Abstract
Cysteine S-nitrosylation is a posttranslational modification by which nitric oxide regulates protein function and signaling. Studies of individual proteins have elucidated specific functional roles for S-nitrosylation, but knowledge of the extent of endogenous S-nitrosylation, the sites that are nitrosylated, and the regulatory consequences of S-nitrosylation remains limited. We used mass spectrometry-based methodologies to identify 1011 S-nitrosocysteine residues in 647 proteins in various mouse tissues. We uncovered selective S-nitrosylation of enzymes participating in glycolysis, gluconeogenesis, tricarboxylic acid cycle, and oxidative phosphorylation, indicating that this posttranslational modification may regulate metabolism and mitochondrial bioenergetics. S-nitrosylation of the liver enzyme VLCAD (very long acyl-CoA dehydrogenase) at Cys238, which was absent in mice lacking endothelial nitric oxide synthase, improved its catalytic efficiency. These data implicate protein S-nitrosylation in the regulation of β-oxidation of fatty acids in mitochondria.
Introduction
Protein S-nitrosylation is a major nitric oxide-derived reversible posttranslational modification that regulates enzymatic activity, protein localization and stability (1-9), and that also plays a role in nitric oxide-mediated signaling (1-3). Although functional roles for S-nitrosylation have been documented for individual proteins, a global analysis of S-nitrosylation and the S-nitrosylation sites under physiological conditions in vivo remain limited. To this end, we implemented a mass spectrometry-based proteomic approach, which allows for the site-specific identification of S-nitrosocysteine residues in complex mixtures (10). The method is based on selective enrichment either of S-nitrosocysteine peptides or intact S-nitrosylated proteins with organomercury compounds. The peptides are released with performic acid, which oxidizes the cysteine to sulfonic acid enabling precise detection of the modified peptides by mass spectrometry. Alternatively proteins can be eluted intact and probed with antibodies against specific proteins enabling quantification of the modified protein molecules. To ensure specificity of detection negative controls are generated by pre-treatment of samples with UV-light which eliminates S-nitrosocysteine and analyzed under the same conditions (10). We used these methodologies to identify endogenous S-nitrosylated proteins in six wild-type mouse organs as well as in the same tissues from mice lacking the endothelial nitric oxide synthase (eNOS). We reasoned that a global discovery of the S-nitrosocysteine proteome in mice will reveal potential functional regulation of core biochemical pathways by S-nitrosylation and the changes in this proteome in the absence of one of the major enzymatic sources of nitric oxide.
Results
A mouse S-nitrosocysteine proteome
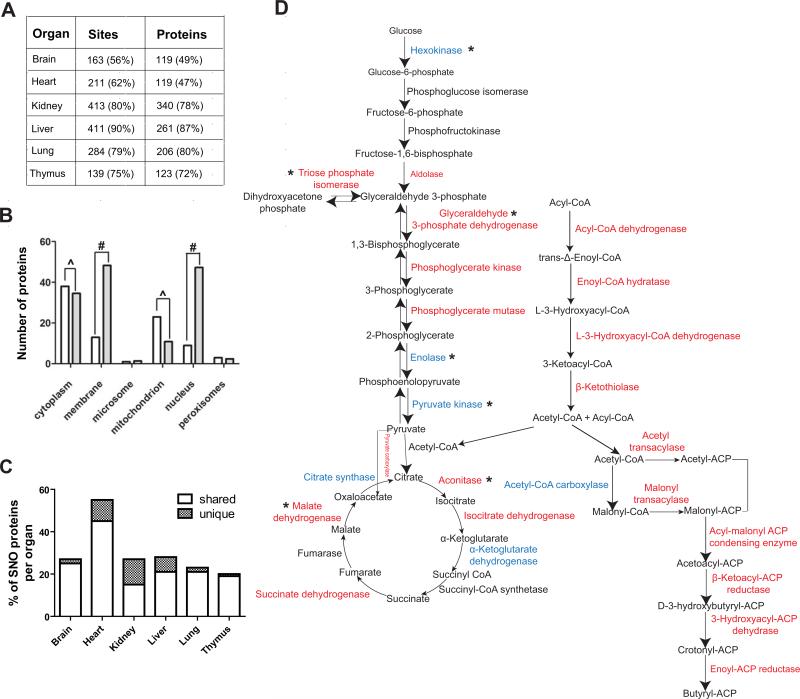
In wild-type mouse brain, heart, kidney, liver, lung and thymus we identified 1011 S-nitrosocysteine containing peptides on 647 proteins (figure 1A and tables S1-S6). Extensive literature searches indicated that this expanded S-nitrosocysteine proteome identified 46 proteins previously reported to be modified under physiological conditions and uncovered 971 previously unknown sites of endogenous S-nitrosylation. In all six organs the number of S-nitrosylation sites exceeded the number of proteins (figure 1A) indicating a potential role of poly S-nitrosylation in the regulation of protein function in vivo (11). Comparison of the proteins identified in the six organs in at least three biological replicates for each organ revealed that on average 72% of the proteins were identified in more than one organ (figure S1A) implying that similar patterns of S-nitrosylation serve global functions in vivo. Moreover, these data indicated that the methodologies employed to acquire these proteomes were accurate and reproducible. Proteins identified only in one organ ranged from 21-46% implying that S-nitrosylation could also serve organ specific roles (figure S1A). The dependency of the sites of S-nitrosylation in vivo to nitric oxide generated by eNOS was explored by analyzing the endogenous sites of modification in eNOS null mice (eNOS−/−) in the same organs. We found that eNOS substantially contributed to the endogenous S-nitrosocysteine proteome because S-nitrosylation of 47-87% of the proteins identified required the presence of eNOS (figure 1A). The brain and the heart had the lowest dependency on eNOS activity compared to the other organs indicating the contribution of other isoforms such as nNOS for S-nitrosylation of proteins in these organs. The absence of a substantial number of S-nitrosocysteine peptides in organs from eNOS−/− mice reinforced the accuracy of the methodologies in identifying this S-nitrosocysteine proteome.
