Summary
Arginylation is an emerging posttranslational modification mediated by Arg-tRNA-protein-transferase (ATE1). It is believed that ATE1 links Arg solely to the N-terminus of proteins, requiring prior proteolysis or action by Met-aminopeptidases to expose the arginylated site. Here, we tested the possibility of Arg linkage to mid-chain sites within intact protein targets and found that many proteins in vivo are modified on the side chains of Asp and Glu by a novel chemistry that targets the carboxy rather than the amino groups at the target sites. Such arginylation appears to be functionally regulated, and it can be directly mediated by ATE1, in addition to the more conventional Ate1-mediated linkage of Arg to the N-terminal alpha amino group. This new type of arginylation implies an unconventional mechanism of ATE1 action that likely facilitates its major biological role.
Introduction
Arginylation is an emerging posttranslational modification mediated by arginyltransferase (ATE1) that transfers Arg from the Arg-tRNA onto proteins. Recent studies have demonstrated an essential role of this modification in key physiological events (Carpio et al., 2013; Karakozova et al., 2006; Lopez Sambrooks et al., 2012; Saha and Kashina, 2011; Varshavsky, 1997; Zhang et al., 2012) and identified a large number of arginylated proteins in vivo (Wong et al., 2007), confirming its important biological role. Despite its significance, very little is known about the mechanisms of ATE1-mediated Arg linkage to proteins. Earlier studies postulated that ATE1 attaches Arg to the N-terminally exposed amino group of a polypeptide chain via a peptide bond, reminiscent of the peptide elongation steps on the ribosome. In vitro and in vivo studies show that ATE1 preferentially targets the N-terminally exposed residues with acidic side chains (Asp and Glu) in proteins, produced via limited proteolysis or N-terminal preprocessing by specific types of aminopeptidases. However very few N-terminally arginylated proteins have been identified in vivo, despite the fact that overall estimates suggest that a large percentage of the proteome may undergo arginylation (Wong et al., 2007). These results leave open the possibility that arginylation may also occur at internal sites within intact proteins, which have not been commonly considered during previous identification strategies.
A recent study identified an in vivo form of the regulatory peptide neurotensin, modified by arginylation on a side chain of an internal Glu via an amide bond with the amino group of Arg (Eriste et al., 2005). Such Arg linkage at the acidic side chain of a mid-chain residue can account for additional arginylated protein substrates in vivo. However, modification of the acidic side chains of internal Asp and Glu, residues ,at first glance would require very different chemistry than “conventional” arginylation chemistry of N-terminal Asp/Glu, leaving an open question whether this alternative modification indeed targets proteins in vivo and whether it can be performed by ATE1.
Here, we report that many proteins in vivo are modified on the side chains of Asp and Glu, and this modification can be directly mediated by ATE1, in addition to its more conventional linkage by N-terminal alpha amino group. This new type of arginylation likely facilitates the major biological role of arginylation in regulation of intact proteins during key physiological processes.
Results
Arginylation in vivo can occur on mid-chain Asp and Glu
To test the possibility that addition of Arg to proteins can happen at internal sites and not on their N-termini, we performed mass spectrometry analysis of cell extracts and subcellular structures described in our prior studies (affinity enriched cell and embryo extracts (Wang et al., 2011), nuclear extracts (Saha et al., 2011), platelets, and myofibril preparations (Kurosaka et al., 2012)), to look for addition of Arg to any Asp, Glu, and Lys (the only three residues that can theoretically accept Arg directly on their side chain), using the search algorithms commonly applied to posttranslational modifications. While Lys searches did not yield any hits, we found a number of previously unidentified sites where Arg was added to mid-chain Asp and Glu (Fig. 1A, Table 1, Supplemental Tables 1, 2, Supplemental Dataset).
