SUMMARY
The low complexity (LC) domains of the products of the fused in sarcoma (FUS), Ewings sarcoma (EWS) and TAF15 genes are translocated onto a variety of different DNA-binding domains and thereby assist in driving the formation of cancerous cells. In the context of the translocated fusion proteins, these LC sequences function as transcriptional activation domains. Here we show that polymeric fibers formed from these LC domains directly bind the C-terminal domain (CTD) of RNA polymerase II in a manner reversible by phosphorylation of the iterated, heptad repeats of the CTD. Mutational analysis indicates that the degree of binding between the CTD and the LC domain polymers correlates with the strength of transcriptional activation. These studies offer a simple means of conceptualizing how RNA polymerase II is recruited to active genes in its unphosphorylated state, and released for elongation following phosphorylation of the CTD.
INTRODUCTION
Numerous forms of cancer result from translocation events wherein the amino-terminal, low complexity (LC) domains of any of three related RNA binding proteins become fused to a variety of different DNA binding domains (Arvand and Denny, 2001; Guipaud et al., 2006; Lessnick and Ladanyi, 2012). The relevant RNA binding proteins include the products of the fused in sarcoma (FUS), Ewings sarcoma (EWS) and TAF15 genes. Collectively these three RNA-binding proteins are referred to as FET (FUS/EWS/TAF15) proteins (Andersson et al., 2008). The proteins encoded by these varied translocation products are causative of transformation (Bertolotti et al., 1999; Crozat et al., 1993; Ichikawa et al., 1999; Rabbitts et al., 1993; Zinszner et al., 1994). They further display dual dependency upon both the DNA-binding domain, which can be represented by members of the homeobox, zinc finger, ETS, or leucine zipper families of DNA-binding domains, as well as the LC domains donated by FUS, EWS or TAF15. The DNA binding domains are understood to direct the cancer-causing fusion proteins to appropriate batteries of genes adequate to facilitate cell growth or survival. By contrast, the LC sequences donated by members of the FET family are understood to function as transcriptional activation domains.
Over the past several decades the fields of biochemistry, biophysics and molecular biology have achieved a concrete understanding of how DNA-binding domains function. The atomic structures of many such domains have been resolved by either X-ray crystallography or NMR spectroscopy, often in complex with their specific DNA substrates. By contrast, far less is known about the manner in which transcriptional activation domains operate at a mechanistic level. Prototypic activation domains are comprised of low complexity sequences that exist in an intrinsically disordered, random coil conformation (Huntley and Golding, 2002; Uversky, 2002). The observation that evolutionary pressure of cancer cell formation has repeatedly led to the selection of the LC domains of FET proteins as the fusion partner to a variety of different DNA binding domains strongly hints that these particular LC sequences may be exceptionally potent transcriptional activation domains.
The LC domain of FUS donated to the translocation product causative of cancer has a highly skewed distribution of amino acids. Of the 220 residues within the FUS LC domain, 84% are comprised of only four amino acids – glycine, serine, glutamine and tyrosine. The domain contains zero representatives of glutamic acid, lysine, arginine, cysteine, histidine, valine, leucine, isoleucine, tryptophan or phenylalanine. By having an amino acid composition dominated by only a four letter code (G, S, Q and Y), the LC domain of FUS would appear to be more like nucleic acids than typical proteins that fold into their ultimate, three-dimensional shape by use of a much wider reliance on all 20 types of amino acid residues. When incubated at high concentrations, the LC domain of FUS has been found to polymerize into uniform, amyloid-like fibers (Han et al., 2012; Kato et al., 2012). Although morphologically similar to pathogenic amyloid aggregates, polymeric fibers formed from the LC domain of FUS are labile to depolymerization, raising the possibility that reversible LC polymerization may engender functional utility. Given two measurable features of the LC domain of FET proteins (transcriptional activation capacity and polymerization propensity), we hereby describe experiments that test the correlative relationship between the two.
RESULTS
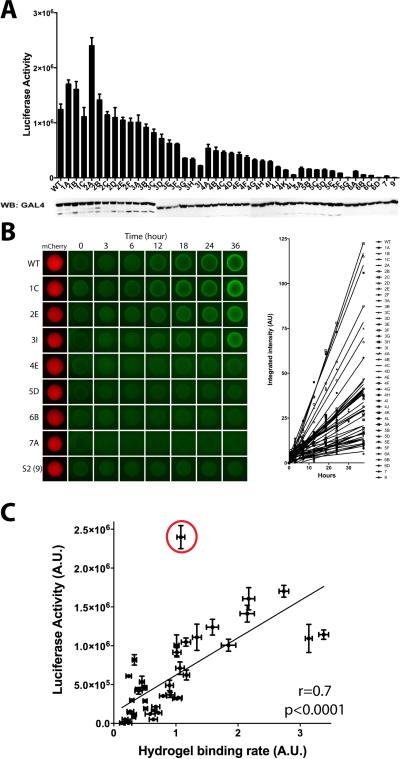
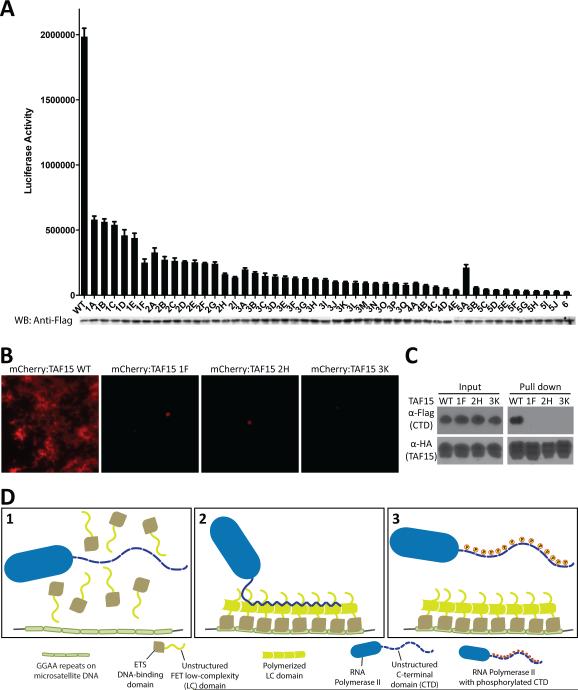
The FUS LC domain contains 27 repeats of the triplet sequence [G/S]Y[G/S], and derivatives carrying 5, 9, 15, and 27 tyrosine-to-serine mutations in randomly chosen triplets showed progressively diminished capabilities of polymerization and stress granule recruitment (Kato et al., 2012). In order to initiate studies of the transcriptional activation capacity of the FUS LC domain, it was linked to the GAL4 DNA binding domain and assayed by transient transfection for activation of firefly luciferase activity driven by a GAL4-dependent reporter gene. Forty-three mutated derivatives were prepared wherein individual tyrosine residues of the triplet sequence within the LC domain were randomly mutated to serine. 3 mutants randomly changed a single tyrosine, 6 changed 2 tyrosines, 9 changed 3 tyrosines, 12 changed 4 tyrosines, 7 changed 5 tyrosines, 4 changed 6 tyrosines, one changed 7 tyrosines, and one changed 9 tyrosines (Table S1). As shown in Figure 1A, concordance was observed between the number of tyrosine residues changed to serine and the degree of impediment upon of transcriptional activation. With a single exception, variants carrying either one or two tyrosine-to-serine changes exhibited transcriptional activation capacity indistinguishable from the GAL4:wild type fusion protein. The exceptional variant (2A) was considerably more active than the wild type LC domain of FUS. Variants carrying three tyrosine-to-serine changes tended to be slightly less active than those carrying zero, 1 or 2 mutations. The trend in loss of activity was observed to track with variants carrying 4, 5 or 6 random tyrosine-to-serine mutations, and mutants carrying 7 or 9 tyrosine-to-serine mutations revealed no detectable capacity to activate expression of the GAL4-responsive luciferase target gene.
