Abstract
Aim:
To develop a pharmacokinetic/pharmacodynamic (PK/PD) model describing the receptor/gene-mediated induction of CYP3A1/2 by dexamethasone (DEX) in rats.
Methods:
A group of male Sprague-Dawley rats receiving DEX (100 mg/kg, ip) were sacrificed at various time points up to 60 h post-treatment. Their blood sample and liver were collected. The plasma concentration of DEX was determined with a reverse phase HPLC method. CYP3A1/2 mRNA, protein levels and enzyme activity were measured using RT-PCR, ELISA and the testosterone substrate assay, respectively. Data analyses were performed using a first-order conditional estimate (FOCE) with INTERACTION method in NONMEM version 7.1.2.
Results:
A two-compartment model with zero-order absorption was applied to describe the pharmacokinetic characteristics of DEX. Systemic clearance, the apparent volume of distribution and the duration of zero-order absorption were calculated to be 172.7 mL·kg−1·h−1, 657.4 mL/kg and 10.47 h, respectively. An indirect response model with a series of transit compartments was developed to describe the induction of CYP3A1/2 via PXR transactivation by DEX. The maximum induction of CYP3A1 and CYP3A2 mRNA levels was achieved, showing nearly 21.29- and 8.67-fold increases relative to the basal levels, respectively. The CYP3A1 and CYP3A2 protein levels were increased by 8.02-fold and 2.49-fold, respectively. The total enzyme activities of CYP3A1/2 were shown to increase by up to 2.79-fold, with a lag time of 40 h from the Tmax of the DEX plasma concentration. The final PK/PD model was able to recapitulate the delayed induction of CYP3A1/2 mRNA, protein and enzyme activity by DEX.
Conclusion:
A mechanism-based PK/PD model was developed to characterize the complex concentration-induction response relationship between DEX and CYP3A1/2 and to resolve the drug- and system-specific PK/PD parameters for the course of induction.
Keywords: drug-drug interactions, dexamethasone, CYP induction, CYP3A1/2, pharmacokinetics, pharmacodynamics, NONMEM
Introduction
Clinical drug-drug interaction (DDIs) is a major problem, largely resulting from the inhibition or induction of drug-metabolizing enzymes (DMEs), particularly cytochrome P450s (CYPs)1. Although CYP induction-mediated DDIs are less frequent and less of a safety concern than CYP inhibition-mediated DDIs2, these interactions can nevertheless reduce both the exposure and the pharmacological effect of a drug; this can occur when the drug is cleared by an enzyme that is induced by a coadministered drug. The induction of CYP3A is particularly important because it is the major CYP isoform and has been estimated to be involved in the metabolism of approximately 50% of marketed drugs3.
The induction of CYPs has been well known for decades. However, the underlying mechanisms of this induction still remain to be understood3. The major mechanism of rat CYP3A1/2 induction is via an increased rate of gene transcription mediated by the pregnane X receptor (PXR)4. Briefly, the mechanism of rat CYP3A1/2 induction involves the binding of an inducer to PXR, the formation of an inducer-bound PXR/RXR (retinoid X-receptor) complex, translocation of the complex to the nucleus, and binding of the complex to DNA-responsive elements in the 5′-regulatory region of target genes (ie, CYP3A1/2) thereby triggering the transcription of these genes. Many drugs that induce CYP3A1/2 are PXR ligands and have demonstrated transactivation in the PXR reporter assay.
Dexamethasone (DEX) has been widely used to treat patients with inflammatory and autoimmune conditions5, 6, 7, and it has been reported to be a remarkable inducer of both human CYP3A4 and rat CYP3A1/28, 9, 10. Recently, multiple in vitro and in vivo studies have demonstrated that the pharmacodynamics (PD) of CYP3A1/2 induction by DEX are dose- and time-dependent11, 12, 13. However, none of these studies explored the quantitative relationship between DEX exposure and the resultant alterations in CYP3A1/2 mRNA levels, protein levels and enzyme activity using a pharmacokinetic/pharmacodynamic (PK/PD) modeling approach.
The objective of the present study was to develop a mechanism-based PK/PD model that could quantitatively characterize the time-dependent relationship among the multiple elements (CYP3A1/2 mRNA, protein and enzyme activity) of CYP3A1/2 induction in rats treated with DEX. The key element in mechanism-based PK/PD modeling is the explicit distinction between parameters that describe drug-specific properties and those that describe biological system-specific properties14. As a result, the potential of unknown compounds to induce CYP3A1/2 could be predicted and simulated by this mechanism-based PK/PD model solely by utilizing knowledge of the in vitro potency (Smax and SC50) and single-dose in vivo pharmacokinetics of these compounds.
