Abstract
Human immunodeficiency virus type 1 (HIV-1) subtype CRF01_AE is transmitted mainly by sexual activity in Guangxi, southwestern China. Other subtypes, including CRF07_BC, CRF08_BC, and subtype B, are also prevalent in this region. Cocirculation of multiple subtypes, as well as a high rate of drug use, creates favorable conditions for the emergence of recombinant viruses in Guangxi. In the present study, we identified a new HIV-1 unique recombinant form (CRF01_AE /08BC) transmitted from the infected index patient to his seronegative sexual partner. This is the first near full-length genome characterization of a CRF01_AE /08BC recombinant virus in Guangxi, and provides an important basis for future analysis on potential new recombinant transmission events.
Guangxi is a heavily epidemic province in China for HIV-1, and the majority of new cases in 2009 were of the CRF01_AE strain, predominantly transmitted through sexual routes.1–3 At the same time, multiple cocirculating subtypes have been identified in this region, including CRF07_BC, CRF08_BC, and subtype B.1,2 In particular, CRF_08BC is prevalent in the central cities and is also mainly transmitted through sexual routes. Cocirculation of subtypes in the same transmission networks creates favorable conditions for viral recombination and circulating recombinant forms (CRFs) involving CRF01_AE and subtype B fragments have been reported from Guangxi.1 However, no full-length genomic sequence had been characterized of any recombinants of CRF01_AE and both subtype B and C fragments.
Here, we identify a new unique recombinant form (URF) involving insertions of subtypes B and C fragments within the CRF01AE backbone. The sample is derived from a transmission event within an HIV-1 serodiscordant couple in Beihai city, near the southern border of Guangxi. HIV-1 nucleic acids were obtained from blood samples of both members of the couple, and the near full-length genome sequence was amplified in the index (initially infected) patient, while only the pol and env genes were amplified for the seroconverted patient. Pairwise genetic distance and epidemiologic records (data not shown) indicate that this transmission event is an intrafamily infection. The index patient's sample GXDY1299 was collected in November 2012 from a male patient living in the village near Beihai city, who was transmitted by sexual contact and later infected his spouse.
