Abstract
The Golgi complex of mammalian cells is composed of interconnected stacks of flattened cisternae that form a continuous membrane system in the pericentriolar region of the cell. At the onset of mitosis, this so-called Golgi ribbon is converted into small tubular–vesicular clusters in a tightly regulated fragmentation process, which leads to a temporary loss of the physical Golgi–centrosome proximity. Mitotic Golgi breakdown is required for Golgi partitioning into the two daughter cells, cell cycle progression and may contribute to the dispersal of Golgi-associated signaling molecules. Here, we review our current understanding of the mechanisms that control mitotic Golgi reorganization, its biological significance, and assays that are used to study this process.
Introduction
The Golgi apparatus fulfills numerous critical functions. As a central organelle of the secretory pathway, it is necessary for the modification, sorting and transport of proteins and lipids, and for the organization of a subset of polarized microtubules (Wilson et al., 2011). Furthermore, Golgi membranes form a centrally localized platform that facilitates the integration of various signaling pathways (Cancino & Luini, 2013; Wilson et al., 2011).
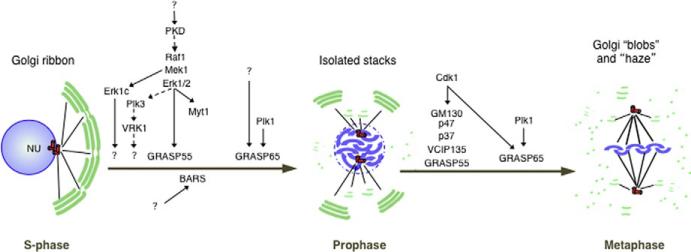
Although many basic Golgi functions are conserved throughout evolution, the structural organization of this organelle varies greatly between species. For example, in the yeast Saccharomyces cerevisiae or the protozoan parasite Toxoplasma gondii, individual Golgi cisternae or stacks of cisternae, respectively, are dispersed throughout the cytosol (Pelletier et al., 2002; Preuss, Mulholland, Franzusoff, Segev, & Botstein, 1992). In mammalian cells, in contrast, the Golgi complex is arranged as a interconnected pericentriolar ribbon in interphase and dispersed tubular–reticular and vesicular elements in mitosis (Misteli & Warren, 1995). This reversible conversion between two very different three-dimensional (3D) Golgi structures is achieved in tightly regulated steps (Fig. 23.1). First, the Golgi ribbon is disconnected to yield separate Golgi stacks (Colanzi et al., 2007). Then, stacks are converted into tubular–reticular membranes, termed “Golgi blobs,” which are dispersed throughout the cytosol (Colanzi, Suetterlin, & Malhotra, 2003; Nelson, 2000). Finally, “Golgi blobs” are broken down by vesiculation to yield the “Golgi haze,” which corresponds to small dispersed vesicles (Colanzi, Suetterlin, et al., 2003). This cell cycle-dependent reorganization leads to a temporary block in Golgi function. For example, in mitosis, protein transport does not occur and the ability of Golgi membranes to nucleate microtubules is significantly downregulated (Maia et al., 2013; Yeong, 2013). Postmitotic Golgi reassembly begins in telophase and involves the formation of two smaller Golgi ribbons that eventually coalesce (Gaietta et al., 2006).
FIGURE 23.1.
The fragmentation of the Golgi ribbon during mitosis is tightly controlled. Schematic representation of the mechanisms leading to breakdown of the Golgi complex in mitosis. The mammalian Golgi complex in S-phase forms a ribbon next to the centrosome (red cylinders) and the nucleus (NU). In G2, BARS, MEK1, the GRASPs, and Plk1 activities are required to convert this interconnected ribbon into isolated stacks. This step is necessary and sufficient for entry into mitosis (prophase). In prophase, the isolated Golgi stacks undergo further disassembly into the Golgi “blobs” and “haze” through Plk1 and Cdk1-mediated phosphorylation of golgins (GM130), and components of the fusion machinery (p47, p37, and VCIP135). The relative fraction of Golgi membranes dispersed as “blobs” or “haze” is cell type specific.
In this review, we will focus on the regulation of cell cycle-dependent Golgi dynamics in mammalian cells. We will describe how this organelle is organized at each stage of the cell cycle and how signaling pathways regulate the conversion of the Golgi ribbon into dispersed tubular–vesicular clusters. In addition, we will discuss why Golgi membranes may fragment during mitosis and describe some assays that have been used to monitor mitotic Golgi reorganization and to identify regulatory components.
23.1 The Organization Of Mammlian Golgi Membranes In Interphase
The characteristic ribbon organization of the mammalian Golgi apparatus depends on a diverse set of proteins. Up to 100 Golgi stacks, each composed of five to eight flattened cisternae are connected by membranous tubular bridges, called the noncompact zones, and are positioned next to the centrosome (Sütterlin & Colanzi, 2010). Golgins are peripheral membrane proteins that are proposed to maintain this 3D arrangement of Golgi membranes by providing a structural skeleton (Munro, 2011). Their involvement in Golgi organization is supported by the finding that the depletion of a golgin, such as Golgin-160, resulted in the conversion of the Golgi ribbon into dispersed, but functional mini-stacks (Yadav, Puri, & Linstedt, 2009). The peripheral Golgi proteins GRASP65 and GRASP55 also contribute to Golgi ribbon formation because RNAi-mediated depletion of either protein resulted in the unlinking of Golgi stacks (Feinstein & Linstedt, 2008; Puthenveedu, Bachert, Puri, Lanni, & Linstedt, 2006). Golgi ribbon organization also depends on an intact micro-tubule and actin cytoskeleton, specialized cytoskeleton-based motors and membrane input from the endoplasmic reticulum (ER) (Marra et al., 2007; Rios & Bornens, 2003; Thyberg & Moskalewski, 1999), but these factors will not be discussed here in detail.
It is not completely understood why mammalian Golgi membranes in interphase are organized into stacks of cisternae and why these interconnected Golgi stacks are positioned in the pericentriolar region of the cell. It is unlikely that Golgi cisternae stacking is important for protein transport because the dispersed Golgi cisternae of S. cerevisiae are able to secrete efficiently (Papanikou & Glick, 2009). Interestingly, a recent study showed that loss of Golgi stacking leads to an increased rate of protein transport and defects in glycosylation (Xiang et al., 2013). Thus, cisternae stacking may increase the accuracy of protein glycosylation.