Materials and methods
Animals and treatments
Normal male Sprague-Dawley rats weighing 200–250 g were purchased from the Department of Laboratory Animal Science at the Peking University Health Science Center. The animals were housed and acclimatized in a temperature- (22 °C) and light-controlled (12 h/12 h light/dark) animal care unit for 10 d before experimentation. They were allowed food and water ad libitum. All of the experiments were conducted in accordance with the European Community guidelines for the use of experimental animals and were approved by the Peking University Committee on Animal Care and Use.
Rats weighing 200–250 g received DEX (Sigma-Aldrich Inc, USA, 100 mg/kg in 5 mL/kg of corn oil) intraperitoneally (ip) after fasting for 12 h prior to the experiments. For the PK analysis, serial blood samples were collected from three treated rats at various times after dosing (0.083, 0.25, 0.5, 0.75, 1, 2, 3, 4, 6, 8, 12, 16, 24, 30, 36, and 48 h). The collected blood was centrifuged at 3000×g at 4 °C for 10 min. Plasma samples were then stored at -80 °C until analysis. In the PD study, 84 rats were randomly sorted into DEX or control (corn oil only) treatment groups. The treated rats were anesthetized with 60 mg/kg pentobarbital sodium and sacrificed at 0, 1, 2, 4, 8, 12, 16, 24, 30, 36, 42, 48, 54, and 60 h (n=3 per time point). For the CYP3A1/2 enzyme activity assays, approximately 4 g of liver was rapidly excised for the preparation of liver microsomes. The remaining liver tissues were flash-frozen in liquid nitrogen and stored at -80°C for RNA extraction.
Plasma concentration assays
The DEX concentrations were determined by a reverse phase high performance liquid chromatography (HPLC) method15. The lower limit of quantification was 0.25 μg/mL. The variability of the assay was <15%, and the inter-day and intra-day coefficients of variation (CV) were less than 10%.
CYP3A1 and CYP3A2 mRNA measurements
RNA isolation and cDNA synthesis
Total RNA was extracted from the rat livers using the TRIzol reagent (Invitrogen, USA), according to the manufacturer's protocol. The amount of purified RNA was determined by UV spectroscopy measurements at 260 nm, and its purity was determined by calculating the ratio of UV activity at 260 nm/280 nm. cDNA was reverse-transcribed from 0.5 μg of total RNA using random primers (25 μmol/L), a dNTP mixture (10 mmol/L), RTase M-MLV (RNase H-) (200 U/μL), M-MLV buffer (5×) and an RNase inhibitor (40 U/μL) (Takara, Dalian, China).
Construction of reference standards
The reference standards were constructed for the absolute quantification of CYP3A1, CYP3A2, and GAPDH (the latter used as a house-keeping control) by real-time PCR. Fragments of these three target genes were cloned by conventional PCR using the appropriate primer sets16, as listed in Table 1. The presence of the specific amplified products was confirmed by electrophoresis using a 1.2% agarose gel with ethidium bromide. The purification of the amplification products was performed using an Agarose Gel DNA Purification Kit (Takara, Dalian, China), according to manufacturer's recommendations. The DNA manipulation techniques, which included the insertion of the amplification products into the pMD19-T vector (Takara, Dalian, China), the transformation of the vector into competent Escherichia coli (JM109) (Takara, Dalian, China), screening for positive clones, plasmid DNA preparation and so on, were performed according to standard protocols. The identity of the cloned insert was confirmed by direct sequencing, which was performed using the universal sequencing primer M13F (-47) by GenScript Co (China).
Table 1. Primers used for real-time PCR.
