Summary
The in vivo role of the nuclear receptor SHP in feedback regulation of bile acid synthesis was examined. Loss of SHP in mice caused abnormal accumulation and increased synthesis of bile acids due to derepression of rate-limiting CYP7A1 and CYP8B1 hydroxylase enzymes in the biosynthetic pathway. Dietary bile acids induced liver damage and restored feedback regulation. A synthetic agonist of the nuclear receptor FXR was not hepatotoxic and had no regulatory effects. Reduction of the bile acid pool with cholestyramine enhanced CYP7A1 and CYP8B1 expression. We conclude that input from three negative regulatory pathways controls bile acid synthesis. One is mediated by SHP, and two are SHP independent and invoked by liver damage and changes in bile acid pool size.
Introduction
The synthesis of bile acids from cholesterol in the liver requires the coordinated actions of a dozen enzymes located in every major compartment of the hepatocyte. In this pathway, a soluble end product is formed from cholesterol by the addition of hydroxyl groups and the oxidation of the side chain. The solubility of bile acids facilitates their excretion, and their detergent properties help to solubilize lipid nutrients in the intestine.
The amount of bile acid synthesized by the liver is closely regulated to prevent overaccumulation of bile acids on the one hand, and cholesterol on the other. The regulatory scheme also ensures sufficient bile acids to solubilize biliary cholesterol and to facilitate lipid digestion and absorption. When bile acids accumulate beyond physiologic levels, a negative feedback mechanism is called into play, and synthesis is decreased (Russell and Setchell, 1992). Conversely, bile acid synthesis is increased when the substrate, cholesterol, accumulates. Both of these regulatory responses are transcriptional in nature and are mediated by members of the nuclear hormone receptor family acting on two genes that encode hydroxylase enzymes in the biosynthetic pathways (Chawla et al., 2001).
As summarized in Figure 1, suppression of bile acid synthesis is triggered when the farnesoid X receptor (FXR, NR1H4) binds excess bile acid and activates transcription of a second nuclear receptor termed short heterodimer partner (SHP, NR0B2) (Goodwin et al., 2000; Lu et al., 2000). SHP binds to and inhibits a third receptor, the CYP7A1 promoter binding factor (CPF, LRH, FTF, or NR5A2), which normally activates genes encoding cholesterol 7α-hydroxylase and sterol 12α-hydroxylase (Castillo-Olivares and Gil, 2000; Lee and Moore, 2002; Nitta et al., 1999). Cholesterol 7α-hydroxylase, encoded by the Cyp7a1 gene, catalyzes the rate-limiting step in the major synthetic pathway, and its inhibition leads to a drastically decreased output of bile acids. Sterol 12α-hydroxylase, the product of the Cyp8b1 gene, synthesizes cholic acid, a bile acid that binds to FXR and mediates suppression of Cyp7a1 (J. Li-Hawkins et al., submitted). The decline in cholic acid ultimately restores Cyp7a1 expression and returns the system to homeostasis. Activation of bile acid synthesis is mediated by the liver X receptor α (LXRα, NR1H3), a nuclear receptor that binds oxysterols formed during the synthesis and metabolism of cholesterol (Lu et al., 2001). In conjunction with CPF, LXR stimulates transcription of Cyp7a1 (Chawla et al., 2001).
Figure 1. Regulation of Bile Acid Synthesis by Nuclear Receptors.

A scheme depicting the roles of four nuclear receptors in regulating bile acid synthesis is shown. Green arrows indicate positive regulation while the red brake indicates negative regulation.
Although the nuclear receptor pathway involving FXR, SHP, CPF and LXRα explains a majority of the known regulatory responses in bile acid biosynthesis, additional mechanisms may exist that affect bile acid output. These are exemplified by mutations in the mouse hepatic nuclear factor 1α, hepatic nuclear factor 4, and pregnane X receptor genes (all of which cause alterations in bile acid metabolism [Hayhurst et al., 2001; Shih et al., 2001; Staudinger et al., 2001; Xie et al., 2001]) and by regulatory responses mediated by serine/threonine kinases (Fabiani et al., 2001; Gupta et al., 2001), cytokines (Miyake et al., 2000), and members of the fibroblast growth factor family (Yu et al., 2000). In these cases, it has proven difficult to distinguish between receptor-dependent and receptor-independent regulation due in part to the complexity of the biological system and the large number of genes involved.
The current study employs a line of mice deficient in SHP to determine the role of this nuclear receptor in the regulation of bile acid synthesis and to search for alternate regulatory pathways that affect cholesterol disposal. Characterization of bile acid metabolism in SHP knockout mice reveals that under normal conditions, the inputs of FXR and SHP are sufficient to mediate negative feedback regulation of bile acid synthesis. When the organism is stressed, for example by damage to the liver, or when the bile acid pool is decreased, alternate pathways of regulation come into play that lead to changes in bile acid synthesis.