Amplification and sequencing procedures were performed as described previously.4 Briefly, the near full length genome of GXDY1299 (9049 bp) was amplified from plasma RNA by reverse transcriptase polymerase chain reaction (RT-PCR). PCR products were sequenced by a total of 26 internal primers after purification. All of the sequences were cleaned and assembled using Sequencher v4.9 (Gene Codes, Ann Arbor, MI). A BLAST search was performed to exclude the possibility of cross-contamination.4 The final 9,049 bp sequence was aligned with HIV-1 reference subtypes (A–D, F–H, J, K) and select CRFs (CRF01_AE, CRF08_BC) obtained from the Los Alamos HIV Database (http://hiv-web.lanl.gov/) using Clustal W, followed by manual editing in BioEdit Sequence Alignment Editor (version 7.1). Phylogenetic trees were constructed with MEGA 4.0 by using the neighbor-joining method, under the Kimura two-parameter substitution model, with 1,000 bootstrap replications.5
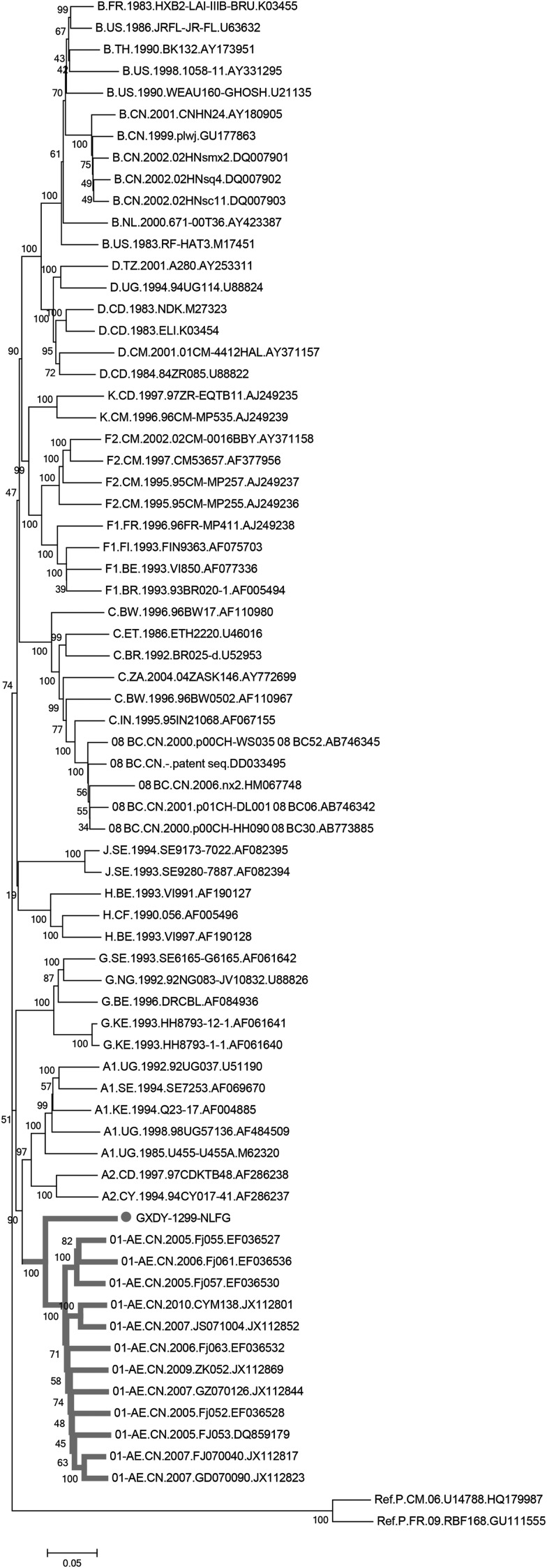
As shown in Fig. 1, the GXDY 1299 sequence is closely related to CRF01_AE reference sequences, but clustered outside of the monophyletic branch (Fig. 1).
FIG. 1.
Phylogenetic analysis of the near full-length genome. A neighbor-joining tree was created with the GXDY1299 strain and standard reference strains representative of different HIV-1 group M subtypes (http://hiv-web.lanl.gov/). HIV-1 group P served as an outgroup. Some circulating recombinant forms (CRFs) were also included, including CRF01_AE, and CRF08_BC. The stability of the nodes was assessed by bootstrap analysis with 1,000 replications. The scale bar represents 5% genetic distance (0.05 substitutions per site).
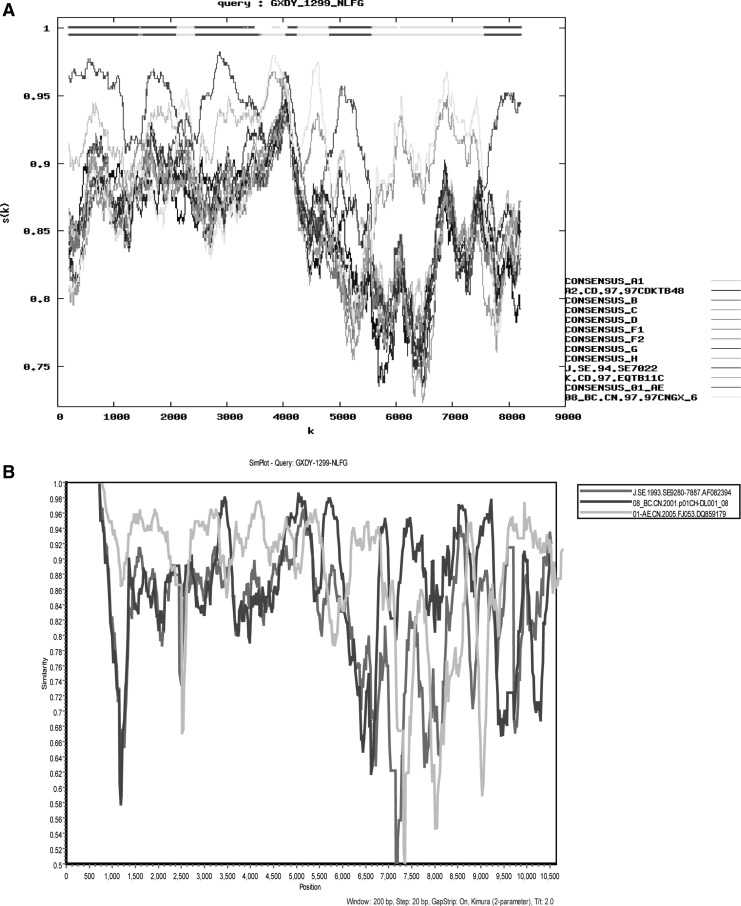
The recombination analysis is performed by RIP (www.hiv.lanl.-gov/content/sequence/RIP/RIP.html) using all default settings except the window size of 300 (Fig. 2A). There is a dip in similarity to CRF01 and CRF08 between positions 1204 and 1371 of the gaps tripped alignment that is more similar to CRF01 than other references. The result shows that the recombinant form consists of CRF01_AE and CRF08_BC. Similarity plot analysis was performed with SimPlot (version 3.5.1)6 using several closely related sequences of the reference sequence set described above. SimPlot results verified that the genome was composed of CRF08_BC and CRF01_AE (Fig. 2B).
