history approach suggests that environments characterized by resource disturbance and high rates of cell mortality will likely select for fast life history proliferation-promoting hallmarks, while more stable but resource limited environments will select for slow life history survival-promoting hallmarks. However, it is important to consider that these cell characteristics can in turn influence environmental conditions and affect life history selection pressures. For example, angiogenic capacity may initially evolve under slow life history selection, but lead to selection for fast life histories with a new abundance of (and likely fluctuations in) blood flow.
Tumor heterogeneity
Tumors are complex ecosystems containing multiple evolving populations29–33. We know surprisingly little about the basic population biology parameters of in vivo neoplastic cells including death rates, proliferation rates, cell turnover rates, nutrient cycling, energetics and longevity. In many cases, it is not even clear what resources are limiting.
It is likely that both the quiescent tortoises and proliferative hares exist in a heterogeneous tumor population34, 35. Tumors are mosaics of different microenvironments. Regions of low but stable resource availability (e.g., hypoxia) promote strong competitor neoplastic cells (tumor interior), while regions with high or fluctuating resource availabilities allow for the coexistence of the cells with traits for inefficient but rapid proliferation (e.g., edge of the tumor)36. Life history phenotypes in cancers should in general reflect proximity to blood flow37, the availability of resources, fluctuations in these availabilities, and extrinsic sources of mortality such as immune predation and chemotherapy.
The spatial heterogeneity in most tumors is apparent from variable enhancement of tumor regions in radiographic imaging following a contrast injection that enhances visible differences among regions with differential blood flow and cell density. (FIG. 2). Additionally, temporal variation in blood flow to the same tumor region has been well documented in experimental systems. Blood flow and nutrients in tumors change over seconds to hours38, 39. These temporal variations in resources should select for cells that proliferate quickly, over-exploit their environments and have higher rates of dispersal19, 20. The coexistence of both stable and fluctuating microenvironments should both select for and permit the coexistence of both fast and slow life history phenotypes within the same tumor36. Tradeoffs between quick colonization (i.e., rapid division and migration into areas of unutilized resources) and effective competition (i.e., investment in survival) have been associated with coexistence and the evolution of slow and fast life histories in some ciliate protists40. While heterogeneity in blood flow is the most obvious source of variations in extrinsic mortality and resources, other factors such as immune response, fibroblast infiltration and hormone or growth factor availability may further contribute to divergent selective forces on the life history phenotypes of neoplastic cells.
Figure 2. Tumor heterogeneity.
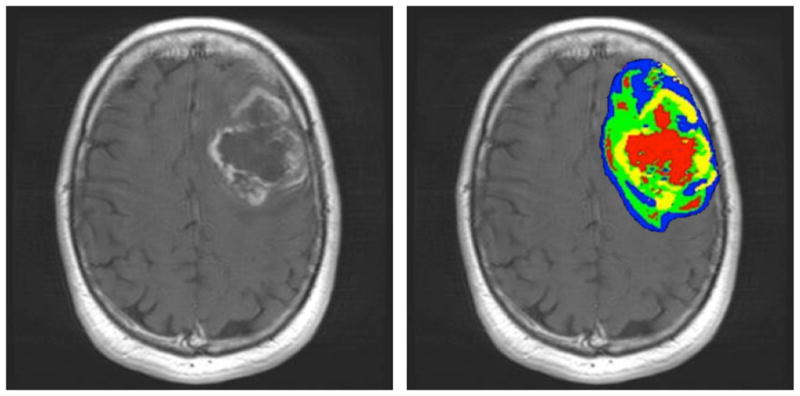
The left image is an MRI scan of a glioblastoma multiforme following gadolinium injection at Moffitt Cancer Center. The gray scale image on the left shows blood flow with white corresponding to highest flow and black to little or no flow. This will result in significant variations in many components of the tumor microenvironment including oxygen, glucose, acid, and serum-derived growth factors. In the right image, distinct intratumoral habitats are identified using combinations of images that correspond to vascular flow and cellular density, revealing heterogeneity with regard to resource availability and space competition within the tumor (red = high blood flow, low cell density; blue = low blood flow, high cell density; yellow = high blood flow, high cell density; green = low blood flow, low cell density).
Cancer progression
The “first law of ecology”41 states that all populations have the capacity to grow exponentially under ideal conditions. In terms of life history theory, this selects for fast life history strategies16. The second law of ecology recognizes limits to growth: no population can grow exponentially forever without reaching some resource limitation, which would then select for slow life history strategies.
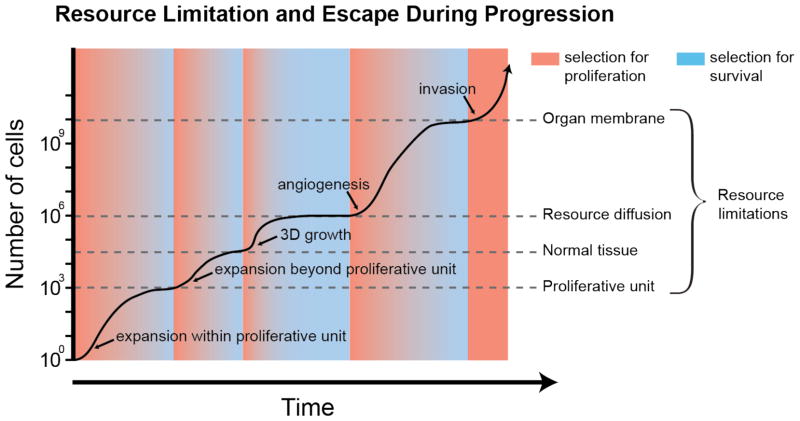
Selection for life history strategies change over both space and time as cells encounter resource limitations and gain the capacity to escape from those limitations. The specific details of fluctuating periods of selection for fast and slow life history strategies accompanying these resource limitations and escapes will vary for different tissues and neoplasms, but there are a number of resource limitations that are similar across many neoplasms, some of which are shown in FIG. 3. The exact order of these events likely varies across neoplasms.
Figure 3. Resource limitation and escape during progression.
The second law of ecology states that an exponentially growing population will eventually reach some limit to its growth. In neoplastic progression, there appear to be a series of limitations to the growth of neoplastic cell populations. During progression, the neoplastic cell population evolves mechanisms for escaping each limitation, temporarily releasing the neoplastic cell population with a burst of proliferation such that mutant cells with fast life history strategies have a competitive advantage (red background). However, when those populations reach a new resource limitation, selection shifts from fast to slow life history strategies (blue background), such that cells that can best compete, sequester resources, and avoid death, have an advantage.