Figure 1.
Overview of the cysteine S-nitrosoproteome of the mouse. (A) The number of sites and proteins identified across six organs in wild-type mouse and their dependency to eNOS activity as percentage of the wild type is indicated in parenthesis. Three biological replicates from each organ from wild type and eNOS−/− mice were analyzed. (B) Sub-cellular localization of the S-nitrosoproteome (white bars) and comparison with the rest of the mouse proteome (grey bars); ^ indicate overrepresentation and # underrepresentation, with both indicating p<0.0001. (C) Percentage of S-nitrosylated mitochondrial proteins. The white bar indicates proteins shared with at least another organ. The hatched bar indicates unique proteins for a particular organ. (D) S-nitrosylated enzymes present in wild-type mouse liver that were absent in eNOS−/− liver and that are involved in glucose metabolism, TCA cycle and fatty acid metabolism are marked in red. S-nitrosylated proteins that are present in eNOS−/− liver are depicted in blue. Asterisk (*) indicates proteins that have been reported to be S-nitrosylated. For all the proteins and sites identified see tables S1 to S6.
An overview of the subcellular localization of the S-nitrosoproteome revealed a tissue wide significant enrichment for cytosolic and mitochondrial proteins as compared to the entire mouse proteome (figure 1B). S-nitrosylation sites in proteins in cellular membranes and nucleus were under-represented (figure 1B). The under representation of membrane proteins may reflect methodological issues because the tissue homogenization method was not optimized for extraction of membrane proteins. S-nitrosylation of proteins also occurs in the nucleus and the discovery of additional sites in nuclear proteins reinforces the potential importance of S-nitrosylation in signaling and transcriptional regulation (5). 20-25% of the S-nitrosoproteome in the brain, kidney, liver, lung and thymus consisted of mitochondrial proteins whereas 56% of the modified proteins were localized to mitochondria in the mouse heart (figure 1C). The mitochondrial proteomes were more than 70% dependent on eNOS activity with the exception of the heart where only 36% of the proteome requires eNOS-derived nitric oxide for S-nitrosylation (figure S1B). The lower dependency of S-nitrosylation on eNOS activity in heart mitochondria implies the presence of another functional NOS isoform. Functional classification of the S-nitrosoproteome revealed key metabolic pathways in which S-nitrosylated enzymes play a central role (figure S1C). We found that a substantial number of enzymes that regulate glycolysis, gluconeogenesis, pyruvate metabolism, TCA cycle, oxidative phosphorylation, amino acid metabolism, ketone body formation and fatty acid metabolic pathways were S-nitrosylated (figure S1C), the majority of which were not identified as S-nitrosylated in eNOS−/− mice (figure 1D and tables S1-S6). This finding is consistent with previous reports indicating that the endothelial nitric oxide synthase null mice have impaired metabolic activity (12,13). Therefore, it appears that protein S-nitrosylation provides the mechanistic link that couples eNOS activity and regulation of metabolism.
Regulation of the enzymatic activity of very long chain acyl-coA dehydrogenase by S-nitrosylation
Clustering of proteins that participate in fatty acid metabolism was apparent in this analysis. We have previously reported that in the mouse liver, S-nitrosylated proteins are clustered in a network that encompasses liver responses to the hormone leptin (10). In this study we found that very long chain acyl-coA dehydrogenase (VLCAD), which catalyzes the rate limiting step in the β-oxidation of fatty acids, was S-nitrosylated in wild-type mouse liver but not in the livers of mice lacking leptin (ob/ob) or eNOS−/− mice (figure S2). We further investigated the biological effect of S-nitrosylation on VLCAD activity and fatty acid metabolism in the liver, which is a major metabolic organ.
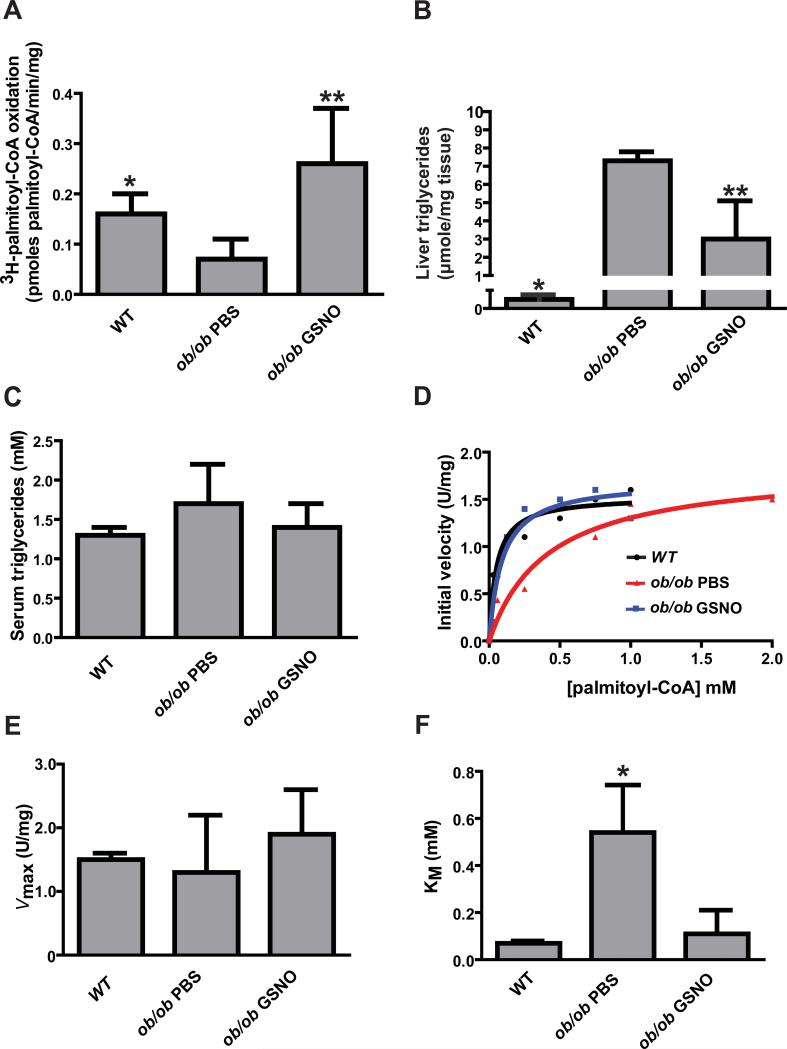