Figure 1. Correlation Between Transcriptional Activation and Hydrogel Binding of Native and Mutated Derivatives of the LC Domain of FUS.
(A) The LC domain of FUS was linked to the DNA binding domain of GAL4 and assayed for activation of a GAL4 reporter gene in transiently transfected U20S cells. Activity of the native LC sequence was compared with 43 variants wherein different number of tyrosine residues were randomly mutated to serine (see text). Identities of specific tyrosine-to-serine changes in each mutant are shown in Table S1. Expression levels for all test protein were assayed by western blotting as shown below histograms. (B) The GFP-linked LC domains of FUS carrying the same mutations as (A) were exposed to mCherry:FUS hydrogels (left) and initial binding rates were measured (right). (C) A correlation plot between the transactivation activity and hydrogel binding rate of the individual LC mutants. Note that there is but one significant outlier indicated by a red circle. This is the “2A mutant” and described in more detail in the text and Figure 5.
A simple assay for the propensity of LC domains to polymerized into amyloid-like fibers has been described previously (Kato et al., 2012). Briefly, upon prolonged incubation of a hybrid protein linking the LC domain of FUS to mCherry, the protein polymerizes into a hydrogel-like state. Microscopic gel droplets can be formed in chamber slides, exposed to soluble test protein, then scored for trapping of the test protein via confocal microscopy. The molecular basis of trapping has been deduced, by TIRF microscopy, to result from co-polymerization of the GFP-tagged test protein into existing polymeric fibers in the mCherry:FUS hydrogel droplets (Kato et al., 2012). Forty out of the forty-three variants described above were fused to GFP and tested for their ability to be trapped by hydrogel droplets composed of mCherry linked to the wild type LC domain of FUS. As shown in Figure 1B, variants carrying one or two tyrosine-to-serine changes were trapped by the mCherry:FUS hydrogel droplets in a manner similar to the protein linking the wild type FUS LC domain to GFP. Sequential diminishment in hydrogel trapping was observed for variants carrying 3, 4, 5, 6, 7 or 9 tyrosine-to-serine mutations. The scatter plot shown in Figure 1C directly compares the transcriptional activation and hydrogel trapping capacities of all forty mutants that were scored in both assays. With the exception of the 2A mutant, which activates transcription considerably better than the wild type LC domain of FUS, a strong correlation was observed between the two activities under study (Pearson's r = 0.7 with a P value of less than 0.0001). These data tentatively indicate that transcriptional activation capacity of the LC domain of FUS may be dependent upon its ability to polymerize.
B-isox Microcrystals Precipitate Nuclear Proteins
Having observed a correlative relationship between fiberization and transcriptional activation capacity, we prepared nuclear extracts from HEK293 cells and subjected the lysate to precipitation by the biotinylated isoxazole (b-isox) chemical. As described previously, the b-isox chemical crystallizes in cold aqueous buffer at concentrations between 30 and 100 μM (Kato et al., 2012). X-ray diffraction studies at high resolution revealed the b-isox crystal surface to contain a wavy repetition of peaks and troughs separated by 4.7 Å, and it has been hypothesized that the troughs are properly disposed to capture proteins containing significant stretches of random coil polypeptide. These disordered, LC sequences are thought to adopt an extended β-strand conformation having dimensions that match the trough of the b-isox crystal surface, thereby facilitating selective precipitation of proteins endowed with substantive regions of disordered polypeptide.
Given that earlier studies were limited to cytoplasmic lysates, we prepared nuclear lysates from cultured mammalian cells, precipitated with b-isox microcrystals, separated the precipitated proteins on SDS polyacrylamide gels, and identified precipitated proteins by shotgun mass spectrometry (Experimental Procedures). Roughly 580 nuclear proteins were identified as being precipitated by b-isox microcrystals (Table S2). This survey revealed overlap with many of the proteins precipitated from cytoplasmic extracts by the b-isox chemical, including many hnRNP proteins. Additionally, however, these efforts revealed the identities of a number of nuclear proteins not observed in earlier experiments that had interrogated cytoplasmic lysates. Prominent among the newly identified, b-isox precipitated nuclear proteins were TAF15, the largest subunit of RNA polymerase II, numerous subunits of the mediator complex (including Med4, Med6, Med10, Med13L, Med 14, Med 15, Med 17 and all subunits of CDK8), a variety of enzymes involved in epigenetic modification of histones, both co-activator and co-repressor proteins known to control the function of nuclear hormone receptors, and many distinct SR proteins known to be involved in pre-mRNA splicing. Parallel studies were performed on nuclear extracts prepared from Saccharomyces cerevisae, leading to the identification of roughly 260 yeast nuclear proteins significantly precipitated by the b-isox chemical (Table S3). We again observed b-isox-mediated precipitation of the largest subunit of RNA polymerase II. We also observed the list of precipitated proteins to contain multiple subunits of the mediator complex, and a number of gene-specific transcription factors endowed with glutamine and asparagine-rich, low complexity sequences.
With respect to nuclear proteins precipitated by the b-isox chemical, our initial focus fell on TAF15 and the largest subunit of RNA polymerase II. Among the 580 mammalian proteins selectively precipitated by b-isox microcrystals, TAF15 registered the second highest number of spectral counts, and the largest subunit of RNA polymerase II registered the third highest number of spectral counts (Table S2). B-isox precipitation of yeast nuclear extracts also revealed the largest subunit of RNA polymerase II as displaying an exceptionally high number of spectral counts (Table S3).
TAF15 represents the third paralog of the FET family of RNA binding proteins, and is endowed with an N-terminal LC domain very similar to the LC domains found in FUS and EWS. Unlike FUS and EWS, TAF15 has been identified as a sub-stoichometric component of the TFIID complex, which has been extensively characterized as an important assembly in transcriptional initiation by RNA polymerase II (Bertolotti et al., 1999). Attention to the largest subunit of RNA polymerase II was prompted by the fact that it contains an intrinsically disordered region of roughly 380 residues at its C-terminus. This C-terminal domain (CTD) of mammalian RNA polymerase II contains 52 heptad repeats of the sequence Y1S2P3T4S5P6S7 (Corden et al., 1985). Extensive work over the past two decades has confirmed the importance of the CTD in the transcription cycle (Buratowski, 2009; Egloff et al., 2012; Phatnani and Greenleaf, 2006). Included among a variety of seminal advancements, the field now understands that in its initial state of recruitment to a promoter, the CTD exists in the unphosphorylated state. A series of obligatory phosphorylation events on serine residues S2, S5 and S7 of the CTD heptad repeats accompanies release of RNA polymerase from the pre-initiation complex so that it can begin transcriptional elongation (Egloff and Murphy, 2008). When aligned contiguously, the CTD contains 52 repeats of the triplet sequence S7Y1S2 (or close variations thereof). These repeats form a subset of the [G/S]Y[G/S] repeats that we have found to be critical for polymerization of the LC domain of FUS (Kato et al., 2012).
Phosphorylation of the CTD Blocks B-isox Precipitation
Using antibodies specific to either the unphosphorylated form of the CTD of RNA polymerase II (8WG16), or antibodies to S2 phosphorylation (3E10), S5 phosphorylation (3E8), or S7 phosphorylation (4E12) (Chapman et al., 2007), we observed that b-isox microcrystals preferentially precipitate only the unphosphorylated form of the yeast CTD (Figure 2). Parallel analyses of b-isox precipitates from mammalian cells also revealed selective precipitation of only the unphosphorylated form of the largest subunit of RNA polymerase II (Figure S1A). We tentatively hypothesize that the CTD is the determinant of the largest subunit of RNA polymerase II that facilitates b-isox precipitation, and that phosphorylation on residues S2, S5 or S7 somehow impedes the ability of the CTD to fit within the 4.7 Å surface troughs of b-isox microcrystals.