There is increasing support for the functional significance of the pericentriolar positioning of the Golgi apparatus. For example, pericentriolar Golgi membranes are proposed to control cell polarization and migration. Interfering with Golgi positioning by depleting Golgin-160 or GMAP210 had no effect on general protein transport to the cell surface, but there were defects in directional protein secretion, which is necessary for cell migration (Yadav et al., 2009). In addition, this specific Golgi position may facilitate functional Golgi–centrosome interactions, such as the regulation of centrosome organization and function from the Golgi (Kodani, Kristensen, Huang, & Sütterlin, 2009). Finally, Golgi fragmentation, which leads to a temporary loss of pericentriolar Golgi positioning, is required for entry into mitosis (Sütterlin, Hsu, Mallabiabarrena, & Malhotra, 2002). In conclusion, ribbon organization, cisternae stacking, and pericentriolar positioning of the Golgi complex may add another level of regulation for mammalian cell-specific processes.
23.2 Mechanism of Mitotic Golgi Disassembly
23.2.1 Severing the Golgi ribbon into stacks
In the first step of mitotic Golgi disassembly, the interconnected Golgi ribbon is converted into isolated stacks, which requires the cleavage of the membrane tubules of the noncompact zones (Fig. 23.1). While these morphological changes appear minor and can only be detected with sensitive approaches, such as FRAP and electron microscopy (Colanzi et al., 2007), they are of functional importance because they are necessary and sufficient for entry of cells into mitosis (Colanzi et al., 2007; Feinstein & Linstedt, 2007). The severing of Golgi stacks depends on at least four factors, which include the fission-inducing protein CtBP1-S/BARS (referred to as BARS) (Hidalgo Carcedo et al., 2004), the protein kinase MEK1 (Acharya, Mallabiabarrena, Acharya, & Malhotra, 1998; Feinstein & Linstedt, 2007), the peripheral Golgi proteins GRASP65 and GRASP55 (Sütterlin et al., 2002; Xiang & Wang, 2010), and the mitotic kinase Plk1 (Sütterlin et al., 2001). Each of these proteins has a specialized role in Golgi ribbon breakdown and their combined activities produce isolated stacks in G2. It is possible that these same factors also control the breakdown of isolated Golgi stacks into the “Golgi blobs,” which have been detected in metaphase (Fig. 23.1).
23.2.1.1 BARS
This membrane fission factor controls the disassembly of Golgi stacks by severing the tubular network of the noncompact zones (Colanzi et al., 2007). Interfering with BARS activity in a semi-intact Golgi fragmentation assay resulted in groups of large tubular–vesicular–saccular networks of Golgi membranes, which were continuous and localized in the pericentriolar region (Colanzi et al., 2007). Interestingly, BARS is required for several membrane-trafficking steps (Bonazzi et al., 2005; Valente et al., 2012; Yang et al., 2011), but Golgi membranes are only fragmented late in G2 indicating that BARS is specifically activated in G2 to promote Golgi ribbon severing. It is likely that this regulation involves the phosphorylation of BARS, which is a known substrate for several protein kinases (Liberali et al., 2008; Valente et al., 2012). BARS phosphorylation may facilitate the recruitment of specific, still unknown binding partners, which then mediate the cutting of the Golgi ribbon.
23.2.1.2 MEK1
The dual specificity kinase MEK1 controls Golgi disassembly by activating several regulatory kinases (Fig. 23.1). MEK1 is a central player of the MAP kinase signal transduction pathway, which normally functions as a tripartite signaling module (Robinson & Cobb, 1997). Loss of MEK1 activity prevented Golgi fragmentation in intact and semi-intact cells (Acharya et al., 1998; Feinstein & Linstedt, 2007).
Two MEK1 activators have been identified that function upstream of MEK1 in Golgi fragmentation. Raf1 is known to directly activate MEK1 and its activity was necessary for mitotic Golgi fragmentation and entry into mitosis (Colanzi, Sutterlin, & Malhotra, 2003). There is also an indirect role for PKD1 and PKD2 in the activation of Raf1/MEK1 in mitosis (Kienzle et al., 2013). Interfering with PKD activity blocked Raf1/MEK1 activation and prevented entry into mitosis. Loss of PKD activity was rescued by expression of active MEK1, indicating that these proteins function in the same regulatory pathway.
MEK1 regulates Golgi breakdown through several effectors. ERK1/2, the best characterized MEK1 substrate, controls mitotic Golgi breakdown through phosphor-ylation of GRASP55 (Feinstein & Linstedt, 2008). Plk3, another MEK1 substrate localizes to the spindle and the Golgi complex (Ruan et al., 2004), where it activates VRK1 through phosphorylation (Lopez-Sanchez, Sanz-Garcia, & Lazo, 2009). Interfering with VRK1 function prevented MEK1-induced Golgi breakdown (Lopez-Sanchez et al., 2009). The ERK1 splice variant ERK1c also associates with the Golgi complex, and its depletion reduced the efficiency of mitotic Golgi fragmentation and entry into mitosis (Shaul & Seger, 2006). As Plk3, VRK1, and ERK1c function downstream of MEK1, each of these components could be involved in the severing of the Golgi ribbon or the conversion of mini-stacks into “Golgi blobs” (Fig. 23.1); however, their targets and specific contributions to Golgi fragmentation remain to be determined.
Myt1 is an MEK1 effector that has been implicated in the control of mitotic Golgi fragmentation. This protein kinase, a known negative regulator of Cdk1 activity, associates with the Golgi complex and the endoplasmic reticulum (Liu, Stanton, Wu, & Piwnica-Worms, 1997). RNAi studies in Drosophila and HeLa cells have revealed a requirement for Myt1 in mitotic Golgi dynamics (Cornwell, Kaminski, & Jackson, 2002; Nakajima et al., 2008). In addition, a recent study reported that Myt1 depletion caused accelerated entry into mitosis, and there was a small but significant increase in cells that were in G2 and that had fragmented Golgi membranes (Villeneuve, Scarpa, Ortega-Bellido, & Malhotra, 2013). The idea that Myt1 function downstream of MEK1 was confirmed by the result that Myt1 is inactivated by MEK1-mediated phosphorylation and that its depletion was able to bypass a requirement for MEK1.