Leptin deficient mice develop liver steatosis spontaneously under normal chow diet starting at four to five weeks of life that is characterized by a diminished rate of palmitate oxidation and accumulation of fat droplets in the form of triglycerides within the hepatocytes (14, 15). The rate of 3H-labeled palmitoyl-CoA oxidation by liver homogenates in ob/ob mice was 47% of the rate in wild-type mice (figure 2A). This significant reduction in the rate of palmitoyl-CoA oxidation indicated a deficiency in mitochondrial β-oxidation of fatty acids. Based on previous observations that showed reversal of hepatic steatosis in ob/ob mice by delivery of S-nitroso-N-acetyl cysteine (15), we injected five week old ob/ob mice intraperitoneally with S-nitrosoglutathione (GSNO). We used GSNO to deliver nitric oxide equivalents because of the extensive use of this pharmacological agent in cellular models to modify proteins by S-nitrosylation despite limited cellular permeability (16,17). GSNO-injected ob/ob mice exhibited a similar palmitoyl-CoA oxidation rate as wild-type mice and a significantly increased rate over PBS-injected ob/ob mice (figure 2A). Moreover the restoration of palmitoyl-CoA oxidation in GSNO-injected ob/ob mice was associated with a significant reduction of the concentration of liver triglycerides. The liver triglyceride concentration in ob/ob mice injected with PBS was significantly higher than in wild-type mice, and treatment of ob/ob mice with GSNO significantly lowered the liver triglyceride concentration (figure 2B). The serum triglyceride concentrations were similar in wild-type mice, PBS-treated ob/ob mice, and GSNO-treated ob/ob mice (figure 2C). The restoration of palmitoyl-CoA oxidation in GSNO-injected ob/ob mice was also corroborated by histological evaluation of liver slices, which showed fewer fat deposits in GSNO-injected than in PBS-injected ob/ob mice (figure S3).
Figure 2.
(A) Rate of 3H-labeled palmitoyl-CoA oxidation in liver homogenates from wild-type and ob/ob mice.* p<0.05 by ANOVA with Bonferroni's post hoc test between wild-type and ob/ob PBS-treated mice (N=4 mice). **p<0.01 by ANOVA with Bonferroni's post hoc test between ob/ob PBS and ob/ob GSNO treated mice (N= 4 mice). (B) Liver triglyceride measurements in wild-type, PBS-treated ob/ob mice, and GSNO-treated ob/ob mice. *p<0.0001 by ANOVA with Bonferroni's post hoc test between wild-type and ob/ob PBS-treated mice (N=4 mice), **p<0.005 by ANOVA with Bonferroni's post hoc test between ob/ob PBS and ob/ob GSNO treated mice (N= 4 mice). (C) Serum triglyceride measurements in wild-type, PBS-treated ob/ob mice, and GSNO-treated ob/ob mice. No statistical difference (N=4 mice). (D) Representative measurements of VLCAD initial velocity measured as a function of palmitoyl-CoA concentration in liver homogenates from wild-type (black line), ob/ob PBS (red line) and ob/ob GSNO (blue line) mice. (E and F) Kinetic analysis of VLCAD enzymatic activity reveals similar Vmax but significantly higher KM in PBS-treated ob/ob mouse liver as compared to wild-type and GSNO-treated ob/ob mouse liver. *p<0.05 by ANOVA with Bonferroni's post hoc test. N=3 biological replicates.
MS/MS analysis localized the site of VLCAD modification to Cys238 in both wild-type and GSNO-injected ob/ob mice (figures S2A and S2B). We also confirmed that VLCAD was not modified by S-nitrosylation in eNOS−/− mice but was readily modified ex vivo at cysteine residue Cys238 by treating liver homogenates with GSNO (figure S4).
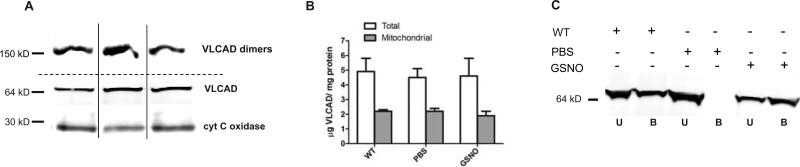
The S-nitrosylation of VLCAD was associated with an increase in acyl dehydrogenase-mediated oxidation of palmitoyl-CoA in GSNO-treated ob/ob mouse liver (figure 2D, S5A and S5B). This increase was sensitive to UV photolysis and was abolished by the inclusion of specific antibodies against VLCAD (figure S5A). Analysis of Michaelis-Menten kinetics (figure 2D) revealed that the Vmax of VLCAD was similar in wild-type, PBS-injected and GSNO-injected ob/ob mice (figure 2E). However, the apparent KM value measured in homogenates from PBS-injected ob/ob mice was more than 5-fold higher than in those from wild-type mice and GSNO-injected ob/ob mice (figure 2F). The KM for VLCAD enzymatic activity also increased 5-fold in eNOS−/− liver homogenates treated ex vivo with GSNO compared to untreated eNOS−/− liver homogenate (figure S4A). Quantification of the abundance of VLCAD protein in liver homogenates and enriched mitochondria preparations indicated no differences in abundance among wild-type, PBS and GSNO injected ob/ob mice (figure 3A and 3B). Furthermore, native gel electrophoresis revealed equal abundance of VLCAD dimers in the mitochondria fractions from all three groups of mice (figure 3A, upper panel). Therefore these data suggest that S-nitrosylation decreases the KM of VLCAD.