Most epithelial tissues are subdivided into proliferative units. A mutant cell population may expand exponentially until it fills the niche of a proliferative unit. While constrained within this proliferative unit, there is selection for slow life histories that compete well for limited space and proliferative opportunities. If a new mutation allows a clone to escape a single proliferative unit, perhaps by stimulating crypt fission, as occurs in the stomach and intestine32, 42, 43, or through invasion into neighboring proliferating units, as seems to occur in the skin44, the neoplastic cell population can grow exponentially again. This will lead to a new phase of fast life history selection until the next resource limitation, that of epithelial tissue architecture, is reached.
As long as the epithelial tissue architecture is intact, a mutant clone that can escape its original proliferative unit is still constrained to the two-dimensional structure of an epithelial sheet, and it will eventually reach a new phase of slow life history selection. Thus, proliferative units and the other constraints of tissue architecture act as powerful tumor suppressors45. When their cellular phenotype becomes capable of three-dimensional growth, neoplastic cells can proliferate or survive without requiring attachment to the basement membrane (e.g. through becoming resistant to anoikis or inducing autophagy46) as is seen in ductal carcinoma in situ47. Three-dimensional growth leads to a new phase of fast life history selection as neoplastic cells proliferate exponentially once again. However, this exponential growth phase ends once the neoplasm begins to reach the limits of oxygen diffusion at approximately 2mm3. This leads to slow life history selection once again, as cells compete for survival under resource-limited conditions (i.e., the limits of diffusion from the existing capillary network).
Evolution of angiogenesis again allows escape from resource constraints and a period of fast life history selection. However, an angiogenic tumor will typically reach space limitations imposed by organ membranes, leading to slow life history selection again. Once the neoplastic cell population escapes the final resource limitation with the capacity for invasion and metastasis, they become difficult to manage clinically and often result in host death48.
The clinical lethality of metastatic cancer may partly be due to the fact that metastatic cells have evolved the capacity to break through resource constraints such as those shown in FIG. 3. When cancer cells colonize new tissues, they may be preadapted to quickly break through some resource constraints, but not others. To the extent that the metastatic site affords similar environmental features and constraints as the primary site in which the cells initially evolved, they will not be held back by resource limitations and may therefore be able to maintain a fast proliferation rate in a new tissue. If the metastatic sites are sufficiently different from the primary site, the growth of metastases may be slowed as the cell populations may encounter novel resource limitations that they are not preadapted to overcome, such as different growth and survival factors in the microenvironment. It is not currently known whether the growth and survival factors in typical secondary sites are similar to those in the primary site. This may help explain why certain tumors are predisposed to metastasize to some sites and not others: if resource constraints and affordances in a metastatic site are similar to the primary tumor, cells that land at that similar site may be much more likely to grow into secondary tumors. If these metastatic cells are able to grow at all in the new tissue, it is likely that they will be able to grow exponentially without encountering as many limitations since they already have the capacity for 3D growth, angiogenesis and invasion. This perspective may partially explain why metastatic cancer progresses very quickly in some cases and more slowly in others28, 49.
There is evidence that in some cases cancer single cell dissemination may occur prior to the primary tumor showing collective cell invasion that can be recognized histologically50. Genetic analyses suggest single disseminated cells may derive from ancestral clones relatively early in progression51. These early dispersers may have evolved under early periods of fast life history selection (FIG. 3). Ecologists have recognized that there can be different mechanisms of dispersal, responding to different selective pressures, even within the lifespan of an organism (e.g., juvenile vs. adult dispersal)52. We predict that early dispersing cells come from lineages that have experienced less selection for increased proliferation and survival compared to cells that disperse later in progression, because they survived fewer phases of fast and slow life history selection. These cells should also be less likely to be pre-adapted to breaking through all the constraints on growing into clinically relevant metastases. The later dispersers, because they have gone through more phases of fast and slow life history selection, may have the capacity for faster proliferation, greater survival, and phenotypic plasticity, making them more stem-like, relative to early dispersers (see below).
Life history tradeoffs may emerge late
We hypothesize that, early in progression, mutations can improve both proliferation and survival without apparent conflict (Box 1 and FIG. 4). This can be achieved by mutations that disrupt the regulatory machinery of the cell that would otherwise be limiting cell proliferation and survival. In normal cells, proliferation and survival are highly constrained to regulate the function (and ultimately promote the fitness) of the multicellular body of which they are a part. During the process of somatic evolution in neoplastic progression, cells may evolve improved proliferation and survival as they disrupt the regulatory machinery that would normally constrain them. However, these early mutations will not necessarily represent adaptations that fully maximize the neoplastic cell’s fitness. These early stage neoplastic cells retain many features that made them useful to the whole organism; namely restrained proliferation, restrained nutrient uptake and metabolism, and additional cellular functions aimed at sustaining the whole organism’s physiology and homeostasis. Later in progression, cancer cells may become subject to cell-level tradeoffs between survival and proliferation, as they can no longer improve one without sacrificing the other.
Box 1. Tradeoffs in trait combinations.
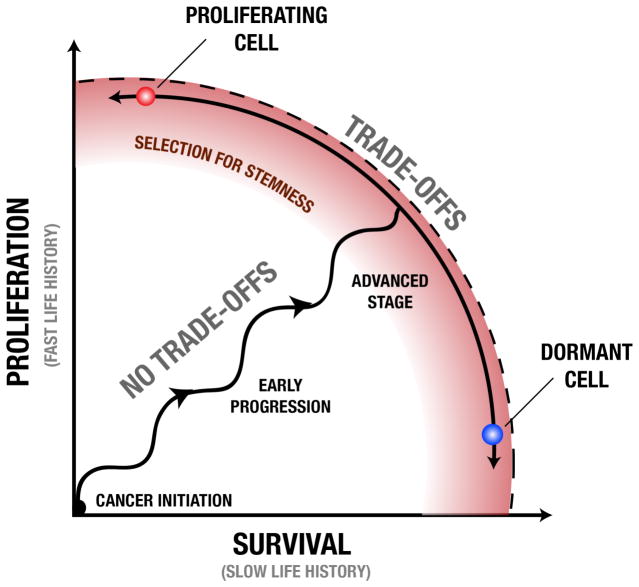
Evolution by natural selection promotes traits that may not have initially been constrained by tradeoffs, but eventually do reach such a constraint. A “fitness set” is defined as all combinations of aptitudes that an organism might have based on its evolutionarily feasible traits109. When an organism is within its fitness set, natural selection can promote improvement in multiple aptitudes. This will push the organism’s traits to the boundary of the fitness set (called the active edge). At this point it is no longer possible to evolve a trait value that simultaneously improves all fitness aptitudes. At the active edge, there is a tradeoff among fitness aptitudes, and selection will move the trait along the active edge until the fitness maximizing balance of aptitudes is achieved (FIG. 4). Tradeoffs between survival and proliferation among cancer cells may be subject to these same constraints. We propose that early in progression, neoplastic cells are within the fitness set and selection simultaneously can improve both cell reproduction and survival. This leads to selection for neoplastic cells that move the population phenotypes towards the active edge (dotted line in FIG. 4). Later in progression, neoplastic cells may be constrained by tradeoffs between survival and reproduction or between competitive quality and reproduction as they have reached the active edge. This may also lead to selection for plasticity or ‘stemness’ along the active edge so that cells can dynamically optimize their proliferative/survival phenotypes for the conditions they experience.