Results
Deletion of Shp Gene
Mutation of the SHP gene (Shp) was accomplished by replacement of 318 bp of exon 1 with a cassette encoding antibiotic resistance (Figure 2A). Homologous recombination in embryonic stem cells, injection into blastocysts, and transmission of the mutation through the mouse germ line (Figure 2B) were carried out with standard methods (Hogan et al., 1995). Two aspects of the introduced mutation were predicted to eliminate Shp expression. First, the loss of ~106 amino acids encoded by exon 1 should remove required portions of the ligand binding and dimerization domains from the SHP protein. Second, the orientation of the inserted neomycin resistance gene was opposite that of Shp and thus should interfere with transcription from the gene. RNA blotting revealed a Shp transcript of ~1.1 kb in the livers of wild-type mice and trace amounts of a truncated mRNA of ~0.8 kb in Shp−/− mice (Figure 2C). Reverse transcriptase PCR and DNA-sequencing experiments indicated that the mRNA transcribed from the mutant allele was a chimera composed of SHP sequences, anti-sense neomycin resistance sequences, and unknown sequences that did not contain an extended translational reading frame (data not shown). We interpreted these results to mean that the replacement mutation produced a null allele of Shp.
Figure 2. Deletion of Mouse SHP Gene.
(A) Schematic representation of the wild-type SHP gene showing the two exon structure (Lee et al., 1998), the targeting vector in which a cassette encoding neomycin resistance replaces 318 bp of SHP exon 1, and the structure of the mutant allele arising after homologous recombination. Arrows below the neomycin resistance cassette indicate the direction of transcription from the gene.
(B) Genotype analysis in mice with different Shp alleles. Genomic DNA was isolated from animals of the indicated genotype and amplified via PCR using allele-specific primers described in Experimental Procedures. Products were separated by agarose gel electrophoresis.
(C) Blot analysis of mouse liver SHP mRNA isolated from wild-type (+/+), heterozygous (+/−), and homozygous knockout (−/−) mice. Poly(A+) RNA was isolated from animals (n = 6) of the indicated Shp genotype, and aliquots (2 μg) were separated by gel electrophoresis, transferred to nylon membranes, and hybridized to a radiolabeled SHP cDNA probe representing exon 1 of the gene (SHP, upper panel). After autoradiography, the filters were stripped of bound radioactivity and reprobed with a radiolabeled cDNA encoding rat cyclophilin (Cyclo, lower panel). The truncated mRNA detected in the Shp−/− sample is composed of portions of SHP exons 1 and 2, sequences from the noncoding strand of the neomycin resistance gene, and sequences of unknown origin.
Mice lacking SHP and maintained on standard laboratory chow were grossly normal, did not experience untimely deaths, and produced expected numbers of male and female offspring. No abnormalities were detected in tissue weights when normalized to total body weight. We were unable to assess whether the absence of SHP caused an obesity phenotype in mice as it does in humans (Nishigori et al., 2001) since the mouse mutation was maintained on a mixed strain background (C57BL/6-129/OlaHsd), and individual weights of both wild-type and knockout animals varied extensively.
Bile Acid Composition
The composition of the bile acid pool reflects in large part the expression of biosynthetic enzymes in the liver. To determine how loss of SHP affected the types of bile acid present in male knockout mice, total bile acids were extracted from the gut, liver, and gallbladder and their structures determined by gas chromatography-mass spectrometry (GC-MS). Shp−/− mice had markedly elevated levels of cholic acid in their bile relative to wild-type mice (Figure 3). The increase in cholic acid was accompanied by a corresponding decrease in the primary bile acid, β-muricholate. Levels of deoxycholic acid, a secondary bile acid derived from cholic acid, were also elevated in the Shp−/− mice, whereas the amounts of five other secondary bile acids were unchanged relative to those in wild-type mice (Figure 3). These data provided an early indication that the absence of SHP altered the expression of enzymes involved in bile acid synthesis, in particular that of sterol 12α-hydroxylase, which is responsible for the synthesis of cholic acid.
Figure 3. Composition of Bile Acid Pool in Male Wild-Type and Shp−/− Mice.
Ethanolic extracts of the intestine, liver, and gallbladder of mice of the indicated genotypes (n = 5) were analyzed by gas chromatography-mass spectrometry. Amounts of individual bile acids are indicated as a percentage of the entire pool. The positions of hydroxyl groups on the ring structures and their stereochemistries are indicated in parentheses. The level of cholic acid increased and that of β-muricholic acid decreased in Shp−/− mice.
Bile Acid Pool Size and Synthetic Rate
The total mass of bile acid present in SHP wild-type and knockout male mice was determined by high-performance liquid chromatography (HPLC). These analyses revealed that the bile acid pool in mutant animals was 32% larger than that in normal controls (Figure 4A, left panel). This increase was statistically significant (p = 0.013).
Figure 4. Parameters of Bile Acid and Cholesterol Metabolism.
(A) The amount of bile acid present in mice of the indicated genotypes (n = 6–7) was determined by high-pressure liquid chromatography. Values graphed in the left panel represent the mean ± SEM and were significantly larger (*, p = 0.013) in Shp−/− mice. Bile acid excretion (right panel) was determined after extraction and enzyme assay. Mutant mice (n = 5) excreted significantly more bile acid (p = 0.010) and thus had higher hepatic synthesis rates than did wild-type controls (n = 5).