Figure 3.
(A) Representative Western blots assessing VLCAD in native gel (top panel) and in SDS-gel (middle panel). Noncontiguous lanes from a single experiment are indicated by black lines. (B) Quantification of abundance of VLCAD in SDS-gels under reducing conditions in total liver homogenates and enriched mitochondria fractions from liver. No statistical difference by ANOVA. N=3 different mice. (C) Representative Western blot for VLCAD in liver homogenates eluted from organomercury resin. The signal intensity was used to determine the abundance of S-nitrosylated VLCAD in the bound (B) fraction and the unmodified VLCAD present in the unbound (U) fraction. The data were repeated in two independent liver homogenates.
Quantification of the fraction of S-nitrosylated VLCAD indicated that in GSNO-treated mice 25 ± 3% of VLCAD molecules in liver were S-nitrosylated. These findings confirm the MS/MS analysis showing S-nitrosylation of VLCAD in wild-type and GSNO-treated mouse liver but not in PBS-treated ob/ob mouse liver (figure 3C). Because the concentrations of VLCAD protein were similar in PBS and GSNO treated ob/ob mice and on average the steady state abundance of S-nitrosylated VLCAD was 25% of the total protein, S-nitrosylation could increase the catalytic efficiency (kcat/KM) (18) of VLCAD 29-fold as compared to the unmodified protein. This substantial increase in catalytic efficiency could in part provide efficient removal of fatty acids in ob/ob mouse liver.
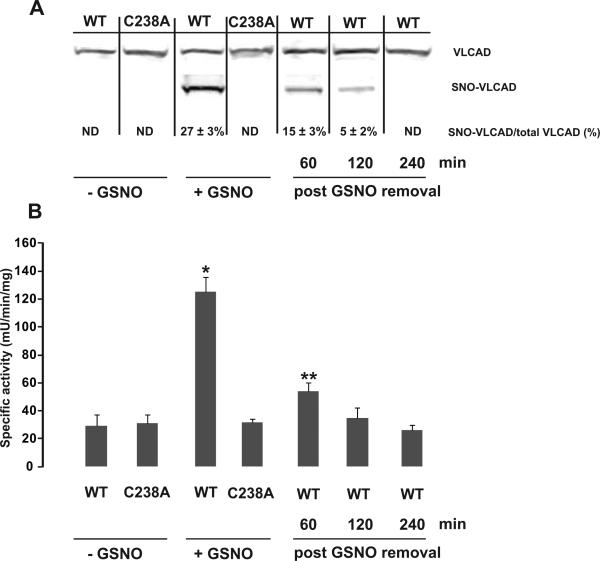
We obtained additional evidence for the functional consequences of S-nitrosylation of VLCAD in mouse hepatocytes transiently expressing wild-type VLCAD or a point mutant that could not be S-nitrosylated (C238A). GSNO treatment resulted in S-nitrosylation of 27 ± 3% of wild-type VLCAD (figure 4A) concomitant with a 5-fold increase of VLCAD specific activity (figure 4B). Under the same experimental conditions the C238A VLCAD mutant was not S-nitrosylated and its basal activity did not change in response to GSNO treatment (figure 4A and 4B). Upon removal of GSNO from the media and extensive washing, both the abundance of S-nitrosylated-VLCAD and the enzymatic specific activity declined over time (figure 4A and 4B). These data indicate that S-nitrosylation of VLCAD at Cys238 is necessary for the regulation of its enzymatic activity and this process is reversible possibly controlled through denitrosylation.
Figure 4.
Hepa 1-6 cells transiently expressing either FLAG tagged wild-type or C238A VLCAD were exposed to GSNO. (A) Representative Western blot analysis of unbound (VLCAD) and bound fractions (SNO-VLCAD) collected after mercury assisted capture in cell lysates. The unbound fraction indicates the abundance of the unmodified protein. The bound fraction indicates the abundance of S-nitrosylated VLCAD. The abundance of both was determined by using a calibrated antibody binding curve using purified VLCAD. The fraction of S-nitrosylated VLCAD as percentage of VLCAD is indicated. ND, not detected. N=3 biological replicates. Noncontiguous lanes from a single experiment are indicated by black lines. (B) The specific activity of VLCAD was significantly higher in GSNO-treated cells expressing wild-type VLCAD but not in GSNO-treated cells expressing equivalent amount of C238A VLCAD mutant protein. *p< 0.001, **p<0.05, by ANOVA with Bonferroni's post hoc test, N=3 biological replicates.
** “This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.”
We used molecular dynamic simulation to gain insight into the effect of S-nitrosylation on VLCAD structure. We initially used quantum mechanics/molecular mechanics (QM/MM) calculations to generate the S-nitrosocysteine residue at position 238 within the protein structure of VLCAD (PDB ID 2UXW) (Figure S6A). Examination of the lowest frequency normal modes, which indicate large global or collective motions of a protein, did not indicate that S-nitrosylation induced large conformational changes. Examination of higher frequency modes to investigate smaller local motions that might occur near the binding site revealed a difference in the movement of atoms in response to S-nitrosylation of Cys238 (Figure S6B). Cys238 resides in a loop, and loops generally represent flexible regions of a protein. Therefore, the data implies that S-nitrosylation enhances loop flexibility, resulting in increased protein movement which in turn facilitates substrate binding and therefore lowers the KM of VLCAD. Cys238 was ≥ 30 Angstroms from the substrate binding site, suggesting that longer range motions can influence the ability of the enzyme to bind substrate.