23.2.1.3 Golgi reassembly and stacking proteins
The two related Golgi reassembly and stacking proteins (GRASPs), GRASP65 and GRASP55, which localize to the cis- or medial/trans Golgi, respectively, are critical regulators of Golgi organization. Their depletion caused Golgi ribbon breakdown and a reduction in the number of cisternae per stack (Feinstein & Linstedt, 2008; Puthenveedu et al., 2006; Sütterlin, Polishchuk, Pecot, & Malhotra, 2005). Interfering with their functions in semi-intact cell assays or in intact cells affected mitotic Golgi fragmentation and mitotic entry (Duran et al., 2008; Preisinger et al., 2005; Sütterlin et al., 2002).
GRASPs have unique structural features with two N-terminal PDZ-like domains and a C-terminal serine/proline-rich regulatory domain (Vinke, Grieve, & Rabouille, 2011). Their Golgi localization is mediated by myristoylation of an N-terminal glycine and interaction with specific golgins (Vinke et al., 2011). GRASPs are phosphor-ylated in a cell cycle-controlled manner, with at least eight phosphorylation sites on GRASP65 and at least five sites on GRASP55 (Duran et al., 2008; Preisinger et al., 2005; Tang, Yuan, Vielemeyer, Perez, & Wang, 2012). These sites are modified by Cdk1 and Plk1, or ERK1/2 in the case of GRASP65 or GRASP55, respectively (Duran et al., 2008; Preisinger et al., 2005; Sengupta & Linstedt, 2010; Tang et al., 2012).
Cell cycle-regulated trans-oligomerization of GRASPs provides a possible mechanism for the control of Golgi reorganization during mitosis. In vitro binding and bead aggregation assays showed that GRASPs form homo-oligomers in trans through their N-terminal GRASP domain and that phosphorylation of specific residues within the C-terminal serine/proline-rich regulatory domain prevents these interactions (Tang, Yuan, & Wang, 2010). In addition, microinjection of GRASP65-specific antibodies into mitotic cells blocked Golgi stacks reformation after cell division (Wang, Seemann, Pypaert, Shorter, & Warren, 2003). These findings suggest that the trans-oligomerization of GRASPs promotes Golgi stacking and ribbon assembly in interphase, and that GRASP phosphorylation in mitosis disrupts these associations, inducing Golgi ribbon disassembly. While this oligomerization-based model can explain how GRASPs may regulate Golgi stack assembly and disassembly, it not clear whether GRASPs are long enough to link individual Golgi stacks into a ribbon.
Live imaging studies with fluorescently tagged GRASP55 support an additional role of GRASP55 in the stabilization of contacts between mini-stacks (Feinstein & Linstedt, 2008). Monitoring Golgi reassembly after nocodazole-induced Golgi fragmentation revealed GRASP55-GFP-positive membrane tubules that formed stable bridges between adjacent mini-stacks. Such bridges were rarely seen in the absence of GRASP55, indicating that GRASP55 may contribute to the formation of these stable contacts between mini-stacks. Thus, the assembly of the Golgi ribbon from dispersed Golgi fragments may require GRASP trans-oligomerization to form mini-stacks and GRASP55-mediated stabilization of tubular contacts to form the interconnected ribbon.
23.2.1.4 Plk1
Several studies have identified a requirement for mitotic kinase Plk1 for mitotic Golgi fragmentation (Sengupta & Linstedt, 2010; Sütterlin et al., 2001). Plk1 phosphorylates GRASP65 on serine 189 in the N-terminal domain that mediates trans-oligomerization (Truschel, Zhang, Bachert, Macbeth, & Linstedt, 2012). Expression of phospho-mimetic GRASP65 mutants prevented cisternae tethering and caused Golgi ribbon disassembly, while expression of nonphosphorylatable mutants prevented Golgi ribbon cleavage during mitosis (Sengupta & Linstedt, 2010). This study led to a model in which Cdk1 first phosphorylates the GRASP65 C-terminal domain, creating a docking site for Plk1. Plk1 then binds to GRASP65 and phosphorylates Ser189, which inhibits the trans-oligomerization properties of GRASP65. However, since the activity of Cdk1 in G2 is low, it is possible that a kinase other than Cdk1 phosphorylates GRASP65 to prime it for Plk1 binding. A likely candidate is the Raf1/MEK1/ERK1/2 pathway, which is active in G2 and which recognizes and modifies the same phosphorylation consensus site as Cdk1 (Yoshimura et al., 2005).
23.2.2 Disassembly of Golgi stacks into “blobs” and “haze”
In prophase, the isolated Golgi stacks that result from Golgi ribbon cleavage are further fragmented into “Golgi blobs” and “Golgi haze.” This additional fragmentation step depends on unstacking of cisternae to disassemble Golgi mini-stacks and vesiculation to fragment Golgi cisternae. Unstacking is mediated by Cdk1 and Plk1, which phosphorylate GRASP65 and thereby disrupt its ability to form trans-oligomers. Once unstacked, Golgi cisternae are broken down by extensive vesiculation through the continuous budding of COPI-coated vesicles (Misteli & Warren, 1994b), and this process is facilitated by the reported block in transport from the ER and inhibition of vesicle fusion with the Golgi complex. The mechanism behind this transport block is incompletely understood, but it may involve the phosphorylation of components of the ER-to-Golgi transport machinery, such as the ARF exchange factor GBF1 (Morohashi, Balklava, Ball, Hughes, & Lowe, 2010). In summary, cell cycle-dependent Golgi organization appears to be controlled by equilibrium between disassembly and reassembly factors. How the functions of these maintenance factors and BARS are coordinated remains to be addressed in future studies.
23.3 Mechanism of Postmitotic Golgi Reassembly
In vitro reconstitution assays have provided great insight into the mechanism of post-mitotic Golgi reassembly. Two AAA ATPases, NSF and p97 (also called VCP for valosin-containing protein), were found to be necessary for the reassembly of small vesicular Golgi elements into short cisternae (Acharya, McCaffery, Jacobs, & Malhotra, 1995; Rabouille, Levine, Peters, & Warren, 1995). Both ATPases utilize specific cofactors, with SNAPs and p115 promoting NSF-mediate fusion, and complexes of p97 with p47 and VCIP135, or with p37 controlling p97 function in Golgi reformation (Kondo et al., 1997; Uchiyama et al., 2006). NSF-mediated fusion is proposed to produce larger vesicles and tubular–reticular elements, which are then fused by a p97-mediatedprocesstogeneratecisternae.Inmitosis,p47,p37,andVCIP135arephosphorylated by Cdk1, which blocks p97-mediated membrane fusion so that Golgi membranes remain disassembled (Kaneko, Tamura, Totsukawa, & Kondo, 2010; Totsukawa, Matsuo, Kubota, Taguchi, & Kondo, 2013; Uchiyama et al., 2003). Thus, mitotic phosphorylation of the membrane fusion machinery can explain the mitotic Golgi phenotype of dispersed tubular–reticular membranes and vesicles in the cytosol (Fig. 23.1).