Figure 2. Biotinylated Isoxazole Preferentially Precipitates Unphosphorylated CTD of RNA polymerase II.
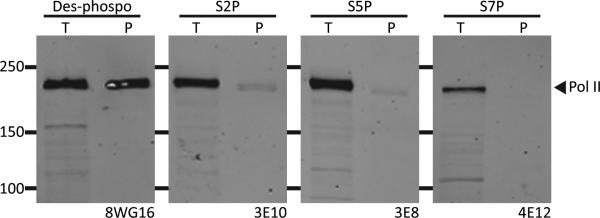
Western blot of total cell extract (T) or b-isox pellet (P) with different CTD antibodies. Bands shown in the four panels correspond in size to the largest subunit of RNA polymerase II (217 kD). The 8WG16 antibody recognizes the unphosphorylated CTD, the 3E10 antibody recognizes serine 2 phosphorylated CTD, the 3E8 antibody recognizes serine 5 phosphorylated CTD, and the 4E12 antibody recognizes serine 7 phosphorylated CTD. B-isox significantly precipitates only unphosphorylated CTD of Pol II. See also Figure S1.
In order to test these ideas more carefully, a GFP fusion protein was prepared linked to heptad repeats 27-52 of the human CTD (Experimental Procedures). The purified, recombinant GFP:CTDC26 fusion protein was effectively precipitated by b-isox microcrystals, yet phosphorylation of the CTD by either CDK7, which phosphorylates S5 and S7 (Akhtar et al., 2009; Kim et al., 2009; Rickert et al., 1999; Roy et al., 1994; Trigon et al., 1998) or CDK9, which phosphorylates S2, S5 and S7 (Czudnochowski et al., 2012; Jones et al., 2004; Ramanathan et al., 2001), prevented b-isox-mediated precipitation (Figure S1B). These observations confirm the CTD as the determinant of the largest subunit of RNA polymerase II responsible for b-isox precipitation, and give evidence that when the CTD is phosphorylated, precipitation is prevented.
Binding of the CTD to Hydrogel Droplets Composed of FET Protein Polymers
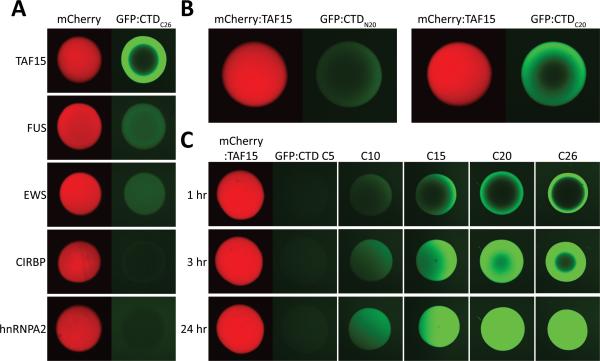
Having recognized that the iterated S7Y1S2 triplet repeats of the CTD correspond to a variant of the [G/S]Y[G/S] triplet repeats diagnostic of the LC domains of FET proteins, we tested whether the GFP:CTD fusion protein might bind to hydrogel droplets formed from the LC domains of the three FET proteins (FUS, EWS and TAF15), as well as hydrogel droplets formed from the LC domains of hnRNPA2 and cold inducible RNA binding protein (CIRBP). As shown in Figure 3A, the GFP:CTD fusion protein was avidly trapped by hydrogel droplets composed of the LC domain of TAF15. No trapping whatsoever was observed when the GFP:CTD test protein was exposed to either hnRNPA2 or CIRBP hydrogel droplets, and weak trapping was observed for FUS and EWS hydrogels.
Figure 3. Selective Binding of GFP:CTD to mCherry:TAF15 Hydrogel Droplets.
(A) Hydrogels composed of LC domains of mCherry:TAF15, mCherry:FUS, mCherry:EWS, mCherry:hnRNPA2 and mCherry:CIRP were incubated with a soluble form of GFP linked to the C-terminal 26 heptad repeats of the CTD of mammalian RNA polymerase II. Little or no retention of the GFP:CTDC26 protein was observed for the mCherry:hnRNPA2 or mCherry:CIRBP hydrogel droplets, and weak binding was observed for mCherry:FUS and mCherry:EWS hydrogels. By contrast, strong retention was observed for mCherry:TAF15 hydrogel droplets. See also Figure S2. (B) The degenerate C-terminal half of the CTD (GFP:CTDC20) binds mCherry:TAF15 hydrogel stronger than the highly conserved, N-terminal half of the CTD (GFP:CTDN20). (C) The binding intensity of the GFP:CTD degenerate repeats to mCherry:TAF15 hydrogel correlates with the number of heptad repeats, with no binding observed for GFP:CTDC5, weak binding for GFP:CTDC10 and strong binding for GFP:CTDC15, GFP:CTDC20 and GFP:CTDC26.
GFP:CTD binding to TAF15 hydrogel droplets was extended in two ways. First, we compared the binding properties of two GFP test proteins, one containing 20 heptad repeats from the highly conserved, N-terminal half of the CTD (repeats 1-20 = GFP:CTDN20) and another containing 20 heptad repeats from the degenerate C-terminal half of the CTD (repeats 33-52 = GFP:CTDC20). As shown in Figure 3B, the GFP test protein appended to the 20 degenerate CTD repeats, GFP:CTDC20, bound TAF15 considerably more avidly than the counterpart fusion containing the 20 conserved CTD repeats (GFP:CTDN20). Second, we compared hydrogel trapping for fusion proteins containing 5, 10, 15, 20 or all 26 of the degenerate CTD repeats (GFP:CTDC5 = CTD repeats 48-52, GFP:CTDC10 = CTD repeats 43-52, GFP:CTDC15 = CTD repeats 38-52, GFP:CTDC20 = CTD repeats 33-52, GFP:CTDC26 = CTD repeats 27-52). As shown in Figure 3C, no trapping by mCherry:TAF15 hydrogel droplets was observed for the fusion protein containing only 5 heptad repeats, intermediate trapping was observed for the fusion containing 10 heptad repeats, and equivalent trapping was observed for fusions containing 15, 20 or 26 repeats.
When evaluating the kinetics of binding of the various GFP:CTD constructs to hydrogel droplets composed of mCherry linked to the LC domains of TAF15, FUS or EWS, it was noticed that gel trapping occurred more rapidly than the homo- or heterotypic trapping of GFP fusion proteins linked to the LC domains of FET proteins. Knowing that the latter trapping occurs via a mechanism of co-polymerization of soluble test proteins to fibrous polymers of which the hydrogel droplets are themselves composed (Kato et al., 2012), we employed fluorescence microscopy to evaluate binding of the GFP:CTDC26 fusion protein to fibrous preparations of mCherry:TAF15. As soon as the mixture of the two protein samples could be applied to the microscope substrate, clear evidence of GFP signal could be seen to coat the entire length of mCherry:TAF15 polymeric fibers (Figure S2). Instead of slow, time-dependent co-polymerization extending from either end of existing fibers, the GFP:CTDC26 protein rapidly bound to the lateral surface of mCherry:TAF15 fibers. These observations offer evidence of a second and distinct means by which LC polymeric fibers are able to trap otherwise soluble test proteins composed, in part, of LC domains. They may further offer mechanistic insight into the perplexing promiscuity of protein:protein interaction of intrinsically disordered proteins (Cumberworth et al., 2013).