Figure 4. Tradeoffs between proliferation and survival during cancer progression.
During progression, neoplastic cell lineages go through periods of selection for increased proliferation (vertical movement) and increased survival (horizontal), possibly in phases as resource constraints are reached (selecting for survival) and broken through (selection for proliferation) Early in progression, proliferation and survival can both increase without significant tradeoffs through destruction of various regulatory systems within the cell that otherwise suppress those functions. Later in progression, the capacity of cells to proliferate and survive becomes limited by fundamental tradeoffs, rather than the regulatory machinery of the cell. While some microenvironments may stably select for a particular point along this active edge, leading to cells that specialize on a fixed life history strategy, temporal changes in the microenvironment or cell migration through different microenvironments may select for cells with phenotypic plasticity. In order to proliferate more quickly without sacrificing survival (or vice versa), cells must be able to alter their phenotypes from reproduction specialist (e.g., a proliferating cell) to a survival specialist (e.g., a dormant cell). This selection for phenotypic plasticity or ‘stemness’ (red zone) later in progression may be explained by the fact that clones adopting a conditional phenotype subject to dynamic life history tradeoffs can achieve higher fitness than those constrained to a phenotype with fixed life history strategy.
Tradeoffs between proliferation and survival may lead to selection favoring extreme phenotypes such a rapidly proliferating cells with poor survival abilities (red cell, FIG. 4) or apparently dormant cells that can survive extremely well (blue cell, FIG. 4). The emergence of these tradeoffs may even lead to selection for cells that can dynamically alter their phenotypes through epigenetic modifications, based on environmental conditions. For example, hypomethylation of oncogene promoters and/or hypermethylation of tumor suppressor promoters may provide a proliferative phenotype, while signals of resource scarcity may induce a quiescence phenotype through hypomethylation of cell cycle inhibitor promoters and/or hypermethylation of pro-apoptotic promoters. In other words, there may be selection for cells that can dynamically alter their phenotypes to proliferate rapidly when resources are temporarily abundant but go dormant when resources are scarce or the environment is otherwise unfavorable53. If the capacity to dynamically switch from a fast to slow life history phenotype provides a fitness advantage, this may lead to selection for cells that are ‘stemlike’ in their multipotent capacities.
Some evidence suggests that life history tradeoffs may change over the course of progression. Slower proliferation rates have been observed in more advanced breast cancer cell lines compared to cell lines taken from earlier stages of progression, but this decrease in proliferation appears to be accompanied by increased detoxification of reactive oxygen species (ROS).11 This apparent tradeoff between proliferation rate and survival (via enhancement of ROS detoxification) in advanced cancer may be explained by competition over NADPH, which is the limiting resource for both proliferation and detoxification11. Whether the emergence of tradeoffs such as these is a fundamental feature of neoplastic progression is an open question.
Cell plasticity and life history tradeoffs
Though the concept of cancer stem cells remains controversial, there is clear evidence of phenotypic heterogeneity that can be regenerated from cells with markers of stemness54, 55. One of the most puzzling aspects of cancer stem cells, and potentially one source of confusion in the literature, is their phenotypic plasticity. Cancer stem cells exhibit the capacity for survival and plasticity under challenging conditions56. Stem cells in normal tissues appear to be capable of generating a wide range of proliferative phenotypes, from rapid proliferation to dormancy57, 58. There is growing evidence that cancer stem cells are also capable of this variation in proliferation rates59, 60. This may allow them to survive and maintain populations across diverse and stressful environmental conditions, including colonization of new (metastatic) microenvironments. In other words, cancer stem cells appear to be capable of generating both a proliferative ‘hare’ phenotype and a quiescent ‘tortoise’ phenotype. This may indicate that cancer stem cells have the ability to conditionally produce cells with fast or slow life history strategies but identical genetic heritage. However, the idea that life history tradeoffs may drive cell plasticity is not dependent on the stem cell hypothesis. Even in the absence of stem cells, phenotypic plasticity may be accomplished through epigenetic modifications61 or gene and non-coding RNA regulation62.
Among organisms, phenotypic plasticity is ubiquitous5, 63 and such changes in life history strategies are typically conditional on reliable environmental cues about the stability (or instability) of the environment. When organisms are likely to encounter a variety of potential environments, they can evolve the capacity for state dependent life history strategies or ‘reaction norms’64. Reaction norms allow individuals to adopt the phenotype that is most likely to maximize their fitness in the particular environment in which they find themselves. Intensely varying environments can induce a change in organismal life history strategies by providing cues that indicate a fast life history strategy would be more effective65. There are many examples of conditional life history strategies in response to predation, where signals of increased predation leads to the organism adopting a faster life history strategy66–68.