In addition to phosphorylation, ubiquitination plays an important role in mitotic Golgi reassembly.The p97 co-factor p47 was found to contain a UBA ubiquitin-binding domain. Upon association with p97, p47 was able to bind ubiquitinated proteins via this UBA domain, and this domain was necessary for Golgi reassembly (Meyer, Shorter, Seemann, Pappin, & Warren, 2000). In addition, VCIP135 was shown to possess deubiquitinase activity, which was necessary for Golgi reassembly in vitro and in intact cells (Wang, Satoh, Warren, & Meyer, 2004). Further support for a role of a ubiquitination–deubiquination cycle in mitotic Golgi dynamics comes from the identification of HACE1, a Golgi-associated ubiquitin ligase, whose activity is required for mitotic Golgi dynamics (Tang et al., 2011). It will be interesting to identify the proteins that are ubiquitinated during Golgi disassembly and deubiquitinated during reassembly.
23.4 The Significance of Golgi Reorganization in Mitosis
Golgi ribbon severing
Although many mechanistic aspects of mitotic Golgi disassembly remain to be defined, it has become clear that entry of cells into mitosis depends on the ability of the Golgi complex to fragment (Hidalgo Carcedo et al., 2004; Sütterlin et al., 2002). This conclusion is based on microinjection and overexpression studies in synchronized cells and implies that Golgi organization and cell cycle progression are tightly linked.
It is incompletely understood how a block in Golgi fragmentation prevents cell cycle progression. As the block in Golgi fragmentation induces a G2 arrest, it is likely to involve cyclin B-dependent kinase 1 (CycB-Cdk1) because this protein kinase is the major regulator of the G2/M transition (Nigg, 2001). The activity of CycB1-Cdk1 activity is known to be controlled by the recruitment and activation of the Ser/Thr kinase Aurora-A at the centrosome (Marumoto et al., 2002). Interestingly, a block in Golgi fragmentation interfered with Aurora-A recruitment to the centrosome in G2 and prevented its activation (Persico, Cervigni, Barretta, Corda, & Colanzi, 2010). This link between Golgi organization in G2 and Aurora-A recruitment and activation provides a first mechanistic insight into how Golgi dynamics may be coordinated with cell-cycle progression.
However, a second cyclin B-dependent, but Aurora-independent mechanism may also contribute to the Golgi-mediated control of mitotic entry. Mammalian cells express two B-type cyclins, CycB1 and CycB2, of which CycB2 associates with the Golgi complex and the ER (Jackman, Firth, & Pines, 1995). Both isoforms cooperate to promote mitotic entry. It is possible that the activation of the Golgi-associated CycB2-Cdk1 complex is regulated by the organization state of the Golgi complex so that impaired Golgi fragmentation could prevent the activation of this central mitotic regulator. In conclusion, Golgi fragmentation may control mitotic entry by simultaneously activating the Aurora-A/Cdk1 pathway as well as the Golgi-associated CycB2-Cdk1 complex.
23.4.2 Golgi stack fragmentation
The reason for Golgi stack disassembly is only beginning to being understood. This process does not seem to be necessary for mitotic progression because cell types that lack a ribbon-like Golgi structure, such plant cells or Drosophila embryos, undergo cell division without breakdown of their stack. In addition, accurate partitioning of an organelle can be achieved without complete fragmentation of this organelle, as has been seen for the ER (Terasaki, 2000). Therefore, the disassembly of isolated stacks into the Golgi haze may achieve goals that are not directly linked to mitotic entry. For example, disassembly of Golgi stacks has been found to lead to inactivation of the small GTPase Arf1 and release of a set of peripheral Golgi proteins (Altan-Bonnet et al., 2006). When Arf1 was kept in an active state, stack disassembly was blocked and cells entered mitosis, but there were severe defects in chromosome segregation and cytokinetic furrow ingression (Altan-Bonnet et al., 2006). Additional Golgi proteins dissociate from the Golgi complex during mitosis to complete Golgi-independent functions (Table 23.1). For instance, the Golgi-associated protein p115 was found to dissociate from the Golgi during mitosis and to partition with the poles of the mitotic spindle, where it controlled spindle formation, chromosome segregation and cytokinesis (Radulescu et al., 2011). Similarly, Miki loses its Golgi association during mitosis and localizes to centrosome to control the recruitment of CG-NAP, a scaffold protein of the g-tubulin ring complex (Ozaki et al., 2012). While Miki relocalization is induced by tankyrase-mediate PARsylation, it cannot be excluded that Golgi fragmentation contributes to Miki dispersal. Thus, complete Golgi vesiculation may function as a regulatory factor in the distribution of these proteins, and it will be interesting to examine whether blocking Golgi “blob” or “haze” formation interferes with the function of these proteins
Table 23.1.
Golgi-associated proteins that acquire new location and function during mitosis
| Protein name | Mitotic localization | Mitotic function | Stimulus for relocalization | Upstream regulator | References |
|---|---|---|---|---|---|
| Arf1 | Cytosol | Unknown | Unknown | Unknown | Altan-Bonnet et al. (2006) |
| p115 | Spindle | Spindle formation/chromosome segregation | Unknown | Unknown | Radulescu, Mukherjee, and Shields (2011) |
| Miki | Centrosome | Spindle formation/chromosome segregation | PARsylation | Tankyrase1 | Ozaki et al. (2012) |
| KBTBD8 | Spindle | Unknown | Unknown | Unknown | Luhrig, Kolb, Mellies, and Nolte (2013) |
| CtBP | Centrosome | Unknown | Unknown | Unknown | Spyer and Allday (2006) |
| Rab6A | Unknown | Dynein/dynactin complex at kinetochores | Unknown | Unknown | Miserey-Lenkei et al. (2006) |
| Nir2 | Cleavage forrow and midbody | Plk1 recruitment | Phosphorylation | Cdk1 | Litvak et al. (2004) |
| ACBD3 | Cytosol | Regulation of Numb signaling | Unknown | Unknown | Zhou et al. (2007) |
| Zw10 | Kinetochores | Mitotic checkpoint | Unknown | Unknown | Vallee, Varma, and Dujardin (2006) |
23.5 Assays to Study Mitotic Golgi Reorganization
23.5.1 Semi-intact cell assay to study mitotic Golgi fragmentation (Acharya et al., 1998)
23.5.1.1 Mitotic cytosol (Nakagawa, Kitten, & Nigg, 1989)
Grow NRK or HeLa cells in 15 cm petri dishes (70% confluency).