Real-time PCR amplification and dissociation curve analysis
Real-time PCR was performed using an ABI PRISM 7300 real-time quantitative PCR system (ABI, USA), using SYBR® Premix Ex Taq™ (Takara, Dalian, China), according to the manufacturer's instructions. Following cycling, dissociation curves were obtained by heating and cooling the samples at the following conditions: 95 °C for 15 s, 60 °C for 30 s, and 95 °C for 15 s17. The standard curves for CYP3A1, CYP3A2, and GAPDH were constructed using standard values obtained by 10-fold serial dilutions, ranging from 1 attomol/μL to 1×106 attomol/μL, of the extract of plasmids harboring the target insert. Reference standard curves were generated for each assay, and the standards and unknown samples were run in triplicate. The controls, which consisted of water and the mRNA control, were used to determine the extent of primer-dimer formation and DNA contamination in the isolated samples, respectively. The amplification plots and dissociation curves of the controls did not show any signal or dissociation product, suggesting the lack of primer-dimer formation and genomic DNA contamination in the RNA samples. The slope, intercept, percent efficiency (E) and R2 values from the three standard curves in each experiment were used to calculate the intra-assay CVs and the inter-assay reproducibility. Low CVs (less than 15%) for both inter- and intra-assay variability implied good reproducibility of the real-time PCR standard curves for CYP3A1, CYP3A2, and GAPDH.
CYP3A1 and CYP3A2 protein measurements
For the CYP3A1/2 protein measurements and the CYP3A1/2 enzyme activity assays, liver microsomes were prepared from fresh livers using a differential centrifugation method, as described previously18. The protein concentration of the microsomal suspension was determined using the BCA assay kit (Biomiga Inc, San Diego, USA), according to the manufacturer's protocol. After determining the protein concentration, the microsomal suspension was diluted with 0.25 mol/L sucrose solution to a final protein concentration of 10 mg/mL and was stored at -80 °C until use. The CYP3A1 and CYP3A2 protein concentrations were determined by a non-competitive enzyme-linked immunosorbent assay (ELISA)19. The amount of microsomal proteins used was 0.1 μg for CYP3A1 and 1 μg for CYP3A2. The isoform protein values are expressed as pmol per milligram of protein.
CYP3A1/2 enzyme activity
The CYP concentrations of rat liver microsomes were determined according to the spectrophotometric method described by Omura and Sato20. The CYP3A1/2 enzyme activities were evaluated by the testosterone substrate assay21. The separation and detection of testosterone and its metabolites were performed using a Bisep™-1100 HPLC system (Unimicro Technologies Inc, USA), according to a previously described method22.
Pharmacokinetic/pharmacodynamic modeling
PK analysis
A two-compartment mammillary model with zero-order absorption was used to describe the plasma PK after administration of 100 mg/kg DEX. The equations describing the model were as follows:
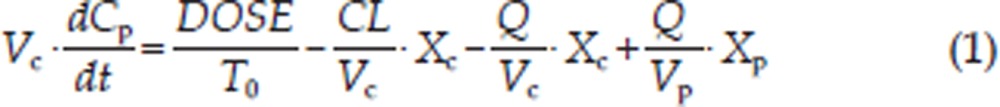 |
 |
where Xc and Xp are the amount of DEX in the central and peripheral compartments, respectively; T0 is the duration of the zero-order phase of absorption; Cp is the plasma concentration; Vc is the central volume of distribution; Vp is the peripheral volume of distribution; CL is the systemic clearance and Q is the inter-compartmental clearance.
PK/PD analysis
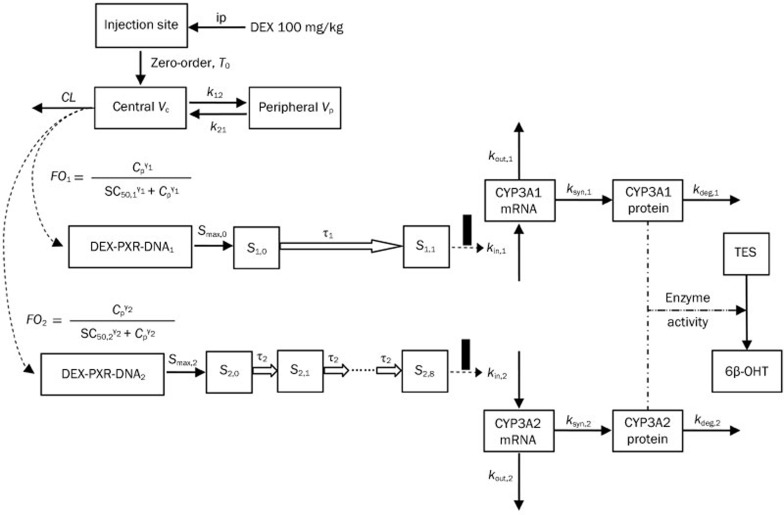
A schematic for the PK/PD model for CYP3A1/2 induction is presented in Figure 1. This model included three elements: CYP3A1/2 mRNA dynamics, CYP3A1/2 protein dynamics and CYP3A1/2 enzyme activity.