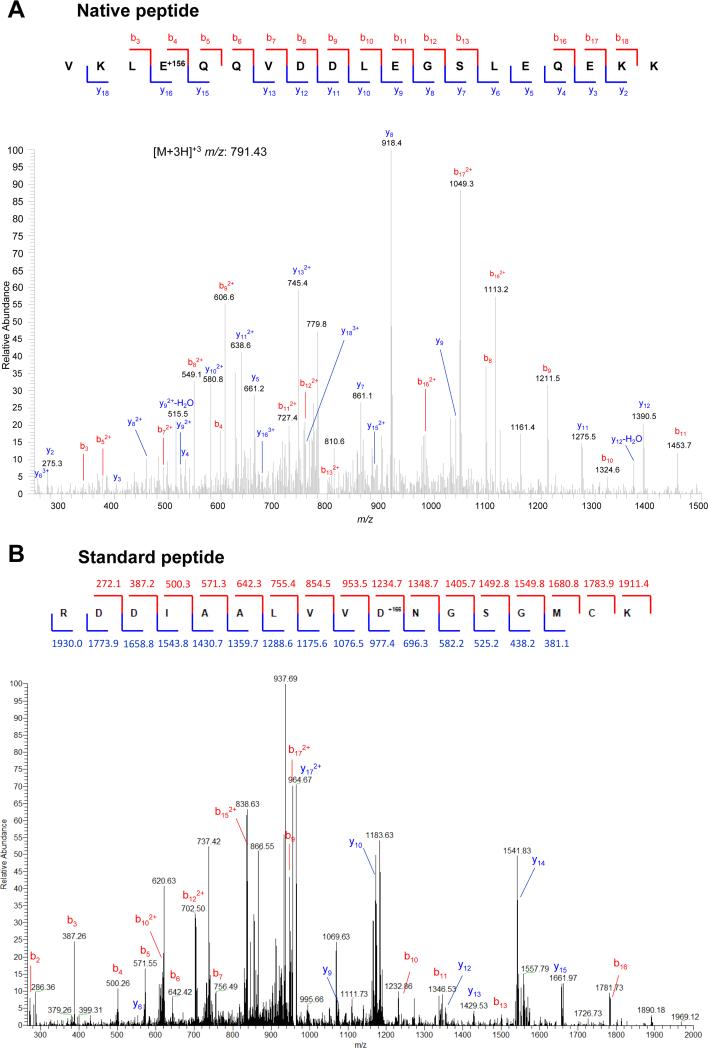
Figure 1. Arginylation on side chains of acidic residues in vivo (top) and in vitro (bottom).
Top, Mass spectrum of a naturally occurring arginylation on the side chain of Asp. Peptide sequence is indicated on top, with the arginylated residue marked by +156 (a mass of Arg). See Table 1 and Supplemental Table 1 for a full list of identified side-chain-arginylated proteins and Supplemental Table 2 for sequences and parameters of the identified peptides. Bottom, mass spectrum of a standard peptide, enzymatically arginylated on the side chain of Asp. Peptide sequence is indicated on top, with the arginylated residue marked by +166 (a mass of stable isotope labeled [13C,15N]-Arg used in the experiment). Numbers indicate the masses of b (red) and y (blue) ions.
Table 1.
Protein sites arginylated in vivo on the side chains of Asp and Glu.
| Accession | Source | Name | modified residue |
|---|---|---|---|
| NP_033738.1 | Myofibrils | actin, alpha cardiac muscle 1 | E102, E169 |
| NP_033736.1 | Platelets | actin, alpha skeletal muscle | E85 |
| NP_033739.1 | Platelets | actin, cytoplasmic 2 | E167 |
| NP_150371.4 | Myofibrils | alpha-actinin-2 dual specificity mitogen-activated protein kinase | E512 |
| NP_036073.1 | Platelets | kinase 6 | E307 |
| NP_031894.1 | Myofibrils | dystrophin | E2570 |
| NP_031953.1 | Platelets | emerin | D127 |
| NP_666232.2 | Platelets | gelsolin precursor | E261, E264 |
| NP_001124340.1 | Platelets | lysyl-tRNA synthetase isoform 1 | D403 |
| NP_081797.1 | Platelets | minor histocompatibility protein HA-1 isoform 2 | D826 |
| NP_034997.2 | Myofibrils | myomesin-1 isoform 1 | E1220 |
| NP_034989.1 | Myofibrils | myosin light chain 3 | E121 |
| NP_034986.1 | Myofibrils | myosin-6 | E1030, E1491, D1654 |
| NP_542766.1 | Myofibrils | myosin-7 | E1578 |
| NP_071855.2 | Platelets | myosin-9 isoform 1 | E1073 |
| NP_032138.3 | Platelets | rab GDP dissociation inhibitor beta | D34 |
| NP_035372.2 | Platelets | RAS guanyl-releasing protein 2 | E408 |
| NP_035782.3 | Myofibrils | titin isoform N2-A | D1068, D14177, E14547 |
| NP_033473.1 | Platelets | tubulin alpha-4A chain [Mus musculus] | E90 |
see Supplemental Table 1 for the list of sites shared by homologous isoforms of these proteins
While we could not detect a primary consensus sequence in the vicinity of the arginylation site, the frequency of occurrence of specific amino acid residues around the arginylation site, compared to their overall frequency in the identified proteins, showed a bias toward His and Pro and against Cys (never found within 5 residues of the arginylated sites and poorly represented within up to 20 flanking residues) (Supplemental Table 3).