CTD Phosphorylation Blocks Hydrogel Binding
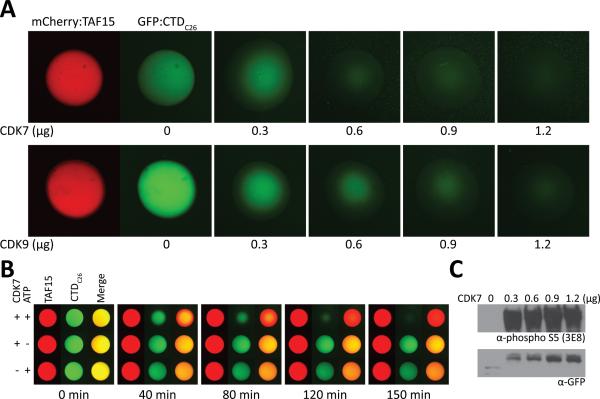
Knowing that phosphorylation of the CTD prevents precipitation by b-isox microcrystals, we evaluated the effects of phosphorylation on trapping of GFP:CTD test protein by TAF15 hydrogels. When phosphorylated by either CDK7 or CDK9, the GFP:CTD fusion protein was fully blocked from binding to hydrogels composed of polymers of the LC domain of TAF15 (Figure S3). Emboldened by this observation, we pre-bound the GFP:CTDC26 test protein to mCherry:TAF15 hydrogel droplets then added either CDK7 or CDK9 in the presence of ATP. In both cases, the GFP signal was lost in an enzyme concentration-dependent manner (Figure 4A). When applied at fixed enzyme levels, the GFP signal diminished in a time and ATP-dependent manner (Figure 4B). A video showing the dynamics of GFP signal loss can be seen in Movie S1. Material released from mCherry:TAF15 gel droplets in response to CDK7 was recovered from the wells of chamber slides and evaluated by western blotting using antibodies to both GFP and the S5 phospho-form of the CTD. As shown in Figure 4C, both GFP and phosphorylated CTD S5 western blotting signals were released from the mCherry:TAF15 hydrogels as a function of enzyme concentration. We conclude from these observations that not only does phosphorylation of the CTD prevent binding to mCherry:TAF15 hydrogels, but pre-bound material is accessible to kinase-mediated phosphorylation and subsequent release from the gels.
Figure 4. CDK7-mediated Release of GFP:CTDC26 from mCherry:TAF15 Hydrogel Droplets.
(A) The GFP:CTDC26 trapped by mCherry:TAF15 hydrogel was released upon addition of CDK7 or CDK9 in the presence of ATP in an enzyme concentration-dependent manner. (B) Hydrogel droplets of mCherry:TAF15 (red) were exposed to GFP:CTDC26 (green). Chamber slides containing individual hydrogel droplets were exposed to CDK7 plus ATP (top row), CDK7 alone (middle row) or ATP alone (bottom row). Samples exposed to both CDK7 and ATP revealed the time-dependent release of GFP:CTDC26. See also Movie S1. (C) Western blot analysis of materials released into the well of chamber slides revealed progressive increases in the presence of soluble, phosphorylated CTD (top) and soluble GFP:CTDC26 (bottom). See also Figure S3.
The 2A Mutant of the FUS LC Domain Displays a Gain-of-function Phenotype
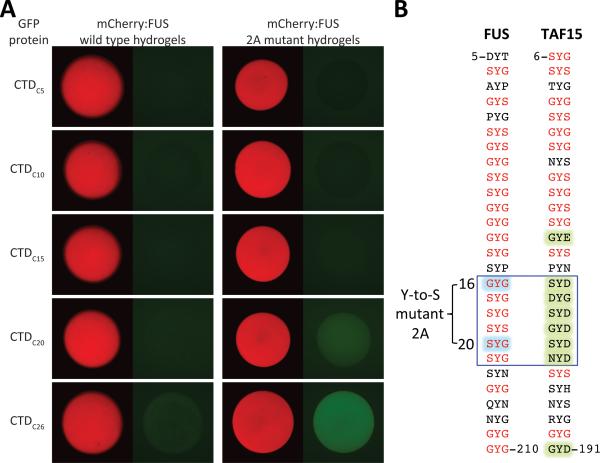
In comparing the properties of mutants of the LC domain of FUS with respect to both fiber polymerization and transcriptional activation (Figure 1), we observed significant discordance for only one mutant. The so called “2A mutant” bound to mCherry:FUS hydrogel droplets with avidity indistinguishable from the wild type protein, yet activated transcription considerably better than the normal LC domain of FUS. Hydrogel droplets were formed from a fusion protein linking the 2A mutant to mCherry, fully analogous to the wild type protein linked to mCherry save for the two tyrosine-to-serine changes. When challenged with the different heptad repeats of GFP:CTD test proteins, significantly stronger gel trapping was observed for hydrogel droplets formed from the 2A mutant LC domain (Figure 5A). We conclude that polymeric fibers formed from the 2A mutant bind the unphosphorylated CTD better than the wild type LC domain of FUS, and offer the hypothesis that this gain-of-function phenotype may explain why the 2A mutant activates transcription better than the wild type LC domain of FUS (Figure 1). Despite the enhanced binding of the 2A mutant to various GFP:CTD test proteins, the latter interactions were blocked by CDK7- or CDK9-mediated phosphorylation (Figure S4A). Moreover, application of either of these protein kinase enzymes, in the presence of ATP, caused pre-bound GFP:CTDC26 protein to be released from hydrogel droplets composed of LC domain prepared from the 2A mutant in a manner indistinguishable of CTD release from hydrogel droplets formed from the wild type LC domain of FUS (Figure S4B).
Figure 5. Hydrogels Formed from the 2A Mutant of FUS LC Domain Display Enhanced CTD Binding.
(A) 2A mutant of FUS LC domain, which exhibits enhanced transactivation activity relative to the wild type protein (Figure 1C), carries tyrosine-to-serine mutations at tyrosine within the 16th and 20th [G/S]Y[G/S] triplet repeats (Figure 5B). The 2A mutant LC domain was fused with mCherry, and hydrogel binding assays were carried out with GFP:CTD fusions carrying different numbers of heptad repeats. Compared with hydrogel droplets formed with the native LC domain of FUS, those formed from the 2A mutant showed enhanced binding to GFP:CTDC20 and GFP:CTDC26. See also Figure S4. (B) Alignment of [G/S]Y[G/S] triplet repeats in low complexity domains of FUS (left) and TAF15 (right). The two tyrosineto-serine mutations causing FUS to suffer a gain-of-function enhancement in transcriptional activation and CTD binding are highlighted in blue. All triplet repeats of TAF15 carrying an aspartic acid or glutamic acid adjacent to tyrosine are highlighted in green. Canonical [G/S]Y[G/S] triplet repeats of both LC domains are shown in red.
An alignment of the 27 [G/S]Y[G/S] triplet repeats located within the LC domains of FUS and TAF15 is shown in Figure 5B. The two tyrosine residues mutated to serine in the 2A, gain-of-function variant, correspond to tyrosine residues embedded within the 16th and 20th [G/S]Y[G/S] triplets of the FUS LC domain. In comparing this region of the two LC domains, FUS that when polymerized binds the CTD weakly, and TAF15 that binds the CTD avidly, we notice that triplet repeats 16-21 of the TAF15 LC domain are unusual. Instead of the central tyrosine residues being flanked by either glycine or serine, six straight repeats bear an aspartic acid residue on one side of the central tyrosine or the other (SYD16, DYG17, SYD18, GYD19, SYD20 and NYD21). As will be discussed, this “16-21 window” may be worthy of close attention with respect to the ability of the LC domains of FET proteins to capture the CTD of RNA polymerase II.
DNA-mediated Enhancement of FET Protein Polymerization
The test tube polymerization of the LC sequences associated with FET proteins is only observed at high concentrations (Kato et al., 2012). One means by which appropriately high concentrations necessary for polymerization could take place in living cells is if FET proteins are iteratively bound to a multivalent substrate. ChIP-chip experiments have given evidence that when the LC domain of EWS is linked to the FLI DNA-binding domain, it binds in living cells to microsatellites carrying iterative repeats of the tetranucleotide sequence GGAA (Gangwal et al., 2008). Since GGAA represents an optimal binding site for the ETS DNA binding domain of the EWS:FLI protein, the fusion protein binds iteratively to these otherwise inert microsatellites. If located within 5-10kb of a target promoter, the translocation product-bound microsatellite is envisioned to become a rogue enhancer. Under such a setting, it is possible that the LC domains of the densely packed EWS:FLI molecules can be prompted to polymerize.