(B) Cholesterol absorption was measured in 14 animals of the indicated genotype by a dual isotope fecal assay and mean values ± SEM plotted as a histogram (left panel). Shp−/− mice absorbed 22% more cholesterol than did wild-type mice; however, the difference did not reach statistical significance. Neutral sterol excretion was significantly lower (*, p = 0.0016) in the knockout mice (right panel, n = 5) and thus confirmed the finding of increased absorption.
Fecal bile acid excretion rates were measured to determine whether the enlarged pool size in the Shp−/− mice was due to an increased rate of synthesis. These measurements indicated that the absence of SHP led to a 33% increase in fecal bile acid excretion (Figure 4A, right panel). Since the mass of bile acid excreted is directly proportional to the amount synthesized in the liver, these data suggested a generalized derepression of bile acid synthesis in the Shp−/− animals.
Cholesterol Absorption and Excretion
Changes in bile acid metabolism in the mouse generally cause corresponding alterations in how the animal handles cholesterol. We determined several parameters of sterol metabolism in Shp−/− mice and found genotype-specific differences in two of them. First, the absorption of cholesterol in the gut was elevated by 22% in knockout male mice relative to wild-type controls (Figure 4B, left panel), and although this difference did not reach statistical significance in this experiment, increased absorption was detected in multiple independent experiments. Second, the excretion rate of neutral sterols was significantly decreased (p = 0.0016) in the mutant mice (Figure 4B, right panel). Inasmuch as increases in cholic acid pool size are known to increase intestinal cholesterol absorption and decrease sterol excretion (Dietschy and Turley, 2002), these data provided an independent confirmation of the experimental outcomes reported for the Shp−/− mice in Figures 3 and 4A.
No differences were detected in plasma cholesterol, triglyceride, or lipoprotein levels between the knockout and wild-type mice. Even though the absorption of dietary cholesterol was enhanced in the Shp−/− animals, there was no decrease in hepatic sterol synthesis (data not shown), presumably because the excess cholesterol was being converted to bile acids. The excretion of total fecal lipid was also unchanged in the mutant mice.
Cholesterol 7α-Hydroxylase Expression
The loss of SHP was predicted to lead to derepression of the cholesterol 7α-hydroxylase gene (Figure 1), and in agreement with this hypothesis, Shp−/− mice maintained on normal rodent chow had modestly elevated levels of CYP7A1 mRNA, protein, and enzyme activity (Figures 5A–5C). The increases in mRNA and protein, which were at most 2- to 3-fold (Figure 5), were less than those observed in mice lacking sterol 27-hydroxylase (Repa et al., 2000; Rosen et al., 1998), FXR (Sinal et al., 2000), or sterol 12α-hydroxylase (J. Li-Hawkins et al., submitted) in which 5- to 7-fold increases in CYP7A1 expression were found. The 2- to 3-fold increase in mRNA and protein caused an increase of only 20%–30% in measured CYP7A1 enzyme activity, which correlated with the 33% increase in bile acid synthesis (Figure 4A). These findings raise the possibility that CYP7A1 activity is subject to posttranslational regulation.
Figure 5. Expression of Cholesterol 7α-Hydroxylase.
(A) CYP7A1 and cyclophilin mRNA levels in livers (n = 9) of wild-type and Shp−/− mice were determined by blot hybridization of poly(A)+-enriched RNA. Densitometric scanning of the resulting autoradiogram indicated that the CYP7A1 mRNA was 2- to 3-fold more abundant in knockout mice.
(B) Enzyme activity was measured in a gas chromatography-based assay using hepatic microsomes isolated from animals (n = 5) of the indicated Shp genotypes. Values in the mutant mice were approximately 20%–30% higher than those in the controls.
(C) Immunodetection of CYP7A1 protein. Aliquots containing approximately 150 μg of microsomal protein were subjected to immunoblotting as described in Experimental Procedures. Signals emanating from Shp−/− samples were approximately 2-fold stronger as judged by densitometry, while those from a control protein (heavy chain binding protein, BIP) were unchanged.
Other Alterations in Gene Expression
To determine how loss of SHP affected the expression of other genes, we first measured mRNA levels of candidate genes involved in bile acid and cholesterol metabolism by blotting. An increased level of sterol 12α-hydroxylase mRNA was detected in the Shp−/− mice (Figure 6A), which is consistent with the elevation in cholic acid found in the bile acid pool (Figure 3). This result was expected, as the expression of the Cyp8b1 gene, which encodes sterol 12α-hydroxylase, is negatively regulated by SHP (Castillo-Olivares and Gil, 2000, 2001). The bile salt export protein (BSEP, ABCB11) mRNA was also elevated in the mutant mice (Figure 6A), presumably due to the increase in cholic acid, which is a potent ligand of FXR, a transcriptional activator of the BSEP gene (Ananthanarayanan et al., 2001). Not all FXR target genes were altered in their expression patterns, since levels of the Na+-taurocholate cotransporting polypeptide 2 (Ntcp2, SLC10A1) mRNA were the same between mice of different Shp genotypes (Figure 6A). The mRNAs specifying other enzymes involved in bile acid synthesis were unchanged, but levels of the scavenger receptor SR-B1 mRNA were approximately 3-fold elevated (Figure 6A).
Figure 6. Gene Expression in Wild-Type and Shp−/− Mice.