Mid-chain Asp arginylation is mediated by ATE1
To test whether ATE1 can directly mediate Arg addition to the side chains of Asp or Glu, we used a test peptide corresponding to the N-terminus of ATE1's natural substrate, beta actin (Karakozova et al., 2006), containing an N-terminal Arg covalently linked into the peptide backbone (RDDIAALVVDNGSGMCK, Fig. 1B). We reasoned that such a peptide would be more likely to facilitate ATE1 recognition than a randomly selected peptide, but should constitute an unfavorable N-terminal target for ATE1, since it already contains an N-terminally linked Arg. In agreement with these predictions, in vitro arginylation of this peptide by purified ATE1-1 (Wang et al., 2011) resulted in its arginylation at Asp9 (Fig. 1B) -- the site where the only possible Arg addition can occur via the acidic side chain of Asp. This reaction, similarly to the N-terminal arginylation, occurred with higher efficiency in the presence of ATE1-1 and ATE1-2, rather than ATE1-3 and ATE1-4 (Wang et al., 2011) (Fig. 2A). Control experiments confirmed that this side chain arginylation is tRNA-dependent, since no Arg incorporation into this peptide could be seen in the absence of tRNA (Fig. S1).
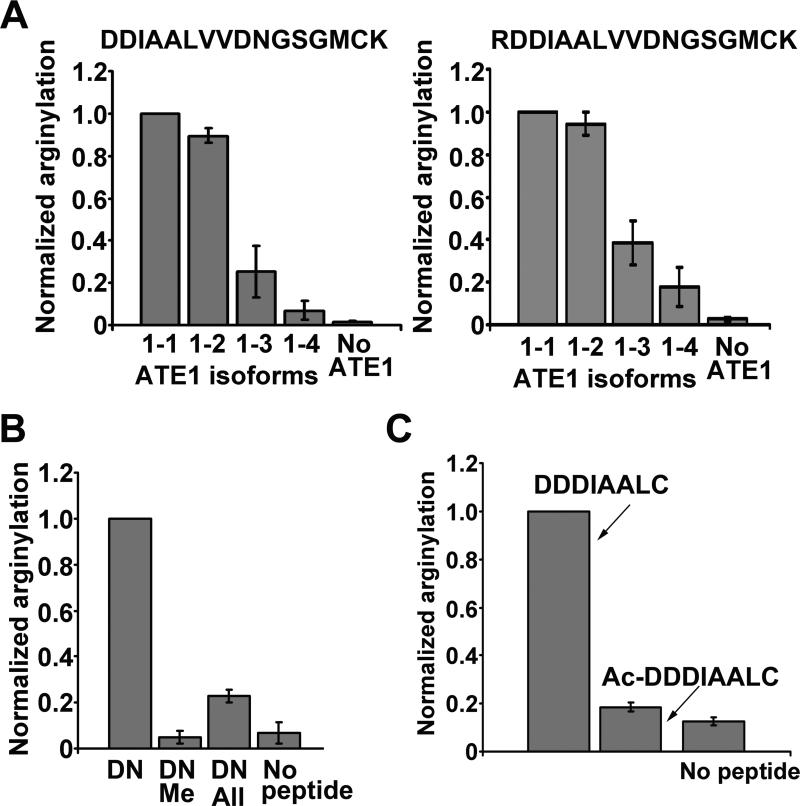
Figure 2. Characterization of side chain and N-terminal arginylation.