As a test of this hypothesis, microsatellite sequences associated with the hNROB1 gene were amplified by PCR from U2OS cells (Experimental Procedures). A PCR product containing 25 repeats of the GGAA tetra-nucleotide was incubated with a recombinant protein linking mCherry to a fusion protein composed of the DNA binding domain of FLI attached to the LC domain of FUS (Experimental Procedures). In the absence of added DNA, the 0.5 μM levels of the mCherry:FUS-FLI protein remained in a largely disaggregated state as deduced by transmission electron microscopy (Figure 6B). By contrast, addition of the PCR product containing the iterative GGAA repeats led to the formation of an extensive network of interwoven fibers (Figure 6A). When evaluated by a gel mobility shift assay, it was apparent that multiple copies of the mCherry:FUS-FLI protein were simultaneously bound to the probe DNA containing iterative repeats of the GGAA tetra-nucleotide (Figure S5). We tentatively conclude that when multiple copies of the mCherry:FUS-FLI protein are simultaneously bound to DNA, polymerization of the LC domain of FUS is enhanced.
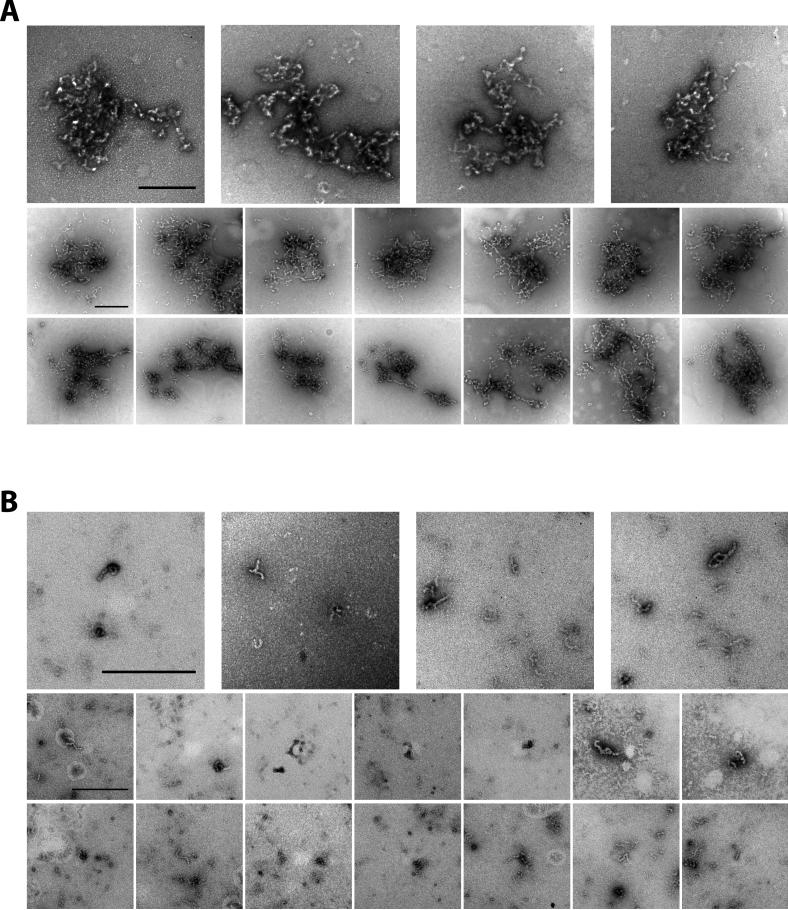
Figure 6. DNA-dependent Enhancement of Fiber Formation of FUS-FLI Fusion Protein.
(A) mCherry:FUS LC domain-FLI DNA-binding domain fusion protein (0.5 μM) was incubated with microsatellite DNA (20 nM) in the presence of 35% glycerol. After 1 hr incubation, materials in the solution were visualized by transmission electron microscopy. FUS-FLI fibers grew long and became large sleave. (B) mCherry:FUS-FLI protein was incubated in the presence of 35% glycerol (no DNA). Small amounts of spontaneous nucleation and fiber growth were observed. All scale bars indicate 0.5 μm. See also Figure S5.
Polymerization-dependent CTD Binding to the LC Domain of TAF15
Hydrogel droplets formed from a fusion protein linking mCherry to the amino terminal 208 residues of TAF15 bind a variety of GFP:CTD constructs, particularly those housing 15 or more of the degenerate, C-terminal heptad repeats (Figure 3C). A truncated version of the TAF15 LC domain restricted to 80 amino-terminal residues was observed to form hydrogel droplets equally as well as the intact LC domain, yet was only minimally capable of trapping the GFP:CTDC10 and GFP:CTDC20 constructs (Figure S6). Addition of the remaining 128 residues of the LC domain yielded a protein that readily formed hydrogels that trapped the GFP:CTD constructs avidly.
These observations hint to the possibility that the amino terminal half of the TAF15 LC domain may be critical for polymerization, and that the carboxyl terminal half might be critical for CTD binding. Proceeding with this idea, a series of 48 variants was prepared wherein between one and six tyrosine residues of the [GS]Y[GS] triplet repeats were randomly mutated to serine (Table S4). Each mutant was linked to the DNA binding domain of GAL4 and tested for its ability to activate the GAL4-luciferase reporter gene. As shown in Figure 7A, all six variants carrying a single tyrosine-to-serine mutation were less competent in transcription activation capacity than the wild type protein. Variants carrying two, three, four, five or six random tyrosine-to-serine mutations suffered sequentially diminished activity. Three of the 48 mutants were chosen for further study. These include the 1F, 2H and 3K mutants, all of which were among the most debilitated variants within their respective classes. All of the tyrosine residues altered in these particularly deleterious mutants localized to the first 80 residues of the TAF15 LC domain that, on it's own, is capable of fiberization and hydrogel formation. The 1F mutant changes tyrosine 46 to serine, the 2H mutant changes tyrosines 38 and 56 to serine, and 3K changes tyrosines 17, 46 and 63 to serine (Table S4).
Figure 7. Polymerization of the LC Domain of TAF15 is Required for Both Transcriptional Activation and CTD Binding.
(A) 48 tyrosine-to-serine mutations were randomly introduced into the LC domain of TAF15 (Table S4). Mutants were assayed for transcriptional activation capacity as GAL4 fusion proteins. Expression levels for individual test proteins were monitored by Western blotting as displayed below histogram depictions of transcriptional activation measurements. (B) mCherry fusion proteins linked to the native LC domain of TAF15 (WT), the 1F mutant, the 2H mutant and the 3K mutant were incubated under conditions favorable for polymerization (Experimental Procedures). Fluorescence microscopy was employed as an assay for fiberization of the four test proteins. See also Movie S2. (C) Co-immunoprecipitation assay was conducted by mixing Flag-tagged GFP:CTDC26 with HA-tagged versions of mCherry linked to the native form of the TAF15 LC domain, or to the 1F, 2H or 3K tyrosine-to-serine mutants. Following HA-mediated immunoprecipitation, samples were run on a denaturing SDS-PAGE gel and subjected to Western blotting using either anti-Flag or anti-HA antibodies. See also Figure S6. (D) Schematic Concept of RNA Polymerase II Recruitment by LC Domain Polymers. 1) The C-terminal domain (CTD) of RNA polymerase II does not bind to monomers of the unstructured LC domain of FET protein fused to the ETS DNA-binding domain. 2) Once the ETS DNA-binding domain of the fusion protein binds to the GGAA repeats on microsatellite DNA, the LC domains of FET protein form fibrous polymer that can recruit RNA polymerase II via direct interaction with the CTD. 3) Phosphorylation of serine residues 2, 5 and 7 of the CTD heptad repeats by CDK7 or CDK9 facilitates release of RNA polymerase II from the FET LC domain polymer.