(A) Aliquots (3 μg) of poly(A+)-enriched RNA isolated from the liver or small intestine (n = 5 mice per genotype) were analyzed by blot hybridization with cDNA probes for the indicated mRNAs. A signal for cyclophilin was used to normalize relative expression levels.
(B) RNA was prepared from pooled livers dissected from wild-type and Shp knockout animals (n = 5) and converted into cDNA in the presence of a deoxynucleoside triphosphate derivatized with a fluorescent dye. Different dyes were used to label the wild-type and knockout cDNA probes, which were subsequently hybridized to a microarray containing ~400 cDNAs whose encoded proteins are involved in lipid or carbohydrate metabolism. Genes with the largest-fold increases in the chip hybridization experiments were then quantitated by real-time PCR. GPAT, glycerol phosphate acyltransferase; FAS, fatty acid synthetase.
In a second series of experiments, we assessed how SHP loss affected the relative mRNA levels of 400 genes involved in lipid and carbohydrate metabolism. A cDNA microarray was probed with differentially labeled cDNA transcribed from hepatic mRNA of wild-type and Shp−/− mice and the results analyzed according to stringent criteria. Real-time PCR was then used to confirm the results of the chip hybridization assay. Representative mRNAs whose expression was found to be increased using both assays are listed in Figure 6B. In agreement with previous findings indicating that SHP inhibits several different members of the nuclear receptor family (Seol et al., 1996), a majority of mRNAs whose expression changed in the knockout mice increased in amount. Only a few mRNAs were less abundant, and in these cases, the decreases were modest (<50%, data not shown). Genes increased in expression encoded ABC transporters (ABCA1, ABCG5, ABCG8, and ABCG1), a cholesterol esterification enzyme (ACAT1), and other products involved in fat metabolism. The levels of mRNAs found to be elevated in the RNA blotting experiments (Figure 6A) were also increased when measured by real-time reverse transcriptase PCR. Whether these changes in gene expression are attributable directly to the loss of the negative regulator, SHP, or whether they are secondary to the widespread changes in cholesterol and bile acid metabolism remains to be determined.
Regulation by FXR Agonists
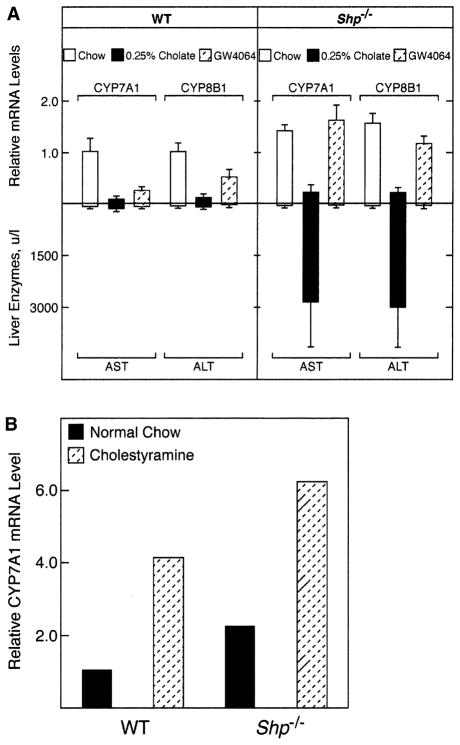
A key prediction regarding the regulatory role of SHP in bile acid metabolism concerns the response of the knockout animals to excess bile acids. If, as proposed, SHP is responsible for the inhibition of cholesterol 7α-hydroxylase and sterol 12α-hydroxylase gene transcription (Figure 1), then these genes should no longer be repressed in response to dietary bile acids in Shp−/− mice. To test this hypothesis, wild-type and mutant mice (n = 4–6) were fed diets supplemented with 0.25% cholic acid for 5 days, and hepatic mRNA levels were measured by real-time reverse transcriptase PCR. In the same experiment, separate groups of animals were treated by gavage with a potent agonist of FXR, GW4064 (Good-win et al., 2000), at 100 mg/kg body weight. We anticipated that the results obtained with this compound would mimic those obtained with cholic acid feeding.
As expected, dietary cholic acid decreased CYP7A1 and CYP8B1 mRNA levels in wild-type mice, as did GW4064 (Figure 7A, left panel). Unexpectedly, bile acid feeding decreased the levels of the hydroxylase mRNAs in Shp−/− mice, but treatment with the FXR agonist did not (Figure 7A, right panel). In some experiments, diets supplemented with 0.25% or 0.5% cholic acid were lethal to Shp−/− mice, but not to wild-type mice. These observations prompted us to assess liver damage by measuring the levels of hepatic enzymes in the plasma. As shown by the data in the lower panels of Figure 7A, neither cholic acid feeding nor GW4064 treatment elevated plasma levels of hepatic aspartate aminotransferase (AST) or alanine aminotransferase (ALT) in wild-type mice; however, bile acid, but not GW4064, caused an enormous elevation in the circulating levels of these enzymes in Shp−/− mice. Together these results suggested that the downregulation of cholesterol 7α-hydroxylase and sterol 12α-hydroxylase mRNAs by bile acid in the knockout mice was a consequence of hepatotoxicity.
Figure 7. Responses to Dietary Bile Acids, Cholestyramine, and FXR Agonist GW4064.