One example of phenotypic plasticity in response to environmental conditions is the protist Tetrahymena vorax, which is able to take on two distinct morphotypes called a microstome and a macrostome. The microstome simply consumes bacteria and small microbes. The macrostome consumes large prey such as other ciliates and protists. A microstome can morph into a macrostome in response to environmental cues of low bacterial abundance and high abundance of other protists. The macrostome, when undergoing cell division, can maintain its macrostome morphology or return to a microstome morphology, again in response to cues of its resource environment69. The ability to maintain distinct morphotypes as a generalist with a flexible “behavioral” strategy leads to cells that are genetically identical but manifest a diversity of environmentally contingent traits. The molecular mechanisms underlying this transition are unknown70. However, research on the phenotypic plasticity in social insects, i.e. honeybees (Apis mellifera) provide evidence that both allelic variation and gene expression (likely from epigenetic modifications71) can produce colony members with very different morphology and behavior 72. It has been suggested that life history traits contribute to the phenotypic plasticity among social insects (i.e., developmental diet of female honeybee larvae determines worker vs. queen)73. Under such an adaptation, the diversity of traits seen in the population emerges from a single flexible “species” rather than the coexistence of genetically distinct specialist species or clones36. Cancer stem cells may provide such a dynamic adaptation where their generalist strategy creates morphological diversity but not the genetic heterogeneity that would be found from the coexistence of tumor cell lineages with specialist traits for either a fast or slow life history strategies.
This concept is supported by experimental evidence that cancer stem cells can differentiate into a diversity of non-stem cell phenotypes when engrafted into an immune-compromised mouse74, 75. Recent evidence even suggests that non-stem-like cancer cells can dedifferentiate back into a cancer stem cell phenotype76 and that cancer stem cells can take on a variety of phenotypes associated with differentiation but still maintain their stem-like properties77.
The high mortality and disruption to resource availability characteristic of chronic wounding, dysregulated angiogenesis and the administration of cytotoxic therapies should provide reliable cues for neoplastic cells to calibrate their phenotypes towards a fast life history strategy. In contrast, when resources are relatively stable but limited, as might be the case for non-angiogenic micrometastasis, this may cue neoplastic cells to calibrate their pheontypes for a slow life history strategy. If these changes in the environment occur on a time scale that is shorter than the generation time of the neoplastic cells (which is likely for migratory cells), then cells that can change their phenotype will have an advantage over cells with fixed life history strategies. This could be accomplished through cells sensing environmental signals and adjusting their transcription profiles. Differentiation on the part of cancer stem cells may be a result of conditional responses to the environment that parallel the ways in which organisms respond to environmental conditions78.
If cancer cells use conditional life history strategies, this might help to explain the apparent microenvironmental control of cancer cell phenotypes79. A number of exciting studies have shown that in different microenvironments, neoplastic cells can be driven towards a benign or more aggressive phenotype. In other words, environmental cues may shift these cells from a fast life history phenotype to a slow life history phenotype or vice versa. Thus, inflammatory microenvironments, for example, may promote neoplastic progression80–84 through environmental signals of high cell mortality that shift the neoplastic cells to a faster life history strategy. Because cell plasticity is considered an aspect of stemness, it is likely that the capacity for plasticity in cell life history strategies may be associated with stem-like cells. However, it is possible that a cell that does not carry stem cell markers could also shift life history strategies in response to environmental conditions.
Thus, the apparent capacity of cancer stem cells to fluidly shift among different phenotypic states based on environmental cues may, by permitting rapid adaptation to a wide range of environmental conditions, confer a fundamental evolutionary advantage on some neoplastic cell populations. It is clear that the microenvironment plays a critical role in neoplastic progression79. However, if there is selection on neoplastic cells for the capacity to adaptively shift phenotypes in response to environmental cues, this would represent a novel component of cancer biology.
Life history characteristics may emerge at the level of cell lineages or clones as well as within a single cell. For example, a cell lineage with high levels of telomere maintenance may proliferate less rapidly initially, but have greater long-term survival of the clone. Similarly, a stem cell lineage that possess the equivalent of a soma in the form of differentiated cell progeny may sacrifice rapid expansion of the stem cells in exchange for a survival benefit conferred through some function of the non-stem cells. We have shown that this is a viable explanation for the existence of non-stem cells in neoplasms85. One mechanism of survival benefit may be analogous to the disposable soma hypothesis of aging86. The disposable soma hypothesis posits that aging is a result of the fact that it is costly to build and maintain a body that will be ‘disposable’ after viable offspring have been produced. It may be the case that a stem cell lineage can similarly enhance its competitive fitness by shunting misfolded proteins, deleterious mutations45, and other cytoplasmic debris into a differentiating (i.e., aging) daughter cell through an asymmetric division. Whether this occurs in neoplasms is an open question. In essence, plasticity could be realized temporally, through a cell switching state, or spatially, through a clone expressing multiple cell types. Spatial plasticity may facilitate the manifestation of complex life history strategies at the clone level.
Treatments influence life history
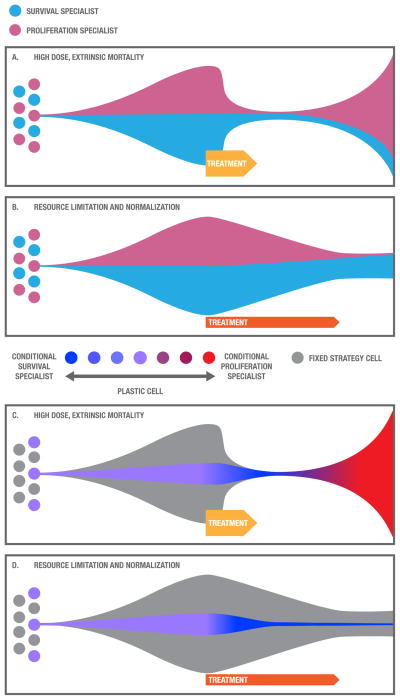
Cancer therapy can influence the life history characteristics of tumors through selection (FIG. 5). A high dose treatment that causes extensive cell mortality (e.g., standard high dose cytotoxic chemotherapy) and disruption of the environment may be initially quite successful, killing the majority of cancer cells. It is likely that high dose therapies first select for slow life history characteristics. For example, in the presence of a cytotoxic agent, cells with slow life history hallmarks may have a survival advantage over their competitors due to survival strategies such efflux pumps, enhanced drug metabolism, enhanced DNA repair, increased cell size and inactivation of apoptotic pathways87, 88. Interestingly (and unfortunately) this selection advantage may only be transient as any surviving cells with a fast life history strategy will have an advantage in the abundance of space and resources following death of the sensitive population (called “competitive release89–91; FIG. 5A). A treatment that limits and normalizes resource availability or otherwise controls the size of a tumor (e.g., adaptive therapy92), on the other hand, may cause less tumor mortality initially but select for cells that specialize in survival and competition, with lower rates of proliferation, enabling long-term cancer control (FIG. 5B). Many targeted therapies are cytostatic93 (preventing proliferation by halting the cell cycle) and are sometimes given in low doses relatively continuously, which should limit the tumor size. This would lead some targeted therapies, in principle, to select for cells with slow life history strategies. However, targeted therapies often do cause massive cell death and if they are given in bursts with just a few high doses, they will be more like a cytotoxic treatment. Dosing and treatment algorithms may have critical effects on the evolution of cancer cells that could be harnessed for slowing progression and the evolution of resistance.