A. Normalized incorporation of [3H]-Arg into a standard peptide corresponding to the N-terminal sequence of beta actin mediating N-terminal arginylation (DDIAALVVDNGSGMCK; left) and the same peptide with an N-terminal Arg inserted into the sequence to facilitate side chain arginylation (RDDIAALVVDNGSGMCK, right) with 4 ATE1 isoforms. Error bars represent SEM (n=3 for DDIAALVVDNGSGMCK and 4 for RDDIAALVVDNGSGMCK). B. N-terminal arginylation requires the presence of both a carboxy and an amino group. Normalized incorporation of [3H]-Arg into a synthetic DN peptide (containing N-terminal Asp and no internal arginylation sites, see text) is greatly inhibited when the amino group of the N-terminal Asp is modified with a methyl group (DN-Me) or when the carboxyl group is an allyl ester (DN-All). Error bars represent SEM (n=3). C. N-terminal acetylation inhibits arginylation. Normalized incorporation of [3H]-Arg into a synthetic peptide corresponding to the actin-s N-terminus (DDDIAALC) is greatly diminished after the N-terminal amino group of this peptide is blocked by acetylation (AcDDDIAALC) despite the presence of multiple side chain carboxyl groups. Error bars represent SEM (n=3).
Both amino and carboxyl group of the N-terminal residue facilitate arginylation
Our finding that ATE1 can add Arg to the side chain of internal residues raises a possibility that, unlike previously believed, ATE1 always targets the carboxyl group of Asp and Glu, even when they are N-terminally exposed. To address this possibility, we tested the requirement of the amino and carboxyl group of the N-terminal acidic residue, by analyzing synthetic peptides derived from the actin's N-terminal sequence without the internal Asp residues (DIAALVHSSGMC, termed DN in Fig.2B), intact or chemically modified by the addition of a methyl group to block the N-terminal amino-group of Asp (DN-Me) or an allyl ester (All) group to block the carboxyl group of the Asp side chain (DN-All). Comparison of the arginylation efficiency of these three peptides showed that while DN was a highly efficient ATE1-1 substrate, blocking either the amino or the carboxyl group of the N-terminal Asp in this peptide inhibited arginylation (Fig. 2B). While a small amount of Arg incorporation was still seen in the DN-All peptide, we detected no arginylated DN-All by MALDI-TOF or MS MS, suggesting that this Arg incorporation could be due to a contaminant in this peptide. Blockage of N-terminal amino group by acetylation in another actin-derived peptide had a similar effect (Fig. 2C). Thus, N-terminal arginylation requires both the carboxy and the amino group at the target site.
N-terminal arginylation of the peptide substrates occurs by the alpha amino group
To directly test which chemical group is involved in the final product of the ATE1-1-mediated N-terminal arginylation, we analyzed in vitro arginylated peptide DDIAALVVDNGSGMCK (corresponding to the N-terminus of beta actin, Fig. 2A) by subtractive Edman sequencing, measuring the mass of the remaining peptide after each Edman cycle by MALDI-TOF. In this test, the majority of the peptide behaved as expected for the conventional Edman sequencing, releasing Arg in the first cycle (see Fig. S2 for the peptide before and after the first Edman cycle). This result was confirmed by conventional Edman sequencing, confirming that ATE1-mediated Arg linkage to the actin's N-terminal peptide in vitro occurred via the alpha amino group.
To further test whether N-terminal Arg-Asp linkage engages amino or carboxyl group of Arg (and, consequently, the carboxy or amino group at the target Asp), we used Nuclear Magnetic Resonance (NMR) to analyze the biological regulatory peptide angiotensin II (DRVYIHPF), enzymatically arginylated with isotopically labeled [13C,15N]-Arg. It has been previously shown that angiotensin II is a good substrate for ATE1 in vitro and accepts Arg on its N-terminal Asp (Soffer, 1975). This was confirmed in our experiments (Fig. 3A, left).