(A) Mice of different Shp genotypes (n = 4–6 each) were fed the indicated diets (normal versus chow supplemented with 0.25% [w/w] cholic acid) or gavaged with the FXR agonist GW4064 (100 mg/kg body weight, q.d.) for 5 days. Livers were dissected, total RNA prepared and pooled, and cholesterol 7α-hydroxylase (CYP7A1) and sterol 12α-hydroxylase (CYP8B1) mRNA levels determined by real-time reverse transcriptase PCR (upper panel) in individual animals. The numbers shown are the averages of triplicate determinations for each mRNA. Hepatic enzyme levels (AST, aspartate aminotransferase; ALT, alanine aminotransferase) were determined in pooled plasmas from the same groups of mice (lower panel).
(B) Animals were fed chow diet supplemented with nothing or cholestyramine (2%, w/w) for 5 days. CYP7A1 and CYP8B1 mRNA levels were quantitated as described in (A).
The effect of reducing the bile acid pool on hydroxylase gene expression was assessed by feeding cholestyramine, a polyanionic polymer that binds bile acids in the small intestine and prevents their return to the liver. Dietary cholestyramine increased the levels of cholesterol 7α-hydroxylase mRNA in normal mice (Figure 7B). An identical response was observed in Shp−/− mice except that the levels of CYP7A1 mRNA were higher than those reached in the wild-type mice (Figure 7B). Hepatic enzymes in the plasma were not elevated in the cholestyraminefed mice (data not shown).
Discussion
The current studies test the hypothesis that the nuclear receptor SHP is responsible in vivo for negative feedback regulation exerted by bile acids on their synthesis. The data show that mice lacking SHP increase the synthesis and accumulation of bile acids and produce more cholic acid than their wild-type counterparts. These increases are caused by a loss of regulation of cholesterol 7α-hydroxylase, the rate-limiting enzyme of bile acid synthesis, and of sterol 12α-hydroxylase, which synthesizes cholic acid, a potent agonist of FXR. When levels of bile acid in Shp knockout mice are raised further by dietary manipulation, cholestasis results, and the synthesis of these enzymes declines. Conversely, when bile acid levels are decreased in Shp−/− mice by cholestyramine feeding, synthesis increases. We conclude from these results that SHP is responsible for maintaining bile acid homeostasis under normal conditions and that at least two SHP-independent regulatory pathways exist to inhibit bile acid synthesis.
A central role for SHP in regulating bile acid metabolism was first proposed from results obtained in cultured cells (Goodwin et al., 2000; Lu et al., 2000). These studies revealed that expression of SHP inhibited transcription from a cholesterol 7α-hydroxylase reporter gene, and they traced the mechanism to the formation of an inhibitory complex between SHP and a second nuclear receptor (CPF/LRH/FTF/NR5A2) required for expression of CYP7A1 (Nitta et al., 1999). By acting in this manner, SHP fulfilled an antireceptor hypothesis put forth when the molecule was isolated (Seol et al., 1996) and emphasized a close functional and structural relationship with a second inhibitory receptor, DAX (Goodfellow and Camerino, 2001). Expression of the mouse SHP gene was further shown to be responsive to FXR in vivo and to FXR and CPF in vitro (Goodwin et al., 2000; Lee et al., 1999; Lu et al., 2000), thus completing a regulatory loop for the control of bile acid synthesis (Figure 1).
The in vivo results presented here confirm and extend these observations. As predicted from the in vitro data, loss of SHP causes increased expression of cholesterol 7α-hydroxylase, and a second CPF target gene, sterol 12α-hydroxylase (Castillo-Olivares and Gil, 2000, 2001). In the absence of SHP, CPF presumably has unopposed access to binding sites in the Cyp7a1 and Cyp8b1 promoters, resulting in enhanced transcription from these target genes. Surprisingly, the levels of CYP7A1 and CYP8B1 mRNAs in the Shp−/− mice were only 2-fold higher than those in wild-type mice (Figures 5 and 6). These elevations were less than the 5- to 7-fold increases observed in animals with reduced bile acid pool sizes (e.g., sterol 27-hydroxylase-deficient mice [Repa et al., 2000]) or with an altered composition of the bile acid pool (e.g., sterol 12α-hydroxylase-deficient mice [J. Li-Hawkins et al., submitted]). The less than expected increases in hydroxylase mRNA levels of Shp−/− mice suggest that another negative regulatory mechanism exists to keep these enzymes in check. This SHP-independent pathway must in some manner sense the bile acid pool given the results in the hydroxylase-deficient animals and the fact that reducing the bile acid pool in Shp−/− mice by feeding cholestyramine raises CYP7A1 and CYP8B1 mRNA levels (Figure 7B).