Figure 5. Effects of treatment on life history strategies.
Different treatments select for and induce different life history strategies. A. Traditional high dose therapies cause high levels of cell mortality, initially selecting for survival specialists (light blue) but, afterwards, with an abundance of resources and a paucity of competitors, proliferating specialists (pink) gain an advantage, which may contribute to recurrence. B. A treatment that normalizes and limits resources selects for cells with slow life history strategies, specializing on survival and competition (pink), which may facilitate long-term cancer control. C. Cells with conditional life history strategies may respond to cytotoxic therapies by shifting first into a survival phenotype (blue) and then back into a rapidly proliferating phenotype (red), which may lead relapse. D. When exposed to a therapy that establishes a constant environment with limited yet stable resources, cells with conditional life history strategies may shift into a slow life history phenotype that invests cellular resources into survival and competition (blue), which may enable long-term cancer control.
Perhaps the most common selection event for cancer evolution is surgical excision. Obviously, surgery is generally designed to remove all of the tumor – essentially an extinction event. However, the surgeon may leave tumor behind either inadvertently or of necessity if it is too extensive to be resected completely. In general, surgical resection targets macroscopically visible neoplastic cells and their neighbors. Neoplastic cells with increased motility that invade individually or in small groups will tend to survive surgery. In addition, the surgery can alter the local microenvironmental conditions in ways that are favorable or unfavorable to tumor growth. For example, an influx of inflammatory cells may increase the immune response to tumors or promote tumor proliferation and motility. In either case, this an interesting and largely unexplored clinical opportunity for application evolutionarily-enlightened local or systemic therapy at the time of surgery that could reduce subsequent tumor growth and slow relapse. For example, this framework suggests that post-operative drugs that inhibit proliferative signals in the microenvironment could slow relapse (though likely at the cost of inhibiting surgical wound healing). Finally, surgery is likely a scattering event in which cancer cells can disseminate locally in the surgical field and systemically through blood and lymphatic vessels. There is a theoretical concern that cells most likely to be “shaken loose” from the primary also may have properties that confer a greater likelihood of proliferating at a distant site. However, the evidence is mixed for whether or not surgery increases the probability of metastases94.
In addition, treatments may influence life history through creating environmental conditions that generate cues that may shift life history strategies of phenotypically plastic (stem-like) cells. For example, a cytotoxic treatment may induce a fast life history phenotype by exposing cells to cues of environmental disruption and high mortality followed by opportunities for proliferation (FIG. 5C). A treatment that limits and normalizes resources might shift cells to a slow life history strategy through exposing them to cues of high cell density and limited resources (FIG. 5D).
The effect of therapy on life history strategies of cancer cells raises the question of whether therapies can be designed to select or induce slow life history strategies. This may be a novel method for long-term treatment of cancer since slow life history strategies are more likely to produce a stable or slowly expanding tumor that does not kill the patient. Thus, an explicit prediction of this approach is that therapeutic strategies that eschew high cell mortality and environmental change and instead promote a stable, predictable environment, and relatively low extrinsic mortality should lead to long-term cancer control. At least one experiment using an ‘adaptive therapy’ protocol suggests this is possible92. Unlike standard chemotherapy, adaptive therapy was designed to maintain a stable tumor size, rather than eradicate the tumor. Adaptive therapy uses a conditional algorithm that adjusts drug dosing according to tumor burden. Mice with xenograft tumors of OVCAR-3 ovarian cancer cells were treated with carboplatin using both adaptive and standard therapy dosing schedules. The host mice in the adaptive therapy group could be kept alive indefinitely, while the ‘standard’ high dose chemotherapy treated mice died in a matter of months92. So far, these results have only been shown in one model system with one drug. In addition, experimental manipulations in mice designed to spatially homogenize the resources in a neoplasm resulted in a dramatic suppression of metastasis 95, 96. We may also be able to select for or induce slow life history strategies through maintaining a constant low dose of a drug (metronomic therapy)97, particularly if we use cell-cycle specific drugs that select against proliferating cells. However, the use of alternative dosing schemes is still in early stages: a recent study of conditional intermittent therapy in prostate cancer showed equivalent overall survival compared to continuous anti-androgen therapy98, making it clear that more research is needed to determine the conditions under which adaptive therapy (and other conditional therapies) work. An understanding of the fundamental dynamics driving life history evolution in neoplasms may help design more effective algorithms for long-term cancer control.
Life history theory may promote dormancy
Many cancer therapies appear to result in residual but dormant neoplastic cells (called “minimal residual disease”), which may lead to recurrence many years later51, 99, 100. In many cases, therapy itself selects for dormancy, by preferentially killing proliferating cells so that only the quiescent cells survive. Furthermore, dormancy may be an effective survival strategy for neoplastic cells when resources are limited. Proliferative cells may experience an internal phenotypic shift from fast to slow life history in response to some extrinsic factors in the microenvironment46, 101. In organismal evolution, lifespan increases among species that hibernate, possibly due to seasonal allocation of resources to reproduction (as opposed to using and requiring resources in all seasons, such as annual plants)102. For neoplastic cells that are dormant following therapy, immediate proliferation will permit initial population growth (fast life history) but only if the environment has sufficient resources. Cells that delay proliferation (slow life history) may be able to survive and maintain their overall quality during lean periods or in locations with sparse resources, but risk missing out on potentially successful cell division opportunities. Thus cancer cell dormancy may represent an example of the third life history tradeoff of offspring now versus offspring later.
This implies that maintenance therapy in patients with minimal residual disease may extend the period of dormancy if it maintains a limitation on resources for the cancer cells or provides other cues that maintain a slow life history phenotype. For example, anti-inflammatory drugs could possibly reinforce dormancy by maintaining a stable microenvironment and inhibit pro-growth cues that are found in wound healing and other inflammatory processes. Non-steroidal anti-inflammatory drugs (NSAIDs) have been found to prevent a wide variety of cancers103, particularly esophageal adenocarcinoma104, 105. We have recently shown that NSAIDs can reduce the rate of mutation in Barrett’s esophagus (a premalignant condition) by an order of magnitude106. This massive reduction in mutation rate per unit time may be due to a shift towards slower life history strategies of cells (i.e, reduction in the rate of cell division), increased investment in DNA repair (i.e., a reduction in mutation rate per division), or reduced exposure to mutagens (e.g., reactive oxygen species).