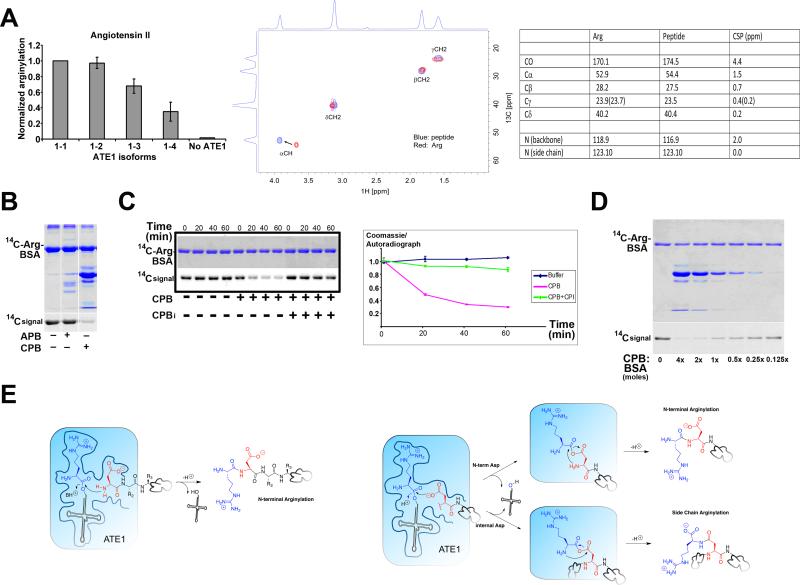
Figure 3. ATE1 mediates different Arg linkage in proteins and peptides.
A. Left, Normalized incorporation of [3H]-Arg into angiotensin II in vitro indicate that it is an efficient ATE1 target. Error bars represent SEM (n=3). Middle, NMR spectrogram of [13C,15N]-Arg linked to angiotensin II indicates a shift at Arg alpha carbon atom, suggesting that this atom is engaged in the linkage to the peptide via the “conventional” peptide bond. Mass shifts are shown in the table on the right. B, Coomassie-stained gel (top) and audioradiograph (bottom) of BSA, in vitro arginylated using [14C]-Arg, incubated for 30 min alone or in the presence of aminopeptidase B (APB) or carboxypeptidase B (CPB) as indicated at the bottom. C, CPB-mediated dearginylation of BSA can be inhibited by CPBi. Coomassie-stained gel (top) and audioradiograph (bottom) of BSA, in vitro arginylated using [14C]-Arg, incubated for increasing intervals of time alone or in the presence of CPB with and without added inhibitor (CPBi). The chart shows the levels of radioactive label divided by BSA protein levels (quantified from gel scans) for each condition at each time point. Error bars represent SEM (n=3). D, CPB-mediated dearginylation of BSA is dose dependent. Coomassie-stained gel (top) and audioradiograph (bottom) of BSA, in vitro arginylated using [14C]-Arg, incubated with decreasing molar ratios of CPB to BSA as indicated. E. Proposed mechanisms for arginylation (see explanations in the text). Protein chains are denoted by freeform circles.
Comparison of the NMR analysis of highly arginylated angiotensin II using ATE1-1 and [13C,15N]-Arg ([13C, 15N]-R-DRVYIHPF) with the free [13C, 15N] Arg revealed that all the carbon and nitrogen atoms from the isotopically labeled Arg in free form or bound to angiotensin II were observed in proton detected 1H, 13C-HSQC and carbon detected C-CAN and C-CACO spectra (Fig 3A, right). The chemical shift analysis showed that the backbone carbonyl carbon of arginylated Arg exhibited the largest chemical shift (4.4 ppm) compared to that of the free Arg, suggesting that alpha carboxyl group of Arg is unambiguously engaged in its linkage to the peptide, as appropriate for a conventional peptide bond.
Arginylation of a test protein substrate occurs largely by carboxyl groups
To test how Arg is linked to proteins rather than peptide substrates, we used BSA as a test substrate, previously found arginylated at two Asp: the N-terminus, exposed by the removal of the signal peptide, and at Asp 135 (Wang et al., 2011). We used [14C]-Arg to arginylate BSA, to enable detection by autoradiography, and treated such radioactively arginylated BSA with either aminopeptidase B (ApB), which removes N-terminal Arg residues from the peptide chain, or carboxypeptidase B (CpB), which removes C-terminal Arg residues (Folk and Gladner, 1958; Hirose et al., 2006). Addition of ApB to the arginylated BSA did not result in detectable reduction of the radioactive label (Fig. 3B), despite the fact that the enzyme preparation was highly active in colorimetric assays with its test substrate (Fig. S3). This observation indicates that the majority of arginylation in BSA does not involve formation of a canonical peptide bond with the N-terminal Asp. In contrast, addition of CpB resulted in a marked removal of radioactivity from BSA without a detectable reduction in the BSA protein level detectable by Coomassie Blue staining (Fig. 3B), although high concentrations of CpB were required. To characterize this further, we tested the dynamics of CpB-mediated Arg removal over time and its dependence on the commercially available CpB inhibitor (CPBi). In these assays, Arg removal from BSA progressed over time and was completely abolished in the presence of CPBi, confirming the specificity of this reaction (Fig. 3C). Dearginylation by CpB was also dose-dependent (Fig 3D). These assays strongly suggest that ATE1-mediated arginylation of BSA occurred to the large extent via Arg addition to the side chain carboxylate groups.