Arrest cells in S-phase with 2 mM thymidine for 10–12 h.
Wash cells to remove thymidine and incubate with 500 ng/ml nocodazole for 10–12 h to arrest cells in metaphase.
Harvest mitotic cells by “shake off.”
Wash mitotic cells with cold PBS and with mitotic extract buffer (MEB: 15 mM PIPES (pH 7.4), 50 mM KCl, 10 mM MgCl2, 20 mM β-mercaptoethanol, 20 mM β-glycerophosphate, 15 mM EGTA, 0.5 mM spermidine, 0.2 mM spermine, 1 mM DTT, 0.1 mM PMSF, 0.2 μg/ml aprotinin, 0.2 μg/ml leupeptin, and 0.2 μg/ml pepstatin).
Resuspend cell pellet in twice the packed cell volume in MEB and allow to swell for 10 min on ice.
Homogenize by passage through an 18-gauge needle and lyse by repeated passages through a 24-gauge needle. Monitor lysis state under the light microscope.
Centrifuge lysate in a table top ultracentrifuge at 48,000 rpm for 45 min using a TLS55 rotor.
Separate high-speed supernatant (approximate protein concentration of 10–12 mg/ml) and freeze as “mitotic extract” in aliquots in liquid nitrogen.
Store at –80 °C.
23.5.1.2 Interphase cytosol
As for mitotic cells, but asynchronous cells were harvested by scraping.
23.5.1.3 Selective permeabilization of tissue culture cells
Grow cells (NRK or HeLa) on coverslips (60% confluency) and incubate with 2 mM thymidine for 12–16 h.
Wash with KHM buffer (25 mM HEPES-KOH [pH 7.2], 125 mM potassium acetate, 2.5 mM magnesium acetate) at room temperature.
Move dish to ice and wash with cold KHM buffer.
Permeabilize plasma membrane by treating cells with 30 μg/ml digitonin in cold KHM for 3–5 min—The exact timing of digitonin treatment has to be determined using antibody staining in the absence of detergent.
Stop permeabilization by washing with cold KHM.
Incubate permeabilized cells with cold KHM, 1 M KCL to remove peripheral membrane proteins for 5 min.
Allow permeabilized cells to warm up during the last 2 min of the salt wash (step 6).
Wash with KHM buffer at RT.
23.5.1.4 Semi-intact cell assay
Prepare 50 μl incubation mixture: 7 mg/ml cytosol, ATP-regeneration system (final concentration 0.5 mM ATP, 0.3 mM UTP, 10 mM creatine phosphate, and 12 IU/ml creatine kinase), and purified recombinant proteins to be tested.
Place on parafilm in a 10 cm tissue culture dish.
Inverse coverslip with permeabilized cells onto reaction mixture and incubate for 60 min at 32 °C.
Fix and analyze by immunofluorescence.
23.5.2 Cell-free assays to study mitotic Golgi fragmentation
(Misteli & Warren, 1994a; Rabouille et al., 1995)
Golgi disassembly and reassembly have been reconstituted in an in vitro assay using Golgi membranes, and mitotic cytosol or purified components. The extent of Golgi fragmentation is evaluated by electron microscopy. As excellent reviews describing this assay have been published, we refer to these for more details (Tang, Xiang, & Wang, 2010; Warren, Levine, & Misteli, 1995).
23.5.3 Microinjection assay in intact cells to study mitotic Golgi fragmentation
Persico et al., 2010; Sütterlin et al., 2002)
23.5.3.1 Cell cycle synchronization of NRK cells and microinjection
Grow NRK cells on fibronectin-coated glass coverslips (60% confluency).
Synchronize cells at the G1/S transition by treating them with 2.5 μg/ml aphidicolin in complete medium for 16 h.
Remove aphidicolin by washing three times 3 min with warm complete medium.
30 min after aphidicolin removal, inject 200 cells with purified recombinant proteins or antibodies (8–12 mg/ml) or Dextran-FITC as a negative control, using an Eppendorf 5246 transjector (Eppendorf, Milan, Italy).
Incubate for 7–8 h, fix, and process for immunofluorescence with antibodies to phospho-Histone H3 to determine mitotic index and with antibodies to detect the injected antibodies or proteins.
Concluding Remarks
The initial observation that Golgi membranes in mammalian cells undergo regulated reorganization in mitosis was made more than three decades ago and has stimulated many studies on the mechanisms of Golgi biogenesis and maintenance. About 10 years ago, these investigations led to the discovery that mitotic Golgi fragmentation is required for entry of cells into mitosis, implying that cell-cycle progression depends not only on the state of the DNA, but also on the organization of an organelle. However, many questions on the link between organelle structure and cell-cycle regulation have remained unanswered. For example, we are only beginning to understand why Golgi membranes undergo such extensive reorganization during mitosis, and why they are positioned next to the centrosome in interphase. This incomplete insight into this process is due to the overall complexity of the process, but also to a lack of appropriate methods. An effective protocol to block the cleavage of the Golgi ribbon is the microinjection of “blocking reagents” (antibodies, recombinant proteins, and peptides), which target factors involved in the process. As a result, there is an acute block of Golgi fragmentation, which then induces a potent and prolonged G2 arrest. However, this approach is limited to the observation of single cells by immunofluorescence and is not amenable to unbiased screens for additional components. Other treatments (kinase inhibitors, RNAi, or protein overexpression) reach every cell within a population, but appear to produce effects that are in general smaller so that results may be difficult to interpret. Thus, it will be essential to develop methods to rapidly inactivate the function of Golgi organization regulators in an entire cell population. Such approaches will help understand the timing and temporal order in which regulatory kinases modify their targets to induce a fragmented Golgi, and to identify regulatory networks involved in this process through global investigations (proteomics, phosphoproteomic).
Acknowledgments
We apologize to those colleagues whose work we were not able to discuss due to space limitation. A. C. acknowledges the Italian Association for Cancer Research (AIRC, Milan, Italy; IG6074) for financial support. C. S. is supported by a grant from the National Institutes of Health (R01 GM089913).