If some cancer cells are able to conditionally shift their life history characteristics in response to environmental cues, it might also be possible to develop screening methods that could identify the presence of conditional life history cells, versus cells with fixed slow life history strategies, to predict the likelihood of recurrence. This may involve detection of cancer cells with stem-cell markers or other markers of cells that have the capacity for plasticity in response to the opportunities and constraints in their microenvironments.
Conclusions
Life history theory provides a framework for understanding several puzzling aspects of cancer, including the mixture of rapidly proliferating and quiescent cells in the same tumor, the phenomenon of tumor dormancy followed by relapse, and the plasticity of so-called cancer stem cells. However, there has been controversy in the evolution and ecology literature over researchers using life history characteristics to classify organisms, rather than focusing on the types of selective pressures shaping a population107, 108. A focus on the selective pressures shaping life history strategies in cancer suggests opportunities for the development of new treatment regimes and prevention strategies that take life history evolution into account. The evolutionary life history approach suggests a potential strategy for prolonging life of the host– not necessarily for eliminating the cancer, but rather for achieving cure via long-term control.
Acknowledgments
We would like to thank Aurora Nedelcu, Aleah Caulin, and AJ Figuredo for thoughtful and thought provoking discussions in the development of these ideas. This work was supported in part by Research Scholar Grant #117209-RSG-09-163-01-CNE from the American Cancer Society, and NIH grants F32 CA144331, R01 CA149566, R01 CA170595, and R01 CA140657, U54 CA143970 and a grant from the McDonnell Foundation 220020270.
References
- 1.Stearns SC. The evolution of life histories. Oxford University Press; New York: 1992. [Google Scholar]
- 2.Stearns SC. Trade-offs in life-history evolution. Functional Ecology. 1989;3:259–268. [Google Scholar]
- 3.Williams GC. Natural selection, the cost or reproduction and a refinement of Lack’s principle. Am Nat. 1966;100:687–690. [Google Scholar]
- 4.Creighton JC, Heflin ND, Belk MC. Cost of reproduction, resource quality, and terminal investment in a burying beetle. Am Nat. 2009;174:673–84. doi: 10.1086/605963. [DOI] [PubMed] [Google Scholar]
- 5.Fabian D, Flatt T. Life History Evolution. Nature Education Knowledge. 2012;3:24. [Google Scholar]
- 6.Partridge L, Prowse N. The effects of reproduction on longevity and fertility in male Drosophila melanogaster. J Insect Physiol. 1997;43:501–512. doi: 10.1016/s0022-1910(97)00014-0. [DOI] [PubMed] [Google Scholar]
- 7.Zakrzewska A, et al. Genome-wide analysis of yeast stress survival and tolerance acquisition to analyze the central trade-off between growth rate and cellular robustness. Mol Biol Cell. 2011;22:4435–46. doi: 10.1091/mbc.E10-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broxterman HJ, et al. Induction by verapamil of a rapid increase in ATP consumption in multidrug-resistant tumor cells. FASEB J. 1988;2:2278–82. doi: 10.1096/fasebj.2.7.3350243. [DOI] [PubMed] [Google Scholar]
- 9.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–9. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 11.Jerby L, et al. Metabolic associations of reduced proliferation and oxidative stress in advanced breast cancer. Cancer Res. 2012;72:5712–20. doi: 10.1158/0008-5472.CAN-12-2215. [DOI] [PubMed] [Google Scholar]
- 12.Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 13.Leroi AM, et al. What evidence is there for the existence of individual genes with antagonistic pleiotropic effects? Mech Ageing Dev. 2005;126:421–9. doi: 10.1016/j.mad.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 14.Campisi J. Cellular senescence and apoptosis: how cellular responses might influence aging phenotypes. Exp Gerontol. 2003;38:5–11. doi: 10.1016/s0531-5565(02)00152-3. [DOI] [PubMed] [Google Scholar]
- 15.Ungewitter E, Scrable H. Antagonistic pleiotropy and p53. Mech Ageing Dev. 2009;130:10–7. doi: 10.1016/j.mad.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeschke JM, Kokko H. The roles of body size and phylogeny in fast and slow life histories. Evol Ecol. 2009;23:867–878. [Google Scholar]
- 17.MacArthur R, Wilson EO. The theory of island biography. Princeton University Press; Princeton, N.J: 1967. [Google Scholar]
- 18.Malthus TR. An essay on the principle of population. Johnson; London: 1798. [Google Scholar]
- 19.Aktipis CA, Maley CC, Pepper JW. Dispersal evolution in neoplasms: the role of disregulated metabolism in the evolution of cell motility. Cancer Prev Res (Phila) 2012;5:266–75. doi: 10.1158/1940-6207.CAPR-11-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Sprouffske K, Huang Q, Maley CC. Solving the puzzle of metastasis: the evolution of cell migration in neoplasms. PLoS One. 2011;6:e17933. doi: 10.1371/journal.pone.0017933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reznick D, Bryant MJ, Bashey F. r- and K- selection revisited: The role of population regulation in life-history evolution. Ecology. 2002;83:1509–1520. [Google Scholar]
- 22.Skutch AF. Life History of Longuemare’s Hermit Hummingbird. International Journal of Avain Science. 1951;93:180–195. [Google Scholar]
- 23.Howe HF, Smallwood J. Ecology of Seed Dispersal. Ann Review of Ecology and Systematics. 1982;13:201–228. [Google Scholar]
- 24.Promislow DEL, Harvey PH. Living fast and dying young: A comparative analysis of life- history variation in mammals. J Zool. 1990;220:417–437. [Google Scholar]
- 25.Ebenman B. Competition between age classes and population dynamics. J Theor Biol. 1988;131:389–400. [Google Scholar]
- 26.Turnbull LA, Rees M, Crawley MJ. Seed mass and the competition/colonization trade-off: a sowing experiment. J Ecol. 1999;87:899–912. [Google Scholar]