Discussion
Our data demonstrate for the first time that ATE1 during posttranslational arginylation can utilize a novel type of chemical linkage of Arg to the side chains of acidic amino acid residues in intact proteins and peptides. Our analysis suggests that many proteins in vivo employ this novel type of Arg linkage, expanding the traditional view that arginylation requires preproteolysis or the action of Met-aminopeptidases to expose Asp and Glu for N-terminal targeting by ATE1. This new type of modification carries a potentially high and independent biological significance and implicates that arginylation, like other regulatory modifications (e.g., protein phosphorylation), can affect side chains of amino acid residues within otherwise intact protein chains and thus can transiently or stably modify these proteins’ structure and facilitate their biological functions.
Our results suggest that ATE1 can mediate two different types of chemical linkages to target residues in a protein, one via the “conventional” N-terminal peptide bond, and one via the carboxyl group of the target residue, presumably linked to the alpha amino group of Arg. While it is uncommon for one enzyme to catalyze the formation of two such different bonds, the dependence of arginylation on both amino and carboxyl group at a target site suggests that both groups may be involved in the formation of an intermediate during the arginylation reaction, and/or participate in ATE1-mediated recognition or catalysis. The formation of an amide bond requires activation of the carboxylic acid for attack by the amine nucleophile. A similar reaction is likely utilized by the enzymes that catalyze glutamylation and use ATP to activate the side chain carboxylates of tubulin and other proteins (Janke et al., 2008; Janke et al., 2005; Rogowski et al., 2009). In the case of arginylation, Arg-tRNA provides the activated carboxylate for ATE-catalyzed arginylation, bypassing the requirement of this reaction for ATP (Wang et al., 2011). N-terminal arginylation could involve direct attack of the free amino group of a N-terminal Asp analogous to protein translation, where the free amino group of the peptide attacks the activated carboxyl group of Arg-tRNA (Figure 3E, left). However, this mechanism cannot account for arginylation of internal Asp residues, which requires activation the Asp β-carboxylate. Therefore we suggest an alternative mechanism whereby N-terminal and side chain arginylation derive from a common intermediate. In this mechanism, the β-carboxylate of Asp first attacks the ester of Arg-tRNA, forming an anhydride intermediate wherein both carboxylate groups are activated (Figure 3E, right). If the Asp is at the N-terminus of a peptide, this intermediate could react with the free amino group to form a canonical peptide bond, creating an N-terminal Arg (Figure 3E, top right). Alternatively, if the Asp is internal, the intermediate would subsequently react with the amino group of Arg to form a noncanonical peptide bond with the side chain of Asp. Similar acyltransfer reactions are found in intein reactions (Volkmann and Mootz, 2013). The resulting adduct would have a C-terminal Arg residue that can be recognized by CpB, albeit with a noncanonical amide bond, which would account for the requirement for high concentrations of CpB.
A number of mid-chain arginylated proteins are involved in the functioning of the actin cytoskeleton. It is possible that arginylation-dependent regulation of these proteins (predominantly mid-chain) may facilitate a different biological role of ATE1 in regulating protein-protein interactions and facilitating their assembly and biological activity, in contrast to its previously described role in the N-end rule pathway of protein degradation, which is exclusive to N-terminal arginylation. We hypothesize that these two types of Arg linkage lead to different types of downstream effects of arginylation as a global biological regulator. These possibilities constitute exciting directions of further studies.
Experimental Procedures
Escherichia coli strains, Miscellaneous Reagents and Animal Care
BL21-CodonPlus® (DE3)-RIL and BL21(DE3) were purchased from Stratagene (USA). The recombinant mouse ATE1 isoforms and Escherichia coli Arginy-tRNA synthetase (RRS) were purified as previously described (Wang et al., 2011). The care and treatment of mice were performed in accordance with the relevant National Institutes of Health (NIH) guidelines.