Discussion
Cysteine S-nitrosylation is a nitric oxide-derived posttranslational modification that modulates protein activity, protein-protein interactions, and subcellular localization under physiological and pathological conditions (1-9). Applying site specific mapping of S-nitrosocysteine residues in wild-type mouse tissue we uncovered widespread modification of proteins participating in metabolic pathways and a significant localization of modified proteins in the mitochondria. This selective localization is consistent with several previous reports indicating functional roles for nitric oxide in mitochondrial biology (19-21) and specifically for the heart where protein S-nitrosylation protects the heart from ischemia-reperfusion injury (22-24). Delivery of nitric oxide donors specifically to the heart increases the overall abundance of mitochondrial S-nitrosylated proteins and protects from ischemic injury (22-24). Despite these important biological contributions of nitric oxide in the functional regulation of mitochondria and metabolism the identification of S-nitrosylated mitochondrial proteins is limited. The identification of specific sites of S-nitrosylation in mitochondrial proteins across six different mouse tissues provides a substantial advance that facilitates mechanistic studies to uncover molecular and biochemical pathways for nitric oxide-mediated regulation of bioenergetics and metabolism. Similar to other post-translational modifications, S-nitrosylation is a dynamic process and several pathways that reverse this post-translational modification have been described (3). Therefore the current S-nitrosocysteine proteome may represent a portion of the endogenously modified proteins under normal physiological conditions.
To explore the biological role of S-nitrosylation in vivo we focused on fatty acid metabolism in the liver, a major metabolic organ in the body. Diet, adipose tissue and de novo lipogenesis are the major sources of fatty acids for the liver. Potential fates of fatty acids in the liver include esterification to triglycerides, which can be stored or packaged with ApoB100 for export as part of very low density lipoprotein particles. Fatty acids can be also converted to phospholipids or transformed to acyl-carnitines for transport into mitochondria where they undergo beta oxidation (25). Any process that increases the input or decreases the output or metabolism of fatty acids potentially contributes to the development of liver steatosis (25). Beta oxidation is a four step enzymatic cycle and each turn of the cycle shortens the length of the fatty acid by two carbon atoms, which is critical for efficient utilization of fatty acids. VLCAD catalyzes the first step in β-oxidation, accounting for about 80% of palmitate dehydrogenase activity in human liver and nearly 70% of palmitate oxidation in mouse liver (26-28). VLCAD is a homodimer consisting of 67 kDa subunits and is embedded in the inner mitochondria membrane 26-28). The enzymatic activity of VLCAD appears to be regulated by protein abundance and phosphorylation of Ser586 (29). Our data indicate that S-nitrosylation of VLCAD at Cys238 through eNOS-derived nitric oxide results in reversible activation of enzymatic activity through conformational changes that alter the KM of the enzyme and could substantially influence the in vivo β-oxidation of fatty acids. Overall, the global analysis of S-nitrosylated proteins in mouse tissues revealed that this posttranslation modification could profoundly influence cellular metabolic processes and mitochondria function.
Materials and Methods
Chemicals and Reagents
Palmitoyl coenzyme A lithium salt and ferricenium hexafluorophosphate were obtained from Sigma-Aldrich (St Louis, MO). Mouse monoclonal antibodies against FLAG, GAPDH and cytochrome c oxidase subunit I were from Stratagene (Santa Clara, CA) and Abcam (Cambridge, MA) respectively. Rabbit and goat (G-16 clone) polyclonal antibodies against VLCAD were obtained from Genetex (Irvine, CA) and Santa Cruz Biotechnology (Canta Cruz, CA) respectively. All chemicals and reagents used were of analytical grade.
Isolation of mouse organs and preparation of protein homogenates
All mouse studies were reviewed and approved by the Institutional Animal Care and Use Committee of the Children's Hospital of Philadelphia Research Institute. Wild-type C57BL/6J, Nos3tm1Unc (eNOS−/−) C57BL/6J and Lepob (ob/ob) C57BL/6J adult mice were obtained from Jackson Laboratories. Food intake and body weight were recorded for ob/ob mice during the course of four weeks when the mice were being injected with PBS or 5 mM GSNO every two days. The average food intake was 42 ± 9 and 41 ± 10 g for PBS and GSNO injected ob/ob mice respectively (N=4 mice per genotype). The average body weight change for the same period of time was 28.2 ± 1.0 and 28.1 ± 4.0 for ob/ob PBS injected and ob/ob GSNO injected mice respectively (N=4 mice per genotype). Mice were anesthetized by CO2 and blood was collected before being perfused through the left ventricle. Intact organs were collected, immediately frozen in liquid nitrogen, and stored at −80 °C until use. Tissues were homogenized into 3 ml of lysis buffer (250mM Hepes-NaOH, pH 7.7, containing 1 mM DTPA, 0.1 mM neocuproine, 1% Triton X-100, and protease inhibitors) on ice using a Teflon pestle and a Jumbo Stirrer (Fisher Scientific). The homogenates were then centrifuged at 13,000 × g for 30 minutes at 4 °C. The soluble protein fraction was collected and the protein concentration was determined by the Bradford assay. Sample preparation and generation of the negative control samples were performed as previously described (10).