- 27.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 28.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Gerlinger M, Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer. 2010;103:1139–43. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillies RJ, Verduzco D, Gatenby RA. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat Rev Cancer. 2012;12:487–93. doi: 10.1038/nrc3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–35. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 33.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 34.van Diest PJ, van der Wall E, Baak JP. Prognostic value of proliferation in invasive breast cancer: a review. J Clin Pathol. 2004;57:675–81. doi: 10.1136/jcp.2003.010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreso A, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–8. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orlando PA, Gatenby RA, Brown JS. Tumor evolution in space: The effects of competition colonization tradeoffs on tumor invasion dynamics. Frontiers in Oncology. doi: 10.3389/fonc.2013.00045. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alfarouk KO, Ibrahim ME, Gatenby RA, Brown JS. Riparian ecosystems in human cancers. Evol Appl. 2013;6:46–53. doi: 10.1111/eva.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brurberg KG, Skogmo HK, Graff BA, Olsen DR, Rofstad EK. Fluctuations in pO2 in poorly and well-oxygenated spontaneous canine tumors before and during fractionated radiation therapy. Radiother Oncol. 2005;77:220–6. doi: 10.1016/j.radonc.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Cardenas-Navia LI, et al. The pervasive presence of fluctuating oxygenation in tumors. Cancer Res. 2008;68:5812–9. doi: 10.1158/0008-5472.CAN-07-6387. [DOI] [PubMed] [Google Scholar]
- 40.Limberger R, Wickham SA. Competition-colonization trade-offs in a ciliate model community. Oecologia. 2011;167:723–32. doi: 10.1007/s00442-011-2013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turchin P. Does population ecology have general laws? OIKOS. 2001;94:17–26. [PubMed] [Google Scholar]
- 42.Graham TA, et al. Use of methylation patterns to determine expansion of stem cell clones in human colon tissue. Gastroenterology. 2011;140:1241–1250. e1–9. doi: 10.1053/j.gastro.2010.12.036. [DOI] [PubMed] [Google Scholar]
- 43.Greaves LC, et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci U S A. 2006;103:714–9. doi: 10.1073/pnas.0505903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W, et al. UVB-induced apoptosis drives clonal expansion during skin tumor development. Carcinogenesis. 2005;26:249–57. doi: 10.1093/carcin/bgh300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cairns J. Mutation Selection and the Natural History of Cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 46.Kenific CM, Thorburn A, Debnath J. Autophagy and metastasis: another double-edged sword. Curr Opin Cell Biol. 2010;22:241–5. doi: 10.1016/j.ceb.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–88. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 48.Etzioni R, et al. The case for early detection. Nat Rev Cancer. 2003;3:243–52. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 49.Seliger B. Strategies of tumor immune evasion. BioDrugs. 2005;19:347–54. doi: 10.2165/00063030-200519060-00002. [DOI] [PubMed] [Google Scholar]
- 50.Rhim AD, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–61. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt-Kittler O, et al. From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:7737–42. doi: 10.1073/pnas.1331931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Debarre F, Gandon S. Evolution in heterogeneous environments: between soft and hard selection. Am Nat. 2011;177:E84–97. doi: 10.1086/658178. [DOI] [PubMed] [Google Scholar]
- 53.Wilting RH, Dannenberg JH. Epigenetic mechanisms in tumorigenesis, tumor cell heterogeneity and drug resistance. Drug Resist Updat. 2012;15:21–38. doi: 10.1016/j.drup.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 54.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–9. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 55.Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283–96. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holzel M, Bovier A, Tuting T. Plasticity of tumour and immune cells: a source of heterogeneity and a cause for therapy resistance? Nat Rev Cancer. 2013;13:365–76. doi: 10.1038/nrc3498. [DOI] [PubMed] [Google Scholar]
- 57.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–5. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson A, et al. Dormant and self-renewing hematopoietic stem cells and their niches. Ann N Y Acad Sci. 2007;1106:64–75. doi: 10.1196/annals.1392.021. [DOI] [PubMed] [Google Scholar]
- 59.Biddle A, et al. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res. 2011;71:5317–26. doi: 10.1158/0008-5472.CAN-11-1059. [DOI] [PubMed] [Google Scholar]
- 60.Kusumbe AP, Bapat SA. Cancer stem cells and aneuploid populations within developing tumors are the major determinants of tumor dormancy. Cancer Res. 2009;69:9245–53. doi: 10.1158/0008-5472.CAN-09-2802. [DOI] [PubMed] [Google Scholar]
- 61.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Godlewski J, et al. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol Cell. 2010;37:620–32. doi: 10.1016/j.molcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.West-Eberhard MJ. Developmental Plasticity and Evolution. Oxford University Press, Inc; New York, New York: 2003. [Google Scholar]
- 64.Houston AI, McNamara JM. Phenotypic plasticity as a state-dependent life-history decision. Evol Ecol. 1992;6:243–253. [Google Scholar]
- 65.Gurney WSC, Middleton DAJ. Optimal resource allocation in a randomly varying environment. Functional Ecology. 1996;10:602–612. [Google Scholar]
- 66.Ball SL, Baker RL. Predator induced life history changes: Antipredator behavior costs or facultative life history shifts? Ecology. 1996;77:1116–1124. [Google Scholar]
- 67.Reznick D, Butler MJ, Rodd H. Life history evolution in guppies. VII. The comparative ecology of high and low predation environments. Am Nat. 2001;157:12–26. doi: 10.1086/318627. [DOI] [PubMed] [Google Scholar]
- 68.Chivers DP, Kiesecker JM, Marco A, Wildy EL, Blaustein AR. Shifts in life history as a repsonse to predation in western toads (Bufo boreas) J Chem Ecol. 1999;25:2455–2463. [Google Scholar]
- 69.Buhse HE, Jr, Williams NE. A comparison of cortical proteins in Tetrahymena vorax microstomes and macrostomes. J Protozool. 1982;29:222–6. doi: 10.1111/j.1550-7408.1982.tb04015.x. [DOI] [PubMed] [Google Scholar]
- 70.Ryals PE, Smith-Somerville HE, Buhse HE., Jr Phenotype switching in polymorphic Tetrahymena: a single-cell Jekyll and Hyde. Int Rev Cytol. 2002;212:209–38. doi: 10.1016/s0074-7696(01)12006-1. [DOI] [PubMed] [Google Scholar]