The TAF15 LC domains of these variants were linked to mCherry and compared to the wild type LC domain with respect to hydrogel formation, fiberization and CTD interaction. None of the mutants formed hydrogels when incubated under standard conditions (Movie S2). When incubated under conditions favorable for fiberization, the mCherry variant linked to the wild type LC domain of TAF15, as visualized by fluorescence microscopy, formed obvious tangles of aggregated fibers. Almost no polymerization was observed for the 1F, 2H and 3K mutants. Inspection of the rare occurrence of visible mCherry aggregates in the latter variants revealed small amorphous particles (Figure 7B). Finally, when mixed with the GFP:CTDC26 construct and subjected to co-immunoprecipitation, no interaction whatsoever was observed for anything but the mCherry fusion protein linked to the native LC domain of TAF15 (Figure 7C). These and other data presented herein give evidence that the LC domains of FET proteins obligatorily rely upon polymerization in order to execute their role in transcriptional activation. Whether LC domain polymerization might be more broadly employed in the context of gene regulation in eukaryotic cells remains open to question.
Schematic Concept of RNA Polymerase Recruitment by LC Domain Polymers
Cancer cells expressing the translocation product wherein the LC domain of EWS is linked to the ETS DNA binding domain of FLI depend upon this fusion protein to drive cell growth or survival. The EWS:FLI translocation product is undoubtedly employed as an aberrant transcription factor required to re-program gene expression in a manner beneficial to cancer cells. We hypothesize that upon binding to the relevant sites on DNA, including microsatellites bearing iterative repeats of the tetranucleotide sequence GGAA, the EWS:FLI fusion protein can polymerize via its LC domains. As depicted schematically in panel D of Figure 7, we further hypothesize that fibrous polymers of the LC domain create binding sites for the CTD of RNA polymerase. Finally, we predict that phosphorylation of the CTD by CDK7, CDK9 or a related protein kinase enzyme facilitates release of the polymer-bound RNA polymerase so that it can escape gene promoters and enter into the process of transcriptional elongation.
DISCUSSION
Here we report evidence that the LC domains of FET proteins may require polymerization as a means of directly capturing the CTD of RNA polymerase II. If correct, these data lend credence to the hypothesis advanced by Kato, Han and colleagues (Han et al., 2012; Kato et al., 2012). The thesis of the initial studies on b-isox precipitation, LC sequences, LC polymerization and LC-derived hydrogels offered the concept that intrinsically disordered domains associated with RNA regulatory proteins and transcription factors might reversibly polymerize as a means of helping establish cellular organization and information flow from DNA to RNA to protein. The present study extends earlier work in several ways. First, by having a simple functional read-out (transcriptional activation), we have been able to establish a pattern of significant concordance between the capacity of LC domains to polymerize and their ability to function as transcriptional activation domains (Figure 1). Indeed, the single most discordant mutant (the 2A mutant) was found to suffer a gain-of-function phenotype in transcriptional activation capacity that may be attributable to its enhanced capacity to bind the CTD of RNA polymerase II. Second, we offer the simple conclusion that, when appended to a DNA binding domain as a function of scores of independent cancer-causing translocation events, the LC domains of FET proteins act to directly recruit the CTD of RNA polymerase II (Figure 3). In order to achieve this task, the LC domains of FET proteins must be capable of polymerization (Figure 7). Whereas the hydrogel droplets deployed as our mainstay assay are composed of long fiberous polymers containing thousands of subunits, we have no idea as to many subunits must polymerize as a “point source” to recruit the CTD of RNA polymerase. It is entirely possible that fiberous seeds composed of only a handful of LC domain subunits might be sufficient to nucleate functionally competent organizational puncta.
Evidence that the LC domains of FET proteins are able to interact with the CTD of RNA polymerase II is consistent with recent studies of FUS in its native form (Schwartz et al., 2012). The latter study provided compelling evidence for direct interaction between the CTD of RNA polymerase II and the intact FUS protein. Intriguingly, this interaction was shown to be RNA dependent. In a new and expanded study co-submitted with the present manuscript, Cech and colleagues offer exciting data that may explain RNA dependence (Schwartz et al., manuscript submitted). Since the intact FUS protein is endowed with an RNA-binding domain, addition of an RNA polymer offers the opportunity for FUS to bind multiple times to the RNA substrate, thereby bringing LC domains of multiple protomers into proximity, and thereby enhancing local concentration and the propensity of the LC domains to polymerize.
Similar phenomena may well be in play in the context of translocation products carrying the LC domains of FET proteins fused to different DNA-binding domains. Beautiful ChIP-chip experiments have given evidence that when the LC domain of EWS is linked to the FLI DNA-binding domain, it binds in living cells to microsatellites carrying iterative repeats of the tetranucleotide sequence GGAA (Gangwal et al., 2008). Since GGAA represents an optimal binding site for the ETS DNA binding domain of the EWS:FLI protein, the fusion protein binds iteratively to these otherwise inert microsatellites. If located within 5-10kb of a target promoter, the translocation product-bound microsatellite has been hypothesized to become a rogue enhancer. We are like-minded with the Cech lab in providing evidence that enhanced local concentration via proximal binding of multiple subunits of LC domain containing proteins will represent a critical stimulus for polymerization (Figure 6).
Despite coming to similar conclusions regarding LC polymerization and CTD binding, the Cech studies differ in many ways from those reported herein. Cech and colleagues have employed intact FUS protein and have studied the protein in the context of its normal cellular function. We, instead, have focused on the isolated parts of FET proteins that are translocated in cancer onto a variety of DNA-binding domains. It is relatively straightforward to conceptualize how the latter class of proteins bind DNA, polymerize locally via their LC domains, and thereby achieve the fibrous organizational state required to bind the CTD of RNA polymerase II. How, instead, might one conceptualize the manner in which native, RNA binding forms of FET proteins achieve the same task? One possibility is that native FET proteins are able to bind to non-coding RNAs that remain nascently attached to DNA. Perhaps these complexes might function as “RNA enhancers”, achieving the goals of bringing FET proteins into the proximity of target genes at locally elevated protein concentrations adequate to facilitate LC domain polymerization necessary for the recruitment of RNA polymerase II.
Even if both of these conceptualizations of polymer-dependent CTD recruitment are fundamentally correct, we are undoubtedly only beginning to see the tip of the iceberg. Hundreds of RNA regulatory proteins and transcription factors contain LC domains, offering almost limitless opportunity for complexity in cellular organization – including the possible existence of solid-state polymeric pathways for information flow from “transcription factories” through fibrous nuclear bodies into various forms of cytoplasmic RNA granules to the ultimate sites of mRNA translation. Anticipating that LC domains are almost certainly regulated by post-translational modification, it is clear that we are at the earliest stage of recognizing this new and unconventional way of thinking about cellular organization. Indeed, if we are correct in assuming that phosphorylation of the CTD of RNA polymerase II triggers its release from FET protein fibers, this may serve as an initial paradigm for considering post-translational modification as a means of controlling the dynamic behavior of LC derived polymers.