Peptides
Human angiotensin II (A9525) was purchased from Sigma; Acetylated and non-acetylated actin N-terminal peptide were synthesized by Dr. Henry Zebroski at the Rockefeller University Proteomics Resource Center; non-arginylated long actin N-terminal peptide, DN, DN-Me, and DN-All were synthesized by Genscript.
In vitro arginylation of peptides
In vitro arginylation reaction of peptides by ATE1 was modified from (Wang et al., 2011). See Supplemental Experimental Procedures for further details.
Mass spectrometry and database searches
Sample preparation and tandem MS/MS were performed as previously described (Wong et al., 2007; Xu et al., 2009). See Supplemental Experimental Procedures for further details.
NMR experiments
All NMR spectra were recorded on a Bruker Avance II 600 MHz NMR spectrometer, equipped with a Bruker TCI triple resonance cryoprobe. All NMR samples were prepared by dissolving the lyophilized compounds in a buffer with 50 mM sodium phosphate, 50 mM NaCl, and pH 6.3. The concentrations of the samples were around 0.5 mM. The spectra were recorded with Bruker Topspin standard pulse sequences at 25 °C, and the spectra were processed with Bruker Topspin software based on the manufacture suggestions.
Statistical analysis
Calculation of standard deviation (SD) and standard error of mean (S.E.M.) was based on Student's t-distribution.
Significance
Arginylation is an emerging posttranslational modification mediated by Arg-tRNA-protein-transferase (ATE1) which links Arg predominantly to the acidic residues, Asp and Glu. Prior studies suggested that this linkage must be solely N-terminal, requiring prior proteolysis or action by Met-aminopeptidases to expose the target residue in a protein chain and leading to implications that intact proteins in vivo cannot undergo arginylation. The present study shows for the first time that many proteins in vivo are modified on the side chains of Asp and Glu via a novel chemical linkage that utilizes the carboxyl rather than the amino groups at the target sites and thus does not require any prior pre-processing. Further, we show that this novel reaction can be mediated by ATE1 itself in the absence of cofactors, and that this reaction likely predominates during arginylation of proteins rather than peptide substrates. This finding expands the scope of reactions involving arginylation and demonstrates a novel mechanism of regulation of intact proteins in vivo.
Supplementary Material
Highlights.
Multiple proteins are arginylated in vivo on side chains of Asp and Glu
ATE1 can mediate both N-terminal and mid-chain arginylation of Asp and Glu
Both amino and carboxyl group of the N-terminal Asp facilitate arginylation
ATE1 adds Arg predominantly to protein side chains rather than the N-terminus
Acknowledgements
We thank Dr. Kayoko M. Fukasawa (Matsumoto Dental University, Japan) for the gift of the plasmid pGEX-Ap-B containing cDNA encoding rat aminopeptidase B, members of the Wistar Institute Proteomics facility for performing MALDI-TOF mass spectrometry throughout the project, John B. Warner and Solongo Batjargal at Dr. E. James Petersson lab at the University of Pennsylvania for help with HPLC purification, W.M. Keck Mass Spectrometry Facility at Yale for the conventional round of Edman sequencing, Dr. Y. I. Wolf for help with sequence analysis of the arginylated sites and preparation of the Supplemental Table 3, Dr. Chaoxing Yuan for helpful technical suggestions, and Dr. John Pehrson for helpful discussions throughout the project. This work was supported by NIH grant R01GM104003 and pilot grants from the Mari Lowe Cancer Center and University of Pennsylvania Research Foundation to A.K. and NIH grant R41GM103533 to J. R. Y.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that they have no conflict of interest.