Chemical enrichment and site specific identification of the S-nitrosocysteine proteome
A detailed experimental procedure for the preparation and activation of columns, homogenate preparation for reaction with organic mercury resin has been previously published (10). Three biological replicates from each organ were analyzed. Each sample had a corresponding UV-pretreated negative control analyzed under identical conditions. The false identification rate was <6% for brain and <3% for all other organs. For washes stringent conditions were selected due to the differences in lipid content among the six organs. Columns were initially washed with 50 bed volumes of 50 mM Tris-HCl, pH 7.4, containing 300 mM NaCl, 0.5% SDS, followed by 50 bed volumes of the same buffer containing 0.05% SDS. Columns were washed with 50 bed volumes of 50 mM Tris-HCl, containing 300 mM NaCl pH 7.4, 1% Triton X-100, 1M Urea followed by 50 bed volumes of the same buffer containing 0.1% Triton X-100 and 0.1 M Urea. Finally, columns were washed with 200 bed volumes of water before proteins were eluted with 10 ml of 50 mM β-mercaptoethanol in water. Samples were concentrated and analyzed by Gel-LC-MS/MS analysis. For on column digestion after the final wash with water, the columns were washed with 10 bed volumes of 0.1 M ammonium bicarbonate. Bound proteins were subjected to digestion by the addition of 1μg/ml of trypsin gold (Promega, Madison, WI) in one bed volume of 0.1 M ammonium in the dark for 16 hours at room temperature. The resin was next washed with 40 bed volumes of 1 M ammonium bicarbonate, pH 7.4, containing 300 mM NaCl, followed by 40 volumes of the same buffer without NaCl. Columns were then washed with 40 volumes of 0.1 M ammonium bicarbonate followed by 200 volumes of deionized water. To elute bound peptides, the resin was incubated with one bed volume of performic acid in water (performic acid is synthesized by reacting 1% formic acid and 0.5% hydrogen peroxide for at least 60 minutes at room temperature with rocking in a glass vial shielded from light) for 30 minutes at room temperature (10). Eluted peptides were recovered by washing the resin with one bed volume of deionized water. Eluates were stored at −80 °C overnight followed by lyophilization and re-suspension into 300 μl of 0.1% formic acid. Peptide suspensions were transferred to low retention tubes (Axygen, Union City, CA) and the volume was reduced to 30 μl by speed vacuum. Twenty μl of peptide suspension was transferred to an HPLC vial and submitted for LC-MS/MS analysis.
The details for in gel digestion and the conditions of MS/MS analysis have been provided previously (10,30). Post mass spectrometry analysis to generate the S-nitrosocysteine proteomes (supplementary tables 1-6) has been performed as described previously with the following exception: cysteine containing peptides that were not matched to proteins from the same organ were matched with proteins identified independently in other organs.
Subcellular localization was determined by either existing UniProt annotation (www.uniprot.org) or prediction by BaCelLo (http://gpcr.biocomp.unibo.it/bacello). Functional analysis to identify the biological functions that were most important was performed using Uniprot and Fatigo (www.fatigo.org). The raw MS/MS data are deposited at http://www.research.chop.edu/tools/msms/spectra.pdf.
VLCAD activity assay
VLCAD enzymatic activity was assessed as previously described (31). Briefly, ferricenium ion was used as an artificial electron acceptor for VLCAD-mediated palmitoyl-CoA dehydrogenation. Liver homogenate (final concentration of 0.03 μg/ μl) or cell lysate (final concentration of 0.09 μg/ μl) in 100 mM potassium phosphate buffer pH 7.2 containing 0.2% Triton X-100 and 0.1 mM EDTA was mixed with 150 μM ferricenium ion and the reaction was initiated by the addition of palmitoyl-CoA (the final volume of the assay was 130 μl). The decrease in ferricenium absorbance as a function of time at 300 nm was recorded and the initial velocity (Vo) of the enzyme was determined from the slope of the curve from time 0 (when palmitoyl-CoA was added) to the time that corresponded to 5% of total change of absorbance. At least nine concentrations of palmitoyl-CoA, ranging from 0.015 to 2 mM, were used for the experimental determination of the apparent Vmax and KM of VLCAD for each animal. The experimental data were fitted to non-linear regression to the Michaelis-Menten equation in Graph Prism software. A unit of enzyme activity is defined as the amount of enzyme that causes the reduction of 1 μmole of ferricenium per minute at room temperature (ε= 4.3 mM−1 cm−1 at 300nm) (32). For the experiments to test the specificity of the assay, 10 μg of anti-VLCAD antibodies were pre-incubated with liver homogenate for 20 minutes. When UV photolysis was used to eliminate S-nitrosocysteines, the liver homogenate was illuminated under a conventional UV transilluminator for 10 minutes on ice. The measurement of specific activity of VLCAD in liver homogenates (figure S4) and cell lysates (figure 4) was performed in the presence of 0.25 mM and 0.125 mM of palmitoyl-CoA respectively.
To quantify the fraction of S-nitrosylated VLCAD we captured the protein onto the organomercury resin. After extensive washing the bound proteins were eluted and probed with antibodies against VLCAD. After extensive washing the bound proteins were eluted and probed with antibodies against VLCAD.
Liver histology and Quantification of triglyceride concentrations
Formalin-fixed liver sections were stained with trichrome stain kit (Sigma, St Louis, MO) according to the manufacturer instructions. Triglycerides were extracted from liver according to the Folch method (33). Serum and liver triglyceride concentrations were quantified using a triglyceride quantification kit following the manufacturer instructions (Abcam, Cambridge, MA).
Measurement of β-oxidation of fatty acids
One milligram of protein suspension was added in 1 ml of Krebs-Ringer bicarbonate buffer containing 2.5 mg/ml fatty acid free BSA, 2.5 mM palmitic acid, 10 mM carnitine and 4μCi 9,10-3H-palmitoyl-CoA (Biomedicals, Santa Ana, CA). The mixture was rocked for 2 hours in the dark at 37 °C, followed by Folch based separation of 9,10- 3H-palmitoyl-CoA and 3H2O (33). The aqueous phase was collected and proteins were precipitated by the addition of 10% TCA followed by centrifugation at 8000 × g for 10 minutes at room temperature. Remaining radioactive palmitoyl-CoA was eliminated by strong anion exchange chromatography using AG 1-X8 formate resin (Life Science, Hercules, CA). The effluent containing the 3H2O was collected and used for scintillation counting. Each experiment was coupled with a background measurement, a sample containing no protein. The background counts number was subtracted from each measurement corresponding to for analysis samples.