We close with three perplexing observations. First, why do polymeric fibers formed from the LC domain of TAF15 bind the CTD so much more avidly than the analogous LC domains of FUS and EWS (Figure 3A)? Second, why is it that mutations of two tyrosines in the LC domain of FUS yield a protein with enhanced transcriptional activation capacity and an enhanced ability to bind the CTD of RNA polymerase II (Figures 1 and 5)? Third, why do the degenerate CTD repeats bind the polymeric fibers of FET LC domains better than the consensus repeats (Figure 3B)? Since TAF15 was discovered as a component of the TFIID complex, one might imagine that its function has become more proximally evolved to the transcription initiation process than FUS or EWS, thus consistent with constitutive, avid affinity for the CTD of RNA polymerase II. The region of TAF15's LC domain most distinctly different from the LC domains of FUS and EWS is in the [G/S]Y[G/S] repeats 16-21, precisely the same region of the location of the two tyrosine-to-serine mutations in the 2A gain-of-function mutant of FUS (Figure 5). Six straight repeats of TAF15 contain aspartic acid on one side or the other of the central tyrosine. We speculate that these aspartic acid residues disrupt or in some way modify the TAF15 polymeric fiber in this region, and that such modifications enhance CTD binding. Likewise, we hypothesize that the two tyrosine-to-serine alterations in the 2A mutant of the FUS LC domain also modify the polymeric fiber in a manner favoring CTD binding and transcriptional activation capacity. Perhaps FUS and EWS can be modified by phosphorylation on serine residues flanking the tyrosines in this “16-21 window” of their LC domains, such that fiber disruption and propensity for CTD binding might be controlled in a regulated manner – rather than being “hard-wired” as is possible owing to TAF15's evolutionary accumulation of aspartic acid residues as phosphomimetics.
Finally, we consider why it might be that the polymeric form of the TAF15 LC domain binds the C-terminal degenerate CTD repeats so much more strongly than the N-terminal consensus CTD repeats (Figure 3B). The N-terminal consensus repeats of the CTD have been shown to bind and co-crystallize with components of the mediator complex (Robinson et al., 2012). Likewise, elegant EM studies have also provided evidence of CTD:mediator interaction (Tsai et al., 2013). Perhaps upon evolutionary duplication of the CTD from 26 repeats in yeast to 52 repeats in mammals has allowed the degenerate repeats to adopt a new function molded to the fibrous LC domains of FET proteins. Close inspection of the degenerate repeats may be revealing in this regard. In comparing the degenerate repeats to our most distant evolutionary relative bearing the degenerate repeats, teleost fish from which humans diverged 450 million years ago, it is notable that the pattern of degeneracy is nearly identical. Briefly put, the degenerate changes installed nearly one half billion years ago have been perfectly conserved. The most notable difference between the degenerate and consensus CTD repeats is the presence of lysine residues replacing S7 (Y1S2P3T4S5P6S7 vs Y1S2P3T4S5P6K7) in seven of the degenerate repeats. Although embarrassingly simplistic, we speculate that the positive charge of these seven lysine residues on the immediate N-terminal sides of tyrosines in the degenerate CTD repeats might blend with the negative charge of six consecutive aspartic acids flanking tyrosines in the “16-21 window” of TAF15. If correct, this idea of charge attraction might explain the longstanding enigma of acidic activation domains.
EXPERIMENTAL PROCEDURES
Materials
Synthesis of the biotinylated isoxazole is described previously (Kato et al., 2012). Mammalian pM expression vector was purchased from Clontech Laboratory (USA). CDK7/Cyclin H/MAT1 and CDK9/Cyclin T1 enzyme complexes were purchased from Millipore.
Transcriptional Activation by Tyrosine-to-Serine Mutants of FUS and TAF15 LC domain
Mammalian expression plasmids for human FUS or TAF15 LC domain fused to GAL4 DNA-binding domain were constructed by insertion of a DNA fragment encoding FUS LC domain (residue 2-266) or TAF LC domain (residue 2-208) into the multiple cloning sites of pM vector (Clontech, USA). Tyrosine-to-serine mutants of GAL4-FUS LC and GAL4-TAF15 LC were generated by QuickChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies). To test transcriptional activity of GAL4-fused LC domains, luciferase assays were performed in quadruplicate in 96-well plate. In each well, 50 ng of pM-LC vector and 20 ng L8G5-Luc (Lemercier et al., 2000) were reverse transfected to HEK293T cells (6,500 cells per well) by lipofectamine 2000 (Invitrogen). Luciferase assays were performed after 24 hours of incubation at 37°C, using Bright-Glo Luciferase Assay System (Promega). In order to obtain similar protein levels relative to wild type, 25 ng, 50 ng or 100 ng of pM-LC mutant plasmids were co-transfected with L8G5-Luc, and expression of GAL4-fused LC domains was determined by Western blotting with anti-GAL4 antibody (sc-510, Santa Cruz Biotechnology). The DNA dosage yielding the same expression level of GAL4-FUS mutant proteins to the wild type protein were used for luciferase assays.
Generation of Bacterial Expression Constructs
Expression plasmids for recombinant proteins used in all the biochemical assays were constructed using pHis-parallel vectors (Sheffield et al., 1999) as described before (Kato et al., 2012). The details were described in Supplementary data.
Protein Purification
All recombinant proteins were over-expressed in E. coli strain BL21(DE3). mCherry and GFP fusion LC domains from FUS, EWS, TAF15, CIRBP and hnRNPA2 were expressed and purified as described previously (Kato et al., 2012). GFP:CTD proteins were purified by Ni-NTA (Qiagen, USA) in the same conditions described previously (Kato et al., 2012), and further purified by a Hi-Load Superdex 200 gel filtration column (GF Healthcare, USA). The expression and purification of his-tagged mCherry:FUS LC domain-FLI DNA binding domain fusion protein is described in Supplementary data.
Hydrogel-binding Assays
Hydrogel droplets of mCherry:LC domain of either FUS, EWS, TAF15, CIRBP or hnRNPA2 were prepared as described before (Kato et al., 2012). Briefly, concentrated mCherry fusion proteins (typically ~50 mg/ml) were dialyzed in gelation buffer containing 20 mM Tris-HCl pH7.5, 200 mM NaCl, 20 mM BME, 0.5 mM EDTA, 0.1 mM PMSF over night. Dialyzed protein solution was sonicated, clarified by centrifugation and concentrated again to ~50-80 mg/ml. Small droplets (0.5 μL) of this protein solution were placed on a glass-bottomed dish (MatTek, USA). Hydrogels typically form after incubation for a couple of days at room temperature. For standard hydrogel binding assays, glycerol stocks of the purified GFP fusion test proteins were diluted to 1 μM in 1 mL of the gelation buffer. The GFP test solution was pipetted into the hydrogel dish so as to soak the hydrogel droplets in the GFP solution. After overnight incubation at 4°C, a horizontal section of the hydrogel droplet was scanned with both the mCherry and GFP excitation wavelengths on a Zeiss LSM510 confocal microscope. To measure binding rates of the FUS LC domain mutants carrying different number of tyrosine-to-serine mutations onto the wild type mCherry:FUS hydrogels, immediately after GFP:mutant solution was administered into the hydrogel dish, scans of a horizontal section of the hydrogel droplet were performed at indicated time points. GFP signals of the scanned sections were integrated by the program ImageJ (Rasband, 1997-2011). Initial binding rates were calculated by a linear regression fitting with the program Prism (Graphpad, USA). The scattering plot for calculating correlation between transactivation and hydrogel binding rate of the mutants was made by the program Prism.
Cyclin-dependent Kinase Assay
For release of GFP:CTDC26 from mCherry:TAF15, mCherry:FUS and mCherry:FUS 2A hydrogels by phosphorylation, unphosphorylated GFP:CTDC26 was pre-incubated with the hydrogels overnight. After removing the GFP:CTDC26 solution, the CDK7 or CDK9 reaction mixtures were applied to the GFP:CTDC26-trapped hydrogel droplets. The hydrogel plates were incubated at 30°C for 1 hr. Hydrogels were analyzed by a confocal microscope. For Western blotting of the released protein from the hydrogel droplets, the reaction mixture was carefully removed from the hydrogel plate and mixed with 2 × SDS lysis buffer. After SDS-PAGE, GFP:CTDC26 protein was transferred to a nitrocellulose filter and visualized by Western blotting using anti-GFP antibodies (Chemicon) or the 3E8 phospho-S5 antibody (Millipore).
Biotinylated Isoxazle-mediated Precipitation
Protein precipitation by b-isox chemical from yeast extract and human nuclear extract is described in Supplementary data.