Evidence for a third negative regulatory pathway influencing Cyp7a1 and Cyp8b1 expression is obtained in Shp−/− mice fed bile acids. In this situation, excess bile acids cause liver damage, as judged by the release of hepatic enzymes into the serum, and they decrease hydroxylase gene expression (Figure 7A). The reduction appears to be dependent on liver impairment and independent of FXR, as treatment with an FXR agonist does not cause cholestasis and does not decrease CYP7A1 or CYP8B1 mRNA levels. We speculate that this response represents a specific protective mechanism designed to eliminate bile acid synthesis in the face of liver injury, since the expression of an enzyme in the alternate pathway of bile acid synthesis (the CYP7B1 oxysterol 7α-hydroxylase) is also decreased in this experiment, whereas mRNAs encoding CPF, OATP2, and stearoyl CoA-desaturase are unchanged. Mice fed lithocholic acid, a potent cholestatic agent and ligand for the nuclear receptor PXR (Xie et al., 2001), repress CYP7A1 expression via a PXR-dependent pathway (Staudinger et al., 2001). We believe this regulatory response is different from that observed here, as the expression of other lithocholic acid and PXR responsive genes, including CYP3A and OATP2, is unchanged in Shp−/− mice, even in those fed cholic acid.
The synthesis of bile acids in the mouse is thus controlled by at least four negative regulatory inputs. One is mediated by the nuclear hormone receptor SHP and responds to bile acid pool size and composition and is the major regulatory pathway under normal conditions. A second is independent of SHP and is inactivated in response to reduction of the bile acid pool. A third inhibitory pathway comes into play when the liver is damaged and also does not require SHP. Input from the latter pathway decreases the steady-state levels of the CYP7A1 and CYP8B1 mRNAs, which suggests a transcriptional mechanism of action; however, experiments to rule out more complex mechanisms have not yet been performed. We do not know the identities of the transcription factors that participate in the SHP-independent regulatory pathways. These may include nuclear receptors, such as FXR (which is fully functional in the Shp−/− mice and is also bathed in excess amounts of cholic acid in these animals [Figure 3]), or other transcription factors. A fourth negative regulatory pathway is mediated by the receptor PXR and is induced by lithocholic acid-mediated cholestasis (Staudinger et al., 2001). Future experiments in mice lacking more than one nuclear receptor may reveal the identities of other transcription factors that regulate bile acid synthesis.
In addition to mRNAs encoding bile acid biosynthetic enzymes, the expression of numerous other mRNAs is increased in Shp−/− mice (Figure 6). Enhanced expression may occur by unregulated CPF activity, as is the case for the CYP7A1 and CYP8B1 hydroxylases, or via FXR due to the altered bile acid pool size or composition, as is the case with BSEP. Whether other nuclear receptors are involved in the expression of these genes, as would be predicted from in vitro studies, remains to be determined, as does the role of SHP in peripheral tissues.
Experimental Procedures
Animals and Diets
Deltagen, Inc. generated Shp−/− mice under contract using standard gene-targeting methods (Hogan et al., 1995). To disrupt the Shp locus, a 318 bp fragment corresponding to a segment of exon 1 (Lee et al., 1998) was replaced by a phosphoglycerate kinase promoter-driven geneticin/neomycin resistance cassette in a targeting vector, and this DNA was linearized and electroporated into embryonic stem cells derived from the 129/OlaHsd strain. Positive (geneticin) selection and screening produced multiple cell lines that harbored the desired mutation. Cells from several different lines were injected into recipient C57BL/6J blastocysts to produce chimeras that transmitted the mutation through the germ line. One of the transmitting high-percentage male chimeras was out crossed to C57BL/6J females to produce F1 heterozygotes, which were intercrossed to produce F2 wild-type, heterozygous, and homozygous mice in the expected Mendelian ratios. Genotyping was accomplished by PCR using primer pairs specific for the wild-type Shp allele (ATGAGCTC CGGCCAGTCAGGGGTCT; TTCTTAAGTATACTGGGCACCGGAG) or mutant allele (ATGAGCTCCGGCCAGTCAGGGGTCT; GGGGATCGA TCCGTCCTGTAAGTCT).
Shp−/− mice and wild-type littermates were bred at Tularik, Inc., South San Francisco, CA and at the University of Texas Southwestern Medical Center in Dallas, TX. In both animal facilities, mice were housed in plastic colony cages containing wood shavings in a temperature-controlled room (22°C) with 12 hr light cycling. Animals were fed a cereal-based rodent diet containing 4% (w/w) total lipid and 0.02% (w/w) cholesterol. In some experiments, the powdered form of this diet was supplemented with cholic acid (Sigma Chemical Co.) or cholestyramine (cholestid, Upjohn, Corp.). GW4064, a synthetic FXR agonist (Goodwin et al., 2000), was administered intra-gastrically once daily by oral gavage as a suspension in 1% (w/w) methylcellulose/1% (w/w) Tween 80. All experiments were carried out using male mice 2–4 months of age.
Cholesterol Balance Studies
Cholesterol absorption was measured by a dual-isotope fecal ratio method (Schwarz et al., 2001). The rate of sterol synthesis in liver and extra hepatic organs was measured in vivo by a [3H]H2O incorporation assay (Jeske and Dietschy, 1980). Bile acid pool composition was determined by GC-MS. Briefly, total bile acids from liver, gall-bladder, and small intestine were extracted with ethanol in the presence of 2 mg norcholic acid as internal standard. Aliquots of the sample extract representing ~5 μg internal standard were hydrolyzed, extracted, and methylated (Czubayko et al., 1991). The resulting bile acid methyl esters were trimethyl silylated and analyzed by GC-MS (Rosen et al., 1998). Quantification of individual bile acids was done by linear calibration with commercially available standards (Steraloids, Inc. or Sigma Chemical Co.). Bile acid pool size was measured by high-performance liquid chromatography (Schwarz et al., 1998).