Site directed mutagenesis to generate the C238A VLCAD mutant
The Quikchange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) was used to introduce single amino acid mutations in cDNA encoding VLCAD. A cysteine-to-alanine mutant at amino acid position 238 was generated using Ex-Mm01013-M14 (hereafter referred to as pVLCAD-3xFLAG), a plasmid encoding mouse VLCAD fused to three FLAG tags in the C-terminal region of the protein (Genecopoeia, Rockville, MD), as a template. The forward primer 5’-TCAGCCATACCCAGCCCCGCTGGAAAATATTACACTCTC-3’ and the reverse primer 5’-GAGAGTGTAATATTTTCCAGCGGGGCTGGGTATGGCTGA-3’ were used to substitute an alanine codon for a cysteine codon in pVLCAD-3xFLAG, thereby generating pVLCAD-C238A-3xFLAG. The sequence of both pVLCAD-3xFLAG and pVLCAD-C238A-3xFLAG was verified prior to use in subsequent experiments.
Cell culture, transfection and treatment with GSNO
Hepa 1-6 cells were grown in Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin, and 100 ng/ml streptomycin, at 37°C in air with 5% CO2. Cells were plated at a density of 5 × 104 cells/cm2 and cultured under normal conditions for 24 hours. Cells were transfected with FLAG tagged WT VLCAD or FLAG tagged C238A VLCAD using Lipofectamine™ 2000 reagent (Life Technologies, NY, USA) according to the protocol provided by the manufacturer. Forty eight hours post transfection the growth medium was replaced with serum free DMEM and 250 μM of freshly prepared GSNO were added for 30 minutes after which cells were extensively washed with PBS. Cell lysates were obtained immediately or 60, 120 and 240 minutes after the removal of GSNO. The samples were protected from UV- light exposure. Lysates were assayed for protein concentration and equal amounts of protein per sample was used for organic mercury assisted capture.
Structure generation of S-nitrosylated cysteine and normal mode analysis
The crystal structure of human very long chain acyl-coA dehydrogenase was downloaded from the Protein Data Bank (PDB ID: 2UXW). Disordered or missing residues were completed using the Mutagenesis tool in PyMol (www.pymol.org). S-nitrosocysteine was generated using the “S-nitrosator”` Python script from the Timerghazin laboratory in the Molecular Modeling Toolkit. “S-nitrosator” utilizes the coordinates of thioredoxin and χ3 values calculated using from PCM-ONIOM(PBE0/def2-TZVPPD:AmberFF) calculation of an S-nitrosocysteine residue in an alpha–helix content. ElNemo (34) was used to observe the 100 lowest frequency modes and perturbed models were generated for the first five non-trivial modes of VLCAD. Residue mean square displacement (r2) was used to identify protein movement.
Statistical analysis
Data were analyzed with GraphPad Prism 5.0d software. All normally distributed data were displayed as means ± SD. Groups were analyzed by one-way analysis of variance (ANOVA).
Acknowledgements
We thank Dr. Seeholzer at the Protein and Proteomics Core at the Children's Hospital of Philadelphia Research Institute for the assistance with MS/MS sample analysis, Drs. Roland Dunbrack (Fox Chase Cancer Center), Qadir K. Timerghazin and Marat Talipov (Marquette University) for help with molecular modeling. We are grateful to Drs. Morris Birnbaum and Min Wan (University of Pennsylvania) for their help with liver palmitoyl-CoA oxidation measurement and discussions, Dr. Marc Yudkoff (The Children's Hospital of Philadelphia) for suggestions and comments. Funding: This work was supported by the National Institutes of Health Grants AG13966, HL054926 and National Institute of Environmental Health Sciences Center of Excellence in Environmental Toxicology Grant ES013508. KR was supported by National Institutes of Health Training Grant T32AG000255. H.I. is the Gisela and Dennis Alter Research Professor of Pediatrics.
Footnotes
Author contributions: PTD, MT, JLG and KR performed experiments and analyzed data; PTD, MT and HI designed experiments, analyzed data and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests. **
References and Notes
- 1.Stamler JS, Lamas S, Fang FC. Nitrosylation. the prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 2.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 3.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 5.Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JV, Snowman AM, Law L, Hester LD, Snyder SH. GAPDH mediates nitrosylation of nuclear proteins. Nat. Cell Biol. 2010;12:1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsushita K, Morrell CN, Cambien B, Yang SX, Yamakuchi M, Bao C, Hara MR, Quick RA, Cao W, O'Rourke B, Lowenstein JM, Pevsner J, Wagner DD, Lowenstein CJ. Nitric oxide regulates exocytosis by S-nitrosylation of N-ethylmaleimide-sensitive factor. Cell. 2003;115:139–150. doi: 10.1016/s0092-8674(03)00803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell DA, Marletta MA. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat. Chem. Biol. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- 8.Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mannick JB, Schonhoff C, Papeta N, Ghafourifar P, Szibor M, Fang K, Gaston B. S-Nitrosylation of mitochondrial caspases. J. Cell Biol. 2001;154:1111–1116. doi: 10.1083/jcb.200104008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon DI, Mullins ME, Jia L, Gaston B, Singel DJ, Stamler JS. Polynitrosylated proteins: characterization, bioactivity, and functional consequences. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4736–4741. doi: 10.1073/pnas.93.10.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schild L, Dombrowski F, Lendeckel U, Schulz C, Gardemann A, Keilhoff G. Impairment of endothelial nitric oxide synthase causes abnormal fat and glycogen deposition in liver. Biochim. Biophys. Acta. 2008;1782:180–187. doi: 10.1016/j.bbadis.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Mohan S, Reddick RL, Musi N, Horn DA, Yan B, Prihoda TJ, Natarajan M, Abboud-Werner SL. Diabetic eNOS knockout mice develop distinct macro- and microvascular complications. Lab. Invest. 2008;88:515–528. doi: 10.1038/labinvest.2008.23. [DOI] [PubMed] [Google Scholar]