Cloning of Microsatellite DNA
The microsatellite DNA in the promoter region of the hNROB1 gene was amplified by PCR from U2OS cell genomic DNA as a template. The sequence of the microsatellite DNA is shown in Supplementary data.
Transmission Electron Microscopy
DNA-dependent fiber formation of mCherry:FUS-FLI protein was carried out in a reaction mixture containing 20 mM Tris-HCl pH7.5, 200 mM NaCl, 20 mM BME, 0.5 mM EDTA, 0.1 mM PMSF, and 35% glycerol, with or without 20 nM microsatellite DNA. mCherry:FUS-FLI protein stored in 50% glycerol was added to the reaction mixture at the final concentration of 0.5 μM, and the reactions were incubated at room temperature for 1 hr. The sample solution (10 μL) was spotted on a TEM grid (FCF-200-Cu Copper grid from Electron Microscopy Sciences, USA). After blotting the excess solution, the surface of the grid was washed with 10 μL distilled water twice. After removing the excess water, the grid was stained for 10 s with 10 μL drop of 2% uranyl acetate in water. After the uranyl solution was blotted, the grid was dried in air. TEM images were obtained at 120 kV on Tecnai TEM.
Fiber Extension Assays
The mCherry:TAF15 WT, 1F, 2H or 3K protein solution (100 μM) in the gelation buffer was sonicated to break down the preformed fibers, if any, into short fiber seeds. After spin down at 14K rpm for 5 min at 4°C, the protein solution was transferred to new test tubes. After overnight incubation at 4°C, the proteins were 10-fold diluted in the gelation buffer and the formed fibers were detected by fluorescence microscopy.
Pull Down of GFP:CTDC26 by mCherry:TAF15
HA- or Flag-tag was added respectively to the C-terminus of mCherry:TAF15 or GFP:CTDC26 by PCR amplification. mCherry:TAF15 WT, 1F, 2H or 3K-HA proteins were diluted to 10 μM in gelation buffer with 0.05 % NP-40. After 4 hr of incubation at 4°C, 1 μM of GFP:CTDC26-Flag and 10 μl of HA-magnetic beads (Pierce, USA) were added to the each tube containing mCherry:TAF15 solution. The mixtures were incubated for overnight at 4°C with gentle rotation. The magnetic beads were washed twice with the gelation buffer with 0.05 % NP-40. Protein was recovered by boiling at 95°C for 10 min in 2X SDS lysis buffer. Protein was visualized by Western blotting using Flag or HA antibodies.
Supplementary Material
Article Highlights.
Low-complexity domains of FET proteins form reversible polymeric fibers.
Polymeric fiber formation correlates with transcriptional activation capacity.
The C-terminal domain (CTD) of RNA polymerase II binds to polymeric fibers.
Pre-bound CTD is released from polymeric fibers by phosphorylation.
ACKNOWLEDGEMENTS
We thank the shared resource cores in Microscopic Imaging, Proteomics and DNA Sequencing at UTSWMC for technical support; Dr. Robert Tycko for provision of the electron micrograph of amyloid fibers included in the graphical abstract; and Drs. Bruce Albert, Deepak Nijhawan, Michael Brown and Michael Rosen for helpful comments on the composition of the manuscript. This work was funded by an unrestricted endowment provided S.L.M. by an anonymous donor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Molecular cell. 2009;34:387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MK, Stahlberg A, Arvidsson Y, Olofsson A, Semb H, Stenman G, Nilsson O, Aman P. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC cell biology. 2008;9:37. doi: 10.1186/1471-2121-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvand A, Denny CT. Biology of EWS/ETS fusions in Ewing's family tumors. Oncogene. 2001;20:5747–5754. doi: 10.1038/sj.onc.1204598. [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Bell B, Tora L. The N-terminal domain of human TAFII68 displays transactivation and oncogenic properties. Oncogene. 1999;18:8000–8010. doi: 10.1038/sj.onc.1203207. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Molecular cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- Corden JL, Cadena DL, Ahearn JM, Jr., Dahmus ME. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- Cumberworth A, Lamour G, Babu MM, Gsponer J. Promiscuity as a functional trait: intrinsically disordered regions as central players of interactomes. The Biochemical journal. 2013;454:361–369. doi: 10.1042/BJ20130545. [DOI] [PubMed] [Google Scholar]
- Czudnochowski N, Bosken CA, Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nature communications. 2012;3:842. doi: 10.1038/ncomms1846. [DOI] [PubMed] [Google Scholar]
- Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends in genetics : TIG. 2012;28:333–341. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends in genetics : TIG. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Gangwal K, Sankar S, Hollenhorst PC, Kinsey M, Haroldsen SC, Shah AA, Boucher KM, Watkins WS, Jorde LB, Graves BJ, et al. Microsatellites as EWS/FLI response elements in Ewing's sarcoma. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10149–10154. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guipaud O, Guillonneau F, Labas V, Praseuth D, Rossier J, Lopez B, Bertrand P. An in vitro enzymatic assay coupled to proteomics analysis reveals a new DNA processing activity for Ewing sarcoma and TAF(II)68 proteins. Proteomics. 2006;6:5962–5972. doi: 10.1002/pmic.200600259. [DOI] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Huntley MA, Golding GB. Simple sequences are rare in the Protein Data Bank. Proteins. 2002;48:134–140. doi: 10.1002/prot.10150. [DOI] [PubMed] [Google Scholar]
- Ichikawa H, Shimizu K, Katsu R, Ohki M. Dual transforming activities of the FUS (TLS)-ERG leukemia fusion protein conferred by two N-terminal domains of FUS (TLS). Molecular and cellular biology. 1999;19:7639–7650. doi: 10.1128/mcb.19.11.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Phatnani HP, Haystead TA, MacDonald JA, Alam SM, Greenleaf AL. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. The Journal of biological chemistry. 2004;279:24957–24964. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Suh H, Cho EJ, Buratowski S. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. The Journal of biological chemistry. 2009;284:26421–26426. doi: 10.1074/jbc.M109.028993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemercier C, Verdel A, Galloo B, Curtet S, Brocard MP, Khochbin S. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. The Journal of biological chemistry. 2000;275:15594–15599. doi: 10.1074/jbc.M908437199. [DOI] [PubMed] [Google Scholar]
- Lessnick SL, Ladanyi M. Molecular pathogenesis of Ewing sarcoma: new therapeutic and transcriptional targets. Annual review of pathology. 2012;7:145–159. doi: 10.1146/annurev-pathol-011110-130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Rabbitts TH, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nature genetics. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- Ramanathan Y, Rajpara SM, Reza SM, Lees E, Shuman S, Mathews MB, Pe'ery T. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. The Journal of biological chemistry. 2001;276:10913–10920. doi: 10.1074/jbc.M010975200. [DOI] [PubMed] [Google Scholar]
- Rickert P, Corden JL, Lees E. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene. 1999;18:1093–1102. doi: 10.1038/sj.onc.1202399. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, Bushnell DA, Trnka MJ, Burlingame AL, Kornberg RD. Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17931–17935. doi: 10.1073/pnas.1215241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Adamczewski JP, Seroz T, Vermeulen W, Tassan JP, Schaeffer L, Nigg EA, Hoeijmakers JH, Egly JM. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Ebmeier CC, Podell ER, Heimiller J, Taatjes DJ, Cech TR. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes & development. 2012;26:2690–2695. doi: 10.1101/gad.204602.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein expression and purification. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Trigon S, Serizawa H, Conaway JW, Conaway RC, Jackson SP, Morange M. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. The Journal of biological chemistry. 1998;273:6769–6775. doi: 10.1074/jbc.273.12.6769. [DOI] [PubMed] [Google Scholar]
- Tsai KL, Sato S, Tomomori-Sato C, Conaway RC, Conaway JW, Asturias FJ. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nature structural & molecular biology. 2013;20:611–619. doi: 10.1038/nsmb.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein science : a publication of the Protein Society. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes & development. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.