- 71.Foret S, et al. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc Natl Acad Sci U S A. 2012;109:4968–73. doi: 10.1073/pnas.1202392109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fitzpatrick MJ, et al. Candidate genes for behavioural ecology. Trends Ecol Evol. 2005;20:96–104. doi: 10.1016/j.tree.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 73.Smith CR, Toth AL, Suarez AV, Robinson GE. Genetic and genomic analyses of the division of labour in insect societies. Nat Rev Genet. 2008;9:735–48. doi: 10.1038/nrg2429. [DOI] [PubMed] [Google Scholar]
- 74.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 75.Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 76.Gupta PB, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–44. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 77.Taussig DC, et al. Leukemia-initiating cells from some acute myeloid leukemia patients with mutated nucleophosmin reside in the CD34(−) fraction. Blood. 2010;115:1976–84. doi: 10.1182/blood-2009-02-206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schlichting CD. Origins of differentiation via phenotypic plasticity. Evol Dev. 2003;5:98–105. doi: 10.1046/j.1525-142x.2003.03015.x. [DOI] [PubMed] [Google Scholar]
- 79.Bissell MJ, Labarge MA. Context, tissue plasticity, and cancer: are tumor stem cells also regulated by the microenvironment? Cancer Cell. 2005;7:17–23. doi: 10.1016/j.ccr.2004.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bunt SK, Sinha P, Clements VK, Leips J, Ostrand-Rosenberg S. Inflammation induces myeloid-derived suppressor cells that facilitate tumor progression. J Immunol. 2006;176:284–90. doi: 10.4049/jimmunol.176.1.284. [DOI] [PubMed] [Google Scholar]
- 81.Bunt SK, et al. Reduced inflammation in the tumor microenvironment delays the accumulation of myeloid-derived suppressor cells and limits tumor progression. Cancer Res. 2007;67:10019–26. doi: 10.1158/0008-5472.CAN-07-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–52. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 85.Sprouffske K, et al. An evolutionary explanation for the presence of cancer nonstem cells in neoplasms. Evol Appl. 2013;6:92–101. doi: 10.1111/eva.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kirkwood TB. Evolution of ageing. Mech Ageing Dev. 2002;123:737–45. doi: 10.1016/s0047-6374(01)00419-5. [DOI] [PubMed] [Google Scholar]
- 87.Borst P. Cancer drug pan-resistance: pumps, cancer stem cells, quiescence, epithelial to mesenchymal transition, blocked cell death pathways, persisters or what? Open Biol. 2012;2:120066. doi: 10.1098/rsob.120066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 89.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smith VH, Holt RD. Resource competition and within-host disease dynamics. Trends Ecol Evol. 1996;11:386–9. doi: 10.1016/0169-5347(96)20067-9. [DOI] [PubMed] [Google Scholar]
- 91.Wargo AR, Huijben S, de Roode JC, Shepherd J, Read AF. Competitive release and facilitation of drug-resistant parasites after therapeutic chemotherapy in a rodent malaria model. Proc Natl Acad Sci U S A. 2007;104:19914–9. doi: 10.1073/pnas.0707766104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gatenby RA, Silva AS, Gillies RJ, Frieden BR. Adaptive therapy. Cancer Res. 2009;69:4894–903. doi: 10.1158/0008-5472.CAN-08-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Contractor KB, Aboagye EO. Monitoring predominantly cytostatic treatment response with 18F-FDG PET. J Nucl Med. 2009;50 (Suppl 1):97S–105S. doi: 10.2967/jnumed.108.057273. [DOI] [PubMed] [Google Scholar]
- 94.Coffey JC, et al. Excisional surgery for cancer cure: therapy at a cost. Lancet Oncol. 2003;4:760–8. doi: 10.1016/s1470-2045(03)01282-8. [DOI] [PubMed] [Google Scholar]
- 95.Mazzone M, et al. Heterozygous deficiency of PHD2 restores tumor oxygenation and inhibits metastasis via endothelial normalization. Cell. 2009;136:839–51. doi: 10.1016/j.cell.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robey IF, et al. Bicarbonate increases tumor pH and inhibits spontaneous metastases. Cancer Res. 2009;69:2260–8. doi: 10.1158/0008-5472.CAN-07-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pasquier E, Kavallaris M, Andre N. Metronomic chemotherapy: new rationale for new directions. Nat Rev Clin Oncol. 2010;7:455–65. doi: 10.1038/nrclinonc.2010.82. [DOI] [PubMed] [Google Scholar]
- 98.Crook JM, et al. Intermittent androgen suppression for rising PSA level after radiotherapy. N Engl J Med. 2012;367:895–903. doi: 10.1056/NEJMoa1201546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7:834–46. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Radich JP, Wood BL. In: Leukemia and Related Disorders. Estey EH, Appelbaum FR, editors. Springer; New York: 2012. pp. 251–271. [Google Scholar]
- 101.Lu Z, et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008;118:3917–29. doi: 10.1172/JCI35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wilkinson GS, South JM. Life history, ecology and longevity in bats. Aging Cell. 2002;1:124–31. doi: 10.1046/j.1474-9728.2002.00020.x. [DOI] [PubMed] [Google Scholar]
- 103.Rothwell PM, et al. Effect of daily aspirin on long-term risk of death due to cancer: analysis of individual patient data from randomised trials. Lancet. 2011;377:31–41. doi: 10.1016/S0140-6736(10)62110-1. [DOI] [PubMed] [Google Scholar]
- 104.Corley DA, Kerlikowske K, Verma R, Buffler P. Protective association of aspirin/NSAIDs and esophageal cancer: a systematic review and meta-analysis. Gastroenterology. 2003;124:47–56. doi: 10.1053/gast.2003.50008. [DOI] [PubMed] [Google Scholar]
- 105.Vaughan TL, et al. Non-steroidal anti-inflammatory drugs and risk of neoplastic progression in Barrett’s oesophagus: a prospective study. Lancet Oncol. 2005;6:945–52. doi: 10.1016/S1470-2045(05)70431-9. [DOI] [PubMed] [Google Scholar]
- 106.Kostadinov RL, et al. NSAIDs Modulate Clonal Evolution in Barrett’s Esophagus. PLoS Genet. 2013;9:e1003553. doi: 10.1371/journal.pgen.1003553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Parry GD. The meaning of r- and K-selection. Oecologia (Berlin) 1981;48:260–264. doi: 10.1007/BF00347974. [DOI] [PubMed] [Google Scholar]
- 108.Mueller LD. Density-dependent population growth and natural selection in food-limited environments: The Drosophila model. Am Nat. 1988;132:786–809. [Google Scholar]
- 109.Levins R. Evolution in Changing Environments. Princeton University Press; Princeton, New Jersey: 1968. [Google Scholar]