- 14.Brix AE, Elgavish A, Nagy TR, Gower BA, Rhead WJ, Wood PA. Evaluation of liver fatty acid oxidation in the leptin-deficient obese mouse. Mol. Genet. Metab. 2002;75:219–226. doi: 10.1006/mgme.2002.3298. [DOI] [PubMed] [Google Scholar]
- 15.de Oliveira CP, de Lima VM, Simplicio FI, Soriano FG, de Mello ES, de Souza HP, Alves VA, Laurindo FR, Carrilho FJ, de Oliveira MG. Prevention and reversion of nonalcoholic steatohepatitis in OB/OB mice by S-nitroso-N-acetylcysteine treatment. J. Am. Coll. Nutr. 2008;27:299–305. doi: 10.1080/07315724.2008.10719703. [DOI] [PubMed] [Google Scholar]
- 16.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 17.Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 18.Koshland DE. The application and usefulness of the ratio k(cat)/K(M). Bioorg. Chem. 2002;30:211–213. doi: 10.1006/bioo.2002.1246. [DOI] [PubMed] [Google Scholar]
- 19.Brown GC, Cooper CE. Nanomolar concentrations of nitric oxide reversibly inhibit synaptosomal respiration by competing with oxygen at cytochrome oxidase. FEBS Lett. 1994;356:295–298. doi: 10.1016/0014-5793(94)01290-3. [DOI] [PubMed] [Google Scholar]
- 20.Kobzik L, Stringer B, Balligand JL, Reid MB, Stamler JS. Endothelial type nitric oxide synthase in skeletal muscle fibers: mitochondrial relationships. Biochem. Biophys. Res. Commun. 1995;211:375–381. doi: 10.1006/bbrc.1995.1824. [DOI] [PubMed] [Google Scholar]
- 21.Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie Restriction Promotes Mitochondrial Biogenesis by Inducing the Expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 22.Prime TA, Blaikie FH, Evans C, Nadtochiy SM, James AM, Dahm CC, Vitturi DA, Patel RP, Hiley CR, Abakumova I, Requejo R, Chouchani ET, Hurd TR, Garvey JF, Taylor CT, Brookes PS, Smith RA, Murphy MP. A mitochondria-targeted S-nitrosothiol modulates respiration, nitrosates thiols, and protects against ischemia-reperfusion injury. Proc. Natl. Acad. Sci. U. S. A. 2009;106:10764–10769. doi: 10.1073/pnas.0903250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lima B, Lam GK, Xie L, Diesen DL, Villamizar N, Nienaber J, Messina E, Bowles D, Kontos CD, Hare JM, Stamler JS, Rockman HA. Endogenous S-nitrosothiols protect against myocardial injury. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6297–6302. doi: 10.1073/pnas.0901043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohr MJ, Aponte A, Sun J, Gucek M, Steenbergen C, Murphy E. Measurement of s-nitrosylation occupancy in the myocardium with cysteine-reactive tandem mass tags: short communication. Circ. Res. 2012;111:1308–1312. doi: 10.1161/CIRCRESAHA.112.271320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aoyama T, Souri M, Ushikubo S, Kamijo T, Yamaguchi S, Kelley RI, Rhead WJ, Uetake K, Tanaka K, Hashimoto T. Purification of human very-long-chain acyl-coenzyme A dehydrogenase and characterization of its deficiency in seven patients. J. Clin. Invest. 1995;95:2465–2473. doi: 10.1172/JCI117947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Djordjevic S, Dong Y, Paschke R, Frerman FE, Strauss AW, Kim JJ. Identification of the Catalytic Base in Long Chain Acyl-CoA Dehydrogenase. Biochemistry. 1994;33:4258–4264. doi: 10.1021/bi00180a021. [DOI] [PubMed] [Google Scholar]
- 28.Strauss AW, Powell KC, Hale DE, Anderson MM, Ahuja A, Brackett JC, Sims HF. Molecular basis of human mitochondrial very-long-chain acyl-CoA dehydrogenase deficiency causing cardiomyopathy and sudden death in childhood. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:10496–10500. doi: 10.1073/pnas.92.23.10496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabuyama Y, Suzuki T, Nakazawa N, Yamaki J, Homma MK, Homma Y. Dysregulation of very long chain acyl-CoA dehydrogenase coupled with lipid peroxidation. Am. J. Physiol. Cell. Physiol. 2010;298:C107–113. doi: 10.1152/ajpcell.00231.2009. [DOI] [PubMed] [Google Scholar]
- 30.Keene SD, Greco TM, Parastatidis I, Lee SH, Hughes EG, Balice-Gordon RJ, Speicher DW, Ischiropoulos H. Mass spectrometric and computational analysis of cytokine-induced alterations in the astrocyte secretome. Proteomics. 2009;9:768–782. doi: 10.1002/pmic.200800385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehman TC, Hale DE, Bhala A, Thorpe C. An acyl-coenzyme a dehydrogenase assay utilizing the ferricenium ion. Anal. Biochem. 1990;186:280–284. doi: 10.1016/0003-2697(90)90080-s. [DOI] [PubMed] [Google Scholar]
- 32.Izai K, Uchida Y, Orii T, Yamamoto S, Hashimoto T. Novel fatty acid beta-oxidation enzymes in rat liver mitochondria. I. Purification and properties of very-long-chain acyl-coenzyme A dehydrogenase. J. Biol. Chem. 1992;267:1027–1033. [PubMed] [Google Scholar]
- 33.Folch J, Lees M, Sloane-Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 34.Suhre K, Sanejouand YH. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004;32:W610–614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z, Wang Z,E, Doulias PT, Wei W, Ischiropoulos H, Locksley RM, Liu L. Lymphocyte development requires S-nitrosoglutathione reductase. J. Immunol. 2010;185:6664–6669. doi: 10.4049/jimmunol.1000080. [DOI] [PMC free article] [PubMed] [Google Scholar]