References
- Carpio MA, Decca MB, Sambrooks CL, Durand ES, Montich GG, Hallak ME. Calreticulin-dimerization induced by post-translational arginylation is critical for stress granules scaffolding. The international journal of biochemistry & cell biology. 2013 doi: 10.1016/j.biocel.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Eriste E, Norberg A, Nepomuceno D, Kuei C, Kamme F, Tran DT, Strupat K, Jornvall H, Liu C, Lovenberg TW, et al. A novel form of neurotensin post-translationally modified by arginylation. J Biol Chem. 2005;280:35089–35097. doi: 10.1074/jbc.M502567200. [DOI] [PubMed] [Google Scholar]
- Folk JE, Gladner JA. Carboxypeptidase B.I. Purification of the zymogen and specificity of the enzyme. J Biol Chem. 1958;231:379–391. [PubMed] [Google Scholar]
- Hirose J, Ohsaki T, Nishimoto N, Matuoka S, Hiromoto T, Yoshida T, Minoura T, Iwamoto H, Fukasawa KM. Characterization of the metal-binding site in aminopeptidase B. Biol Pharm Bull. 2006;29:2378–2382. doi: 10.1248/bpb.29.2378. [DOI] [PubMed] [Google Scholar]
- Janke C, Rogowski K, van Dijk J. Polyglutamylation: a fine-regulator of protein function? ‘Protein Modifications: beyond the usual suspects' review series. EMBO Rep. 2008;9:636–641. doi: 10.1038/embor.2008.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM, Temurak N, van Dijk J, Boucher D, van Dorsselaer A, et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- Karakozova M, Kozak M, Wong CC, Bailey AO, Yates JR, 3rd, Mogilner A, Zebroski H, Kashina A. Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science. 2006;313:192–196. doi: 10.1126/science.1129344. [DOI] [PubMed] [Google Scholar]
- Kurosaka S, Leu NA, Pavlov I, Han X, Ribeiro PA, Xu T, Bunte R, Saha S, Wang J, Cornachione A, et al. Arginylation regulates myofibrils to maintain heart function and prevent dilated cardiomyopathy. Journal of molecular and cellular cardiology. 2012;53:333–341. doi: 10.1016/j.yjmcc.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Sambrooks C, Carpio MA, Hallak ME. Arginylated calreticulin at plasma membrane increases susceptibility of cells to apoptosis. J Biol Chem. 2012;287:22043–22054. doi: 10.1074/jbc.M111.338335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowski K, Juge F, van Dijk J, Wloga D, Strub JM, Levilliers N, Thomas D, Bre MH, Van Dorsselaer A, Gaertig J, et al. Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell. 2009;137:1076–1087. doi: 10.1016/j.cell.2009.05.020. [DOI] [PubMed] [Google Scholar]
- Saha S, Kashina A. Posttranslational arginylation as a global biological regulator. Dev Biol. 2011;358:1–8. doi: 10.1016/j.ydbio.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Wong CC, Xu T, Namgoong S, Zebroski H, Yates JR, 3rd, Kashina A. Arginylation and methylation double up to regulate nuclear proteins and nuclear architecture in vivo. Chem Biol. 2011;18:1369–1378. doi: 10.1016/j.chembiol.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffer RL. Enzymatic arginylation of beta-melanocyte-stimulating hormone and of angiotensin II. J Biol Chem. 1975;250:2626–2629. [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule pathway of protein degradation. Genes to cells : devoted to molecular & cellular mechanisms. 1997;2:13–28. doi: 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- Volkmann G, Mootz HD. Recent progress in intein research: from mechanism to directed evolution and applications. Cellular and molecular life sciences : CMLS. 2013;70:1185–1206. doi: 10.1007/s00018-012-1120-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Han X, Saha S, Xu T, Rai R, Zhang F, Wolf YI, Wolfson A, Yates JR, 3rd, Kashina A. Arginyltransferase is an ATP-independent self-regulating enzyme that forms distinct functional complexes in vivo. Chem Biol. 2011;18:121–130. doi: 10.1016/j.chembiol.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CCL, Xu T, Rai R, Bailey AO, Yates JR, Wolf YI, Zebroski H, Kashina A. Global Analysis of Posttranslational Protein Arginylation. PLoS Biology. 2007;5:e258. doi: 10.1371/journal.pbio.0050258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wong CC, Kashina A, Yates JR., 3rd Identification of N-terminally arginylated proteins and peptides by mass spectrometry. Nat Protoc. 2009;4:325–332. doi: 10.1038/nprot.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Saha S, Kashina A. Arginylation-dependent regulation of a proteolytic product of talin is essential for cell-cell adhesion. The Journal of cell biology. 2012;197:819–836. doi: 10.1083/jcb.201112129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.