References
- Acharya U, Mallabiabarrena A, Acharya JK, Malhotra V. Signaling via mitogen-activated protein kinase kinase (MEK1) is required for Golgi fragmentation during mitosis. Cell. 1998;92:183–192. doi: 10.1016/s0092-8674(00)80913-7. [DOI] [PubMed] [Google Scholar]
- Acharya U, McCaffery JM, Jacobs R, Malhotra V. Reconstitution of vesiculated Golgi membranes into stacks of cisternae: Requirement of NSF in stack formation. Journal of Cell Biology. 1995;129:577–589. doi: 10.1083/jcb.129.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altan-Bonnet N, Sougrat R, Liu W, Snapp EL, Ward T, Lippincott-Schwartz J. Golgi inheritance in mammalian cells is mediated through endoplasmic reticulum export activities. Molecular Biology of the Cell. 2006;17:990–1005. doi: 10.1091/mbc.E05-02-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonazzi M, Spano S, Turacchio G, Cericola C, Valente C, Colanzi A, et al. CtBP3/BARS drives membrane fission in dynamin-independent transport pathways. Nature Cell Biology. 2005;7:570–580. doi: 10.1038/ncb1260. [DOI] [PubMed] [Google Scholar]
- Cancino J, Luini A. Signaling circuits on the Golgi complex. Traffic. 2013;14:121–134. doi: 10.1111/tra.12022. [DOI] [PubMed] [Google Scholar]
- Colanzi A, Hidalgo Carcedo C, Persico A, Cericola C, Turacchio G, Bonazzi M, et al. The Golgi mitotic checkpoint is controlled by BARS-dependent fission of the Golgi ribbon into separate stacks in G2. The EMBO Journal. 2007;26:2465–2476. doi: 10.1038/sj.emboj.7601686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colanzi A, Suetterlin C, Malhotra V. Cell-cycle-specific Golgi fragmentation: How and why? Current Opinion in Cell Biology. 2003;15:462–467. doi: 10.1016/s0955-0674(03)00067-x. [DOI] [PubMed] [Google Scholar]
- Colanzi A, Sutterlin C, Malhotra V. RAF1-activated MEK1 is found on the Golgi apparatus in late prophase and is required for Golgi complex fragmentation in mitosis. Journal of Cell Biology. 2003;161:27–32. doi: 10.1083/jcb.200208099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornwell WD, Kaminski PJ, Jackson JR. Identification of Drosophila Myt1 kinase and its role in Golgi during mitosis. Cellular Signalling. 2002;14:467–476. doi: 10.1016/s0898-6568(01)00276-5. [DOI] [PubMed] [Google Scholar]
- Duran JM, Kinseth M, Bossard C, Rose DW, Polishchuk R, Wu CC, et al. The role of GRASP55 in Golgi fragmentation and entry of cells into mitosis. Molecular Biology of the Cell. 2008;19:2579–2587. doi: 10.1091/mbc.E07-10-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein TN, Linstedt AD. Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition. Molecular Biology of the Cell. 2007;18:594–604. doi: 10.1091/mbc.E06-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein TN, Linstedt AD. GRASP55 regulates Golgi ribbon formation. Molecular Biology of the Cell. 2008;19:2696–2707. doi: 10.1091/mbc.E07-11-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaietta GM, Giepmans BN, Deerinck TJ, Smith WB, Ngan L, Llopis J, et al. Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17777–17782. doi: 10.1073/pnas.0608509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo Carcedo C, Bonazzi M, Spano S, Turacchio G, Colanzi A, Luini A, et al. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science. 2004;305:93–96. doi: 10.1126/science.1097775. [DOI] [PubMed] [Google Scholar]
- Jackman M, Firth M, Pines J. Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. The EMBO Journal. 1995;14:1646–1654. doi: 10.1002/j.1460-2075.1995.tb07153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Tamura K, Totsukawa G, Kondo H. Phosphorylation of p37 is important for Golgi disassembly at mitosis. Biochemical and Biophysical Research Communications. 2010;402:37–41. doi: 10.1016/j.bbrc.2010.09.097. [DOI] [PubMed] [Google Scholar]
- Kienzle C, Eisler SA, Villeneuve J, Brummer T, Olayioye MA, Hausser A. PKD controls mitotic Golgi complex fragmentation through a Raf-MEK1 pathway. Molecular Biology of the Cell. 2013;24:222–233. doi: 10.1091/mbc.E12-03-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A, Kristensen I, Huang L, Sütterlin C. GM130-dependent control of Cdc42 activity at the Golgi regulates centrosome organization. Molecular Biology of the Cell. 2009;20:1192–1200. doi: 10.1091/mbc.E08-08-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H, Rabouille C, Newman R, Levine TP, Pappin D, Freemont P, et al. p47 is a cofactor for p97-mediated membrane fusion. Nature. 1997;388:75–78. doi: 10.1038/40411. [DOI] [PubMed] [Google Scholar]
- Liberali P, Kakkonen E, Turacchio G, Valente C, Spaar A, Perinetti G, et al. The closure of Pak1-dependent macropinosomes requires the phosphorylation of CtBP1/BARS. The EMBO Journal. 2008;27:970–981. doi: 10.1038/emboj.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V, Argov R, Dahan N, Ramachandran S, Amarilio R, Shainskaya A, et al. Mitotic phosphorylation of the peripheral Golgi protein Nir2 by Cdk1 provides a docking mechanism for Plk1 and affects cytokinesis completion. Molecular Cell. 2004;14:319–330. doi: 10.1016/s1097-2765(04)00214-x. [DOI] [PubMed] [Google Scholar]
- Liu F, Stanton JJ, Wu Z, Piwnica-Worms H. The human Myt1 kinase preferentially phosphorylates Cdc2 on threonine 14 and localizes to the endoplasmic reticulum and Golgi complex. Molecular and Cellular Biology. 1997;17:571–583. doi: 10.1128/mcb.17.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez I, Sanz-Garcia M, Lazo PA. Plk3 interacts with and specifically phosphorylates VRK1 in Ser342, a downstream target in a pathway that induces Golgi fragmentation. Molecular and Cellular Biology. 2009;29:1189–1201. doi: 10.1128/MCB.01341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhrig S, Kolb S, Mellies N, Nolte J. The novel BTB-kelch protein, KBTBD8, is located in the Golgi apparatus and translocates to the spindle apparatus during mitosis. Cell Division. 2013;8:3. doi: 10.1186/1747-1028-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia AR, Zhu X, Miller P, Gu G, Maiato H, Kaverina I. Modulation of Golgi-associated microtubule nucleation throughout the cell cycle. Cytoskeleton. 