Fecal bile acid excretion rates were measured by an enzymatic method (Turley and Dietschy, 1978). The amount of neutral sterols in fecal extracts and total cholesterol concentrations in liver tissue were measured by gas chromatography (Turley et al., 1994).
Plasma Lipoprotein Cholesterol and Triacylglycerol Concentrations
Mice were exsanguinated from the vena cava after euthanasia with CO2. Plasma was isolated from EDTA-treated blood and pooled within genotype and/or dietary groups. Samples were fractionated by fast protein liquid chromatography using two serial Superose HR6 columns. Cholesterol and triacylglycerol contents of each fraction and of whole plasma were determined enzymatically using commercially available kits (Roche Molecular Biochemicals).
RNA Analyses
Total RNA was extracted from frozen tissue using TRIZOL Reagent (GIBCO-BRL; #15596-018). Equal amounts of total RNA were pooled within genotype and/or dietary groups for the purification of poly(A)+ RNA using an Oligotex mRNA Maxi Kit (QIAGEN; #70061). Aliquots of poly(A)+ RNA (3–5 μg) were analyzed by blotting using standard procedures (Sambrook and Russell, 2000). Real-time reverse transcriptase PCR measurements of individual mRNAs were performed on pools of total RNA using CYBR green dye to measure duplex DNA formation.
Cholesterol 7α-Hydroxylase
Hepatic microsomes were prepared from freshly dissected livers by sequential centrifugation (Schwarz et al., 1997). Equal amounts of microsomal protein were pooled within experimental groups. CYP7A1 protein was analyzed by immunoblotting using a rabbit-generated polyclonal antibody that recognizes amino acids 476–490 of the murine protein. CYP7A1 enzymatic activity was measured in microsomal incubations in the presence of NADPH using endogenous cholesterol as a substrate. Reactions were terminated by the addition of 5 ml ethylacetate and analyzed by GC-MS. The amount of 7α-hydroxycholesterol formed in each sample was quantitated using five matrix-assisted calibration standards containing 3, 10, 30, 100, or 300 ng of 7α-hydroxycholesterol and 30 ng of 7α-hydroxycholesterol.
Acknowledgments
We thank Kevin Anderson, Norma Anderson, Scott Clark, Jeff Cormier, Yolanda Hatter, Melody Kerr, and Stephen Ostermann for excellent technical assistance; Jay Horton for help with in vivo sterol synthesis; Julie Li-Hawkins for assistance with bile acid metabolism; and Mike Brown for critical reading of the manuscript. This research was supported by grants from the NIH (HL20948), the Keck Foundation, and the Perot Family Foundation.
References
- Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem. 2001;276:28857–28865. doi: 10.1074/jbc.M011610200. [DOI] [PubMed] [Google Scholar]
- Castillo-Olivares A, Gil G. α1-fetoprotein transcription factor is required for the expression of sterol 12α-hydroxylase, the specific enzyme for cholic acid synthesis. J Biol Chem. 2000;275:17793–17799. doi: 10.1074/jbc.M000996200. [DOI] [PubMed] [Google Scholar]
- Castillo-Olivares A, Gil G. Suppression of sterol 12α-hydroxylase transcription by the short heterodimeric partner: insights into the repression mechanism. Nucleic Acids Res. 2001;29:4035–4042. doi: 10.1093/nar/29.19.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Czubayko F, Beumers B, Lammsfuss S, Lutjohann D, von Bergmann K. A simplified micro-method for quantification of fecal excretion of neutral and acidic sterols for outpatient studies in humans. J Lipid Res. 1991;32:1861–1867. [PubMed] [Google Scholar]
- Dietschy JM, Turley SD. Control of cholesterol turnover in the mouse. J Biol Chem. 2002;277:3801–3804. doi: 10.1074/jbc.R100057200. [DOI] [PubMed] [Google Scholar]
- Fabiani ED, Mitro N, Anzolovich AC, Pinelli A, Galli G, Crestani M. The negative effects of bile acids and tumor necrosis factor-α on the transcription of cholesterol 7α-hydroxylase (CYP7A1) converge to hepatic nuclear factor-4. J Biol Chem. 2001;276:30708–30716. doi: 10.1074/jbc.M103270200. [DOI] [PubMed] [Google Scholar]
- Goodfellow PN, Camerino G. DAX-1, an “antitestis” gene. EXS. 2001;1:57–69. [PubMed] [Google Scholar]
- Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid synthesis. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- Gupta S, Stravitz RT, Dent P, Hylemon PB. Down-regulation of cholesterol 7-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J Biol Chem. 2001;276:15816–15822. doi: 10.1074/jbc.M010878200. [DOI] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzales FJ. Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- Jeske DJ, Dietschy JD. Regulation of rates of cholesterol synthesis in vivo in the liver and carcass of the rat measured using [3H]water. J Lipid Res. 1980;21:364–376. [PubMed] [Google Scholar]