FIG. 2.
Recombinant Identification Program (RIP) and similarity plot analysis were performed for GXDY1299 to identify parental subtypes. (A) Similarity distance analysis was performed using RIP from the Los Alamos National Laboratory HIV Database with default settings, except for the window size of 300. (B) Similarity plots for GXDY1299 using SimPlot software with subtype reference strains CRF01_AE, CRF08_BC, and J. These reference strains were verified above by RIP, which may contain the segments of GXDY1299.
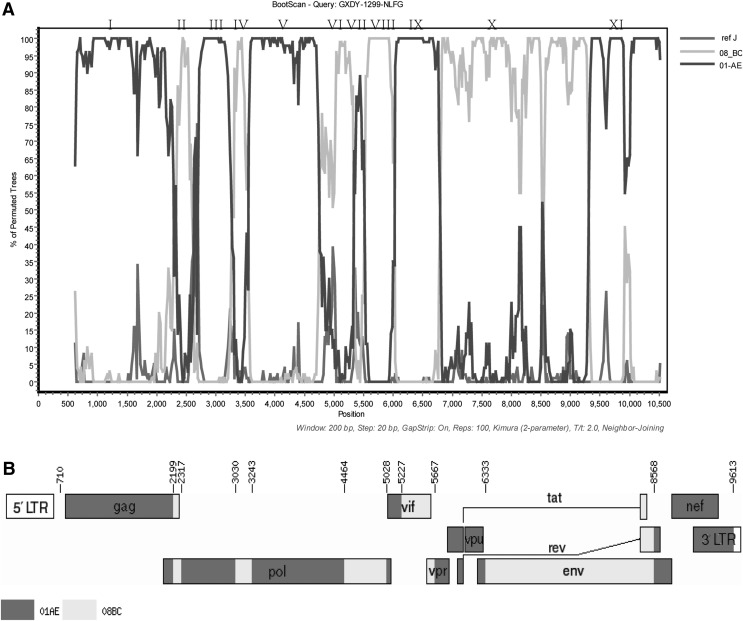
Bootscan analysis was subsequently performed to identify the recombination breakpoint. The query sequence GXDY1299 was bootscanned with reference sequences (CRF01_AE, CRF08_BC as parents, subtype J as outgroup). The parameters were a window size of 200, a step size of 20, and the neighbor-joining method using the Kimura two-parameter model with replicates of 100. Phylogenetic trees were constructed with MEGA 5.0 using the neighbor-joining method with 1,000 bootstrap replications to further trace the close subtype of the mosaic fragments.7
From the bootscan result, GXDY1299 is composed of at least 11 interlaced mosaic segments, including CRF01_AE (I, III, V, VII, IX and XI) and CRF08_BC (II, IV, VI, VIII, and X) (Fig. 3A). Corresponding CRF01_AE, CRF08_BC segments were analyzed by phylogenetic trees for their evolutionary relationships to the respective subtype references (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/aid). From the results of the phylogenetic trees, we found that the B and C segments are closely related to CRF_08BC. Although the bootstrap values of the phylogenetic clades are not high for the shorter segments such as segment II, which seems to be due to just a few point mutations and not a recombination event, the larger segments are all clustered with CRF08_BC with high bootstrap values (Supplementary Fig. S1). These results point toward a close relationship between GXDY1299 and CRF08_BC.