Figure 1.
A schematic picture for the PK/PD model for CYP3A1/2 induction.
CYP3A1 and CYP3A2 mRNA dynamics
Graphical analyses of the CYP3A1 and CYP3A2 mRNA concentrations versus time demonstrated a considerable time delay between CYP3A1/2 mRNA and drug concentration (Figures 2 and 3). Therefore, an indirect response model with a chain of transit compartments, as described previously23, 24, was applied for the transduction of the DEX exposure to the mRNA response. In the absence of the drug, it was assumed that CYP3A1 and CYP3A2 mRNA were produced from their DNA according to a zero-order rate constant, kin, and degraded according to a first-order rate constant, kout. The effect of the DEX concentration on the transcription rates of mRNA was described as a PXR-mediated process. The fractional occupancy (FO) of the DNA responsive elements by the DEX-PXR complex was related to the DEX plasma concentration and was calculated from the DEX concentration using a Hill equation, as reported by Sarangapani25. The time delay accounting for the transcription process of CYP3A1 and CYP3A2 mRNA was characterized by two parallel chains of transit compartments, as shown in Figure 1. The optimal number of transit compartments was assessed by stepwise addition or deletion of one transit compartment and was selected from the inflection point on the objective function value (OFV) versus compartment number curve26, 27. The equations describing this chain of events are as follows:
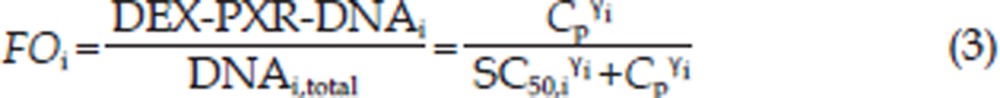 |
 |
 |
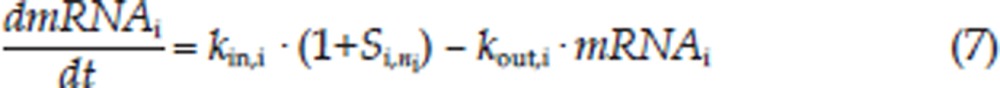 |
The initial condition for CYP3A mRNA is as follows:
 |
where i is the identification for the CYP3A subtype: 1 for CYP3A1 and 2 for CYP3A2; DEX-PXR-DNAi is the concentration of the DNA ligand binding sites bound to the DEX-PXR complex; DNAi,total is the total concentration of binding sites for this complex; Si,0 is the stimulation constant; Smax,i is the maximum stimulation of the transcription rate of CYP3A mRNA (kin,i); kout,i is the degradation rate constant for CYP3A mRNA; SC50,i is the DEX concentration required to reach 50% Smax,i; γi is the Hill coefficient; Si,ni is the nith transit compartment of the stimulation and τi is the mean transit time between each transit compartment.
CYP3A1 and CYP3A2 protein dynamics
CYP3A1 and CYP3A2 mRNA induction was followed by an increase of CYP3A1 and CYP3A2 protein expression. This induction is described by the following equation:
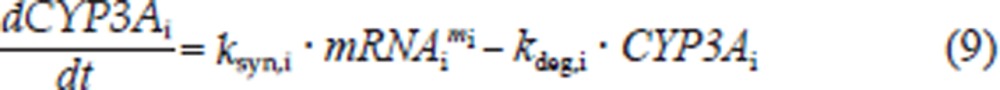 |
The initial condition for each CYP3Ai protein is as follows:
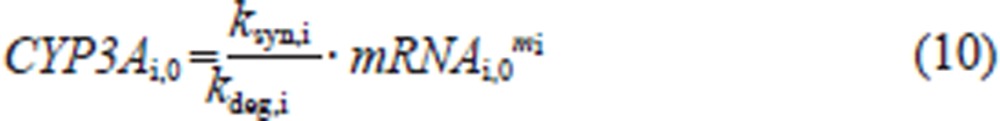 |
where ksyn,i is the synthesis rate constant, assuming that the translation of CYP3Ai protein is proportional to its mRNA level; kdeg,i is the first-order degradation rate constant of CYP3Ai protein and mi is the amplification factor, indicating that one copy of the mRNA can be translated into multiple copies of the protein.