2013;70:32–43. doi: 10.1002/cm.21079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra P, Salvatore L, Mironov A, Jr., Di Campli A, Di Tullio G, Trucco A, et al. The biogenesis of the Golgi ribbon: The roles of membrane input from the ER and of GM130. Molecular Biology of the Cell. 2007;18:1595–1608. doi: 10.1091/mbc.E06-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumoto T, Hirota T, Morisaki T, Kunitoku N, Zhang D, Ichikawa Y, et al. Roles of aurora-A kinase in mitotic entry and G2 checkpoint in mammalian cells. Genes to Cells. 2002;7:1173–1182. doi: 10.1046/j.1365-2443.2002.00592.x. [DOI] [PubMed] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. The EMBO Journal. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miserey-Lenkei S, Couedel-Courteille A, Del Nery E, Bardin S, Piel M, Racine V, et al. A role for the Rab6A’ GTPase in the inactivation of the Mad2-spindle checkpoint. The EMBO Journal. 2006;25:278–289. doi: 10.1038/sj.emboj.7600929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Warren G. COP-coated vesicles are involved in the mitotic fragmentation of Golgi stacks in a cell-free system. Journal of Cell Biology. 1994;125:269–282. doi: 10.1083/jcb.125.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Warren G. Mitotic disassembly of the Golgi apparatus in vivo. Journal of Cell Science. 1995;108(Pt. 7):2715–2727. doi: 10.1242/jcs.108.7.2715. [DOI] [PubMed] [Google Scholar]
- Morohashi Y, Balklava Z, Ball M, Hughes H, Lowe M. Phosphorylation and membrane dissociation of the ARF exchange factor GBF1 in mitosis. Biochemical Journal. 2010;427:401–412. doi: 10.1042/BJ20091681. [DOI] [PubMed] [Google Scholar]
- Munro S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harbor Perspectives in Biology. 2011;3:a005256. doi: 10.1101/cshperspect.a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa J, Kitten GT, Nigg EA. A somatic cell-derived system for studying both early and late mitotic events in vitro. Journal of Cell Science. 1989;94(Pt. 3):449–462. doi: 10.1242/jcs.94.3.449. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Yonemura S, Murata M, Nakamura N, Piwnica-Worms H, Nishida E. Myt1 protein kinase is essential for Golgi and ER assembly during mitotic exit. Journal of Cell Biology. 2008;181:89–103. doi: 10.1083/jcb.200708176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ. W(h)ither the Golgi during mitosis? Journal of Cell Biology. 2000;149:243–248. doi: 10.1083/jcb.149.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nature Reviews. Molecular Cell Biology. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- Ozaki Y, Matsui H, Asou H, Nagamachi A, Aki D, Honda H, et al. Poly-ADP ribosylation of Miki by tankyrase-1 promotes centrosome maturation. Molecular Cell. 2012;47:694–706. doi: 10.1016/j.molcel.2012.06.033. [DOI] [PubMed] [Google Scholar]
- Papanikou E, Glick BS. The yeast Golgi apparatus: Insights and mysteries. FEBS Letters. 2009;583:3746–3751. doi: 10.1016/j.febslet.2009.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, Stern CA, Pypaert M, Sheff D, Ngo HM, Roper N, et al. Golgi biogenesis in Toxoplasma gondii. Nature. 2002;418:548–552. doi: 10.1038/nature00946. [DOI] [PubMed] [Google Scholar]
- Persico A, Cervigni RI, Barretta ML, Corda D, Colanzi A. Golgi partitioning controls mitotic entry through Aurora-A kinase. Molecular Biology of the Cell. 2010;21:3708–3721. doi: 10.1091/mbc.E10-03-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisinger C, Korner R, Wind M, Lehmann WD, Kopajtich R, Barr FA. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. The EMBO Journal. 2005;24:753–765. doi: 10.1038/sj.emboj.7600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D, Mulholland J, Franzusoff A, Segev N, Botstein D. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Molecular Biology of the Cell. 1992;3:789–803. doi: 10.1091/mbc.3.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nature Cell Biology. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Radulescu AE, Mukherjee S, Shields D. The Golgi protein p115 associates with gamma-tubulin and plays a role in Golgi structure and mitosis progression. Journal of Biological Chemistry. 2011;286:21915–21926. doi: 10.1074/jbc.M110.209460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios RM, Bornens M. The Golgi apparatus at the cell centre. Current Opinion in Cell Biology. 2003;15:60–66. doi: 10.1016/s0955-0674(02)00013-3. [DOI] [PubMed] [Google Scholar]
- Robinson MJ, Cobb MH. Mitogen-activated protein kinase pathways. Current Opinion in Cell Biology. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- Ruan Q, Wang Q, Xie S, Fang Y, Darzynkiewicz Z, Guan K, et al. Polo-like kinase 3 is Golgi localized and involved in regulating Golgi fragmentation during the cell cycle. Experimental Cell Research. 2004;294:51–59. doi: 10.1016/j.yexcr.2003.10.022. [DOI] [PubMed] [Google Scholar]
- Sengupta D, Linstedt AD. Mitotic inhibition of GRASP65 organelle tethering involves Polo-like kinase 1 (PLK1) phosphorylation proximate to an internal PDZ ligand. Journal of Biological Chemistry. 2010;285:39994–40003. doi: 10.1074/jbc.M110.189449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul YD, Seger R. ERK1c regulates Golgi fragmentation during mitosis. Journal of Cell Biology. 2006;172:885–897. doi: 10.1083/jcb.200509063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyer M, Allday MJ. The transcriptional co-repressor C-terminal binding protein (CtBP) associates with centrosomes during mitosis. Cell Cycle. 2006;5:530–537. doi: 10.4161/cc.5.5.2524. [DOI] [PubMed] [Google Scholar]
- Sütterlin C, Colanzi A. The Golgi and the centrosome: Building a functional partnership. Journal of Cell Biology. 2010;188:621–628. doi: 10.1083/jcb.200910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sütterlin C, Hsu P, Mallabiabarrena A, Malhotra V. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 2002;109:359–369. doi: 10.1016/s0092-8674(02)00720-1. [DOI] [PubMed] [Google Scholar]