- Lee YK, Moore DD. Dual mechanisms for repression of the monomeric orphan receptor liver receptor homologous protein-1 (LRH-1) by the orphan small heterodimeric partner (SHP) J Biol Chem. 2002;276:2463–2467. doi: 10.1074/jbc.M105161200. [DOI] [PubMed] [Google Scholar]
- Lee HK, Lee YK, Park SH, Kim YS, Park SH, Lee JW, Kwon HB, Soh J, Moore DD, Choi HS. Structure and expression of the orphan nuclear receptor SHP gene. J Biol Chem. 1998;273:14398–14402. doi: 10.1074/jbc.273.23.14398. [DOI] [PubMed] [Google Scholar]
- Lee YK, Parker KL, Choi HS, Moore DD. Activation of the promoter of the orphan receptor SHP by orphan receptors that bind DNA as monomers. J Biol Chem. 1999;274:20869–20873. doi: 10.1074/jbc.274.30.20869. [DOI] [PubMed] [Google Scholar]
- Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- Lu TT, Repa JJ, Mangelsdorf DJ. Orphan nuclear receptors as eLiXiRs and FiXeRs of sterol metabolism. J Biol Chem. 2001;276:37735–37738. doi: 10.1074/jbc.R100035200. [DOI] [PubMed] [Google Scholar]
- Miyake JH, Wang SL, Davis RA. Bile acid induction of cytokine expression by macrophages correlates with repression of hepatic cholesterol 7α-hydroxylase. J Biol Chem. 2000;275:21805–21808. doi: 10.1074/jbc.C000275200. [DOI] [PubMed] [Google Scholar]
- Nishigori H, Tomura H, Tonooka N, Kanamori M, Yamada S, Sho K, Inoue I, Kikuchi N, Onigata K, Kojima I, et al. Mutations in the small heterodimer partner gene are associated with mild obesity in Japanese subjects. Proc Natl Acad Sci USA. 2001;98:575–580. doi: 10.1073/pnas.021544398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta M, Ku S, Brown C, Okamoto AY, Shan B. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7α-hydroxylase gene. Proc Natl Acad Sci USA. 1999;96:6660–6665. doi: 10.1073/pnas.96.12.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repa JJ, Lund EG, Horton JD, Leitersdorf E, Russell DW, Dietschy JM, Turley SD. Disruption of the sterol 27-hydroxylase gene in mice results in hepatomegaly and hypertriglyceridemia: reversal by cholic acid feeding. J Biol Chem. 2000;275:39685–39692. doi: 10.1074/jbc.M007653200. [DOI] [PubMed] [Google Scholar]
- Rosen H, Reshef A, Maeda N, Lippoldt A, Shpizen S, Triger L, Eggertsen G, Bjorkhem I, Leitersdorf E. Markedly reduced bile acid synthesis but maintained levels of cholesterol and vitamin D metabolites in mice with disrupted sterol 27-hydroxylase gene. J Biol Chem. 1998;273:14805–14812. doi: 10.1074/jbc.273.24.14805. [DOI] [PubMed] [Google Scholar]
- Russell DW, Setchell KDR. Bile acid biosynthesis. Biochemistry. 1992;31:4737–4749. doi: 10.1021/bi00135a001. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Schwarz M, Lund EG, Lathe R, Bjorkhem I, Russell DW. Identification and characterization of a mouse oxysterol 7α-hydroxylase cDNA. J Biol Chem. 1997;272:23995–24001. doi: 10.1074/jbc.272.38.23995. [DOI] [PubMed] [Google Scholar]
- Schwarz M, Russell DW, Dietschy JM, Turley SD. Marked reduction in bile acid synthesis in cholesterol 7α-hydroxylase-deficient mice does not lead to diminished tissue cholesterol turnover or to hypercholesterolemia. J Lipid Res. 1998;39:1833–1843. [PubMed] [Google Scholar]
- Schwarz M, Davis DL, Vick BR, Russell DW. Genetic analysis of cholesterol absorption in inbred mice. J Lipid Res. 2001;42:1801–1811. [PubMed] [Google Scholar]
- Seol W, Choi HS, Moore DD. An orphan nuclear receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science. 1996;272:1336–1339. doi: 10.1126/science.272.5266.1336. [DOI] [PubMed] [Google Scholar]
- Shih DQ, Bussen M, Sehayek E, Ananthanarayanan M, Shneider BL, Suchy FJ, Shefer S, Bollileni JS, Gonzalez FJ, Breslow JL, et al. Hepatocyte nuclear factor-1α is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet. 2001;27:375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, Mac-Kenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci USA. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley SD, Dietschy JM. Reevaluation of the 3α-hydroxysteroid dehydrogenase assay for total bile acids in bile. J Lipid Res. 1978;19:924–928. [PubMed] [Google Scholar]
- Turley SD, Herndon MW, Dietschy JM. Reevaluation and application of the dual-isotope plasma ratio method for the measurement of intestinal cholesterol absorption in hamsters. J Lipid Res. 1994;35:328–339. [PubMed] [Google Scholar]
- Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, Waxman DJ, Evans RM. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci USA. 2001;98:3375–3380. doi: 10.1073/pnas.051014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Wang F, Kan M, Jin C, Jones RB, Weinstein M, Deng CX, McKeehan WL. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J Biol Chem. 2000;275:15482–15489. doi: 10.1074/jbc.275.20.15482. [DOI] [PubMed] [Google Scholar]