FIG. 3.
Bootscan plots and schematic representation of the near full-length genome's mosaic structure. (A) Bootscan plots of GXDY1299 using CRF01_AE, CRF08_BC, and reference strain J as subtype references. The parameters of SimPlot bootscan analysis were set as default except for a window size of 200 and step size of 20. The breakpoint positions refer to HXB2 coordinates located by the HIV Sequence Locator (www.hiv.lanl.gov/content/sequence/LOCATE/locate.html). (B) Genomic map of GXDY1299. The mosaic map was generated using the Recombinant HIV-1 Drawing Tool (www.hiv.lanl.gov/content/sequence/DRAW_ CRF/recom_mapper.html).
The breakpoints positions of the near full-length genome refer to HXB2 coordinates, and were located by the HIV Sequence Locator (www.hiv.lanl.gov/content/sequence/LOCATE/locate.html). Thus, the map of the new recombinant virus GXDY1299 is as follows: U/A (632–709 nt), 01AE (710–2,175 nt), 08BC (2,176–2,295 nt), 01AE (2,296–3,029 nt), 08BC (3,030–3,242 nt), 01AE (3,243–4,465 nt), 08BC (4,464–5,027 nt), 01AE (5,028–5,224 nt), 08BC (5,225–5,665 nt), 01AE (5,667–6,332 nt), 08BC (6,333–8,567 nt), and 01AE (8,568–9,613 nt) (Fig. 3B).
The recombinant GXDY1299 was discovered by analyzing a transmission event between a heterosexual couple in a stable marital relationship. From survey data, the index patient was infected via extramarital heterosexual contact with more than three people. The index patient and his spouse have no history of injection drug use, blood transfusion, or other HIV infection risk factors. It is most likely that this was a heterosexual transmission event. Interestingly, a recent molecular epidemiologic survey of Guangxi province identified four HIV-1 URFs using the 5′ half genome, one of which (identification number 2261) has the same breakpoints in the gag and pol segments as the sequence we report here.1 It is highly possible that these sequences are from the same ancestor. Since the URF identified in that survey is not sampled from Beihai city, it is also likely that the novel recombination has become a CRF.
In sum, we have identified a new recombination form in Guangxi province that most likely incorporated elements from two major heterosexually transmitted subtypes, CRF01_AE and CRF08_BC. Although further confirmation is needed for the epidemiologic source of the GXDY1299 recombinant, it is highly likely to have originated through a commercial sexual transmission chain. More research on the recombinant form and transmission network may help provide relevant information for route- and network-specific interventions and subtype-sensitive vaccine designs.
Sequence Data
The nucleotide sequence of GXDY1299 has been submitted to GenBank with the accession number KF541292.
Supplementary Material
Acknowledgments
This research was supported by National Major Projects for Infectious Diseases Control and Prevention (grant 2012ZX10001008, 2012ZX10001002), the National Natural Science Foundation of China (grant 81020108030), and Chinese State Key Laboratory for Infectious Disease Development (grant 2012SKLID103).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Li L, Chen L, Liang S, et al. : Subtype CRF01_AE dominate the sexually transmitted human immunodeficiency virus type 1 epidemic in Guangxi, China. J Med Virol 2013;85(3):388–395 [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Lu L, Ba L, et al. : Dominance of HIV-1 subtype CRF01_AE in sexually acquired cases leads to a new epidemic in Yunnan province of China. PLoS Med 2006;3(11):e443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng H, Sun Z, Liang S, et al. : Emergence of a new HIV type 1 CRF01_AE variant in Guangxi, Southern China. AIDS Res Hum Retroviruses 2012;28(10):1352–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei H, Su L, Feng Y, et al. : Near full-length genomic characterization of a novel HIV Type 1 CRF07_ BC/01_AE recombinant in men who have sex with men from Sichuan, China. AIDS Res Hum Retroviruses 2013;29(8):1173–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall T: BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 1999;41:95–98 [Google Scholar]
- 6.Lole KS, Bollinger RC, Paranjape RS, et al. : Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 1999;73(1):152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamura K, Dudley J, Nei M, and Kumar S: MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007;24(8):1596–1599 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.