- Sütterlin C, Lin CY, Feng Y, Ferris DK, Erikson RL, Malhotra V. Polo-like kinase is required for the fragmentation of pericentriolar Golgi stacks during mitosis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9128–9132. doi: 10.1073/pnas.161283998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sütterlin C, Polishchuk R, Pecot M, Malhotra V. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Molecular Biology of the Cell. 2005;16:3211–3222. doi: 10.1091/mbc.E04-12-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Xiang Y, De Renzis S, Rink J, Zheng G, Zerial M, et al. The ubiquitin ligase HACE1 regulates Golgi membrane dynamics during the cell cycle. Nature Communications. 2011;2:501. doi: 10.1038/ncomms1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Xiang Y, Wang Y. Reconstitution of the cell cycle-regulated Golgi disassembly and reassembly in a cell-free system. Nature Protocols. 2010;5:758–772. doi: 10.1038/nprot.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Yuan H, Vielemeyer O, Perez F, Wang Y. Sequential phosphorylation of GRASP65 during mitotic Golgi disassembly. Biology Open. 2012;1:1204–1214. doi: 10.1242/bio.20122659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Yuan H, Wang Y. The role of GRASP65 in Golgi cisternal stacking and cell cycle progression. Traffic. 2010;11:827–842. doi: 10.1111/j.1600-0854.2010.01055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M. Dynamics of the endoplasmic reticulum and golgi apparatus during early sea urchin development. Molecular Biology of the Cell. 2000;11:897–914. doi: 10.1091/mbc.11.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S. Role of microtubules in the organization of the Golgi complex. Experimental Cell Research. 1999;246:263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]
- Totsukawa G, Matsuo A, Kubota A, Taguchi Y, Kondo H. Mitotic phosphor-ylation of VCIP135 blocks p97ATPase-mediated Golgi membrane fusion. Biochemical and Biophysical Research Communications. 2013;433:237–242. doi: 10.1016/j.bbrc.2013.02.090. [DOI] [PubMed] [Google Scholar]
- Truschel ST, Zhang M, Bachert C, Macbeth MR, Linstedt AD. Allosteric regulation of GRASP protein-dependent Golgi membrane tethering by mitotic phosphor-ylation. Journal of Biological Chemistry. 2012;287:19870–19875. doi: 10.1074/jbc.M111.326256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Jokitalo E, Lindman M, Jackman M, Kano F, Murata M, et al. The localization and phosphorylation of p47 are important for Golgi disassembly-assembly during the cell cycle. Journal of Cell Biology. 2003;161:1067–1079. doi: 10.1083/jcb.200303048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Totsukawa G, Puhka M, Kaneko Y, Jokitalo E, Dreveny I, et al. p37 is a p97 adaptor required for Golgi and ER biogenesis in interphase and at the end of mitosis. Developmental Cell. 2006;11:803–816. doi: 10.1016/j.devcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Valente C, Turacchio G, Mariggio S, Pagliuso A, Gaibisso R, Di Tullio G, et al. A 14-3-3gamma dimer-based scaffold bridges CtBP1-S/BARS to PI(4)KIIIbeta to regulate post-Golgi carrier formation. Nature Cell Biology. 2012;14:343–354. doi: 10.1038/ncb2445. [DOI] [PubMed] [Google Scholar]
- Vallee RB, Varma D, Dujardin DL. ZW10 function in mitotic checkpoint control, dynein targeting and membrane trafficking: Is dynein the unifying theme? Cell Cycle. 2006;5:2447–2451. doi: 10.4161/cc.5.21.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve J, Scarpa M, Ortega-Bellido M, Malhotra V. MEK1 inactivates Myt1 to regulate Golgi membrane fragmentation and mitotic entry in mammalian cells. The EMBO Journal. 2013;32:72–85. doi: 10.1038/emboj.2012.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinke FP, Grieve AG, Rabouille C. The multiple facets of the Golgi reassembly stacking proteins. Biochemical Journal. 2011;433:423–433. doi: 10.1042/BJ20101540. [DOI] [PubMed] [Google Scholar]
- Wang Y, Satoh A, Warren G, Meyer HH. VCIP135 acts as a deubiquitinating enzyme during p97-p47-mediated reassembly of mitotic Golgi fragments. Journal of Cell Biology. 2004;164:973–978. doi: 10.1083/jcb.200401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Seemann J, Pypaert M, Shorter J, Warren G. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. The EMBO Journal. 2003;22:3279–3290. doi: 10.1093/emboj/cdg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G, Levine T, Misteli T. Mitotic disassembly of the mammalian Golgi apparatus. Trends in Cell Biology. 1995;5:413–416. doi: 10.1016/s0962-8924(00)89094-7. [DOI] [PubMed] [Google Scholar]
- Wilson C, Venditti R, Rega LR, Colanzi A, D’Angelo G, De Matteis MA. The Golgi apparatus: An organelle with multiple complex functions. Biochemical Journal. 2011;433:1–9. doi: 10.1042/BJ20101058. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Wang Y. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. Journal of Cell Biology. 2010;188:237–251. doi: 10.1083/jcb.200907132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang X, Nix DB, Katoh T, Aoki K, Tiemeyer M, et al. Regulation of protein glycosylation and sorting by the Golgi matrix proteins GRASP55/65. Nature Communications. 2013;4:1659. doi: 10.1038/ncomms2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Puri S, Linstedt AD. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Molecular Biology of the Cell. 2009;20:1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Valente C, Polishchuk RS, Turacchio G, Layre E, Moody DB, et al. COPI acts in both vesicular and tubular transport. Nature Cell Biology. 2011;13:996–1003. doi: 10.1038/ncb2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeong FM. Multi-step down-regulation of the secretory pathway in mitosis: A fresh perspective on protein trafficking. BioEssays: News and Reviews in Molecular. Cellular and Developmental Biology. 2013;35:462–471. doi: 10.1002/bies.201200144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Yoshioka K, Barr FA, Lowe M, Nakayama K, Ohkuma S, et al. Convergence of cell cycle regulation and growth factor signals on GRASP65. Journal of Biological Chemistry. 2005;280:23048–23056. doi: 10.1074/jbc.M502442200. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Atkins JB, Rompani SB, Bancescu DL, Petersen PH, Tang H, et al. The mammalian Golgi regulates numb signaling in asymmetric cell division by releasing ACBD3 during mitosis. Cell. 2007;129:163–178. doi: 10.1016/j.cell.2007.02.037. [DOI] [PubMed] [Google Scholar]