CYP3A1/2 enzyme activity
Because the measurement of 6β-hydroxy testosterone (6β-OHT) formation is a reliable indicator of CYP3A1/2 activity levels28 and all of the reactions were performed in the linear range, with respect to CYP concentration and incubation time, the formation of 6β-OHT (EA, enzyme activity) was described to be proportional to the CYP3A1 and CYP3A2 protein concentrations:
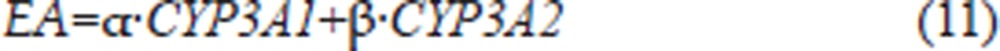 |
where α and β are the rates of formation of 6β-OHT for CYP3A1 and CYP3A2, respectively, and their values were expressed as pmol of 6β-OHT per min per pmol of CYP3A1 or CYP3A2.
Data analysis
The development of the PK/PD model was performed in two steps using a sequential process, as described previously29. First, the pharmacokinetic data were modeled. Then, the PK parameters were fixed, and the predicted DEX concentration-time profile was used as an input function for the entire PK/PD model. All of the three PD models described by Equations (3)–(11) were estimated simultaneously. This approach is computationally much faster than the simultaneous estimation of the PK and PD models and is not expected to compromise the precision and accuracy of the PD parameter estimates, unless the PK model is misspecified29, 30. All of the analyses were performed using a first-order conditional estimate (FOCE) with INTERACTION method in NONMEM version 7.1.2 (Icon Development Solutions, MD, USA).
The animals used in the PK study contributed to several measurements, and the population approach was used to compute the mean population estimates. An exponential error model was selected for modeling the between-subject variability (BSV). In contrast, the PD data were fit using the naïve pool approach because each animal contributed with a single measurement; therefore, it was not possible to distinguish between inter-animal and residual variability. The residual variability for both the PK and PD models was modeled initially with a combined error model; if one of the components (additive or proportional) of the residual was negligible, it was deleted from the model.
The goodness of fit was assessed by model convergence, precision in parameter estimates, decrease in objective function value (χ2=10.83, P<0.001, df=1) and examination of the residuals. The ability of the PK/PD model to describe the observed data was evaluated by a visual predictive check (VPC) based on 1000 data sets that were simulated with the obtained final parameter estimates, and the median and 2.5th and 97.5th percentiles were calculated. Exploratory analyses, graphical displays and other statistical analyses, including the evaluation of the NONMEM outputs, were performed (version 2.12.1 for Windows).
Results
Pharmacokinetics
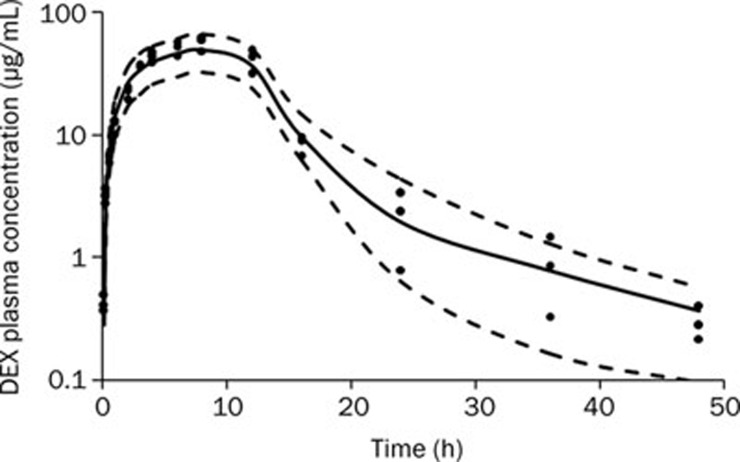
The time course of plasma DEX concentrations after 100 mg/kg ip administration is shown in Figure 2. During an early exploratory analysis, several models (ie, one-, two-, and three-compartment PK models with zero-order or first-order absorption processes were developed to fit to the time course of the DEX plasma concentration. The two-compartment model had a lower OFV than the one-compartment model (123.188 versus 142.226; P<0.001 by χ2 test; df=2), and it showed a more favorable distribution of CWRES over time. The addition of an extra peripheral compartment did not yield a significantly lower OFV. Absorption of DEX from the peritoneal cavity was best characterized by a zero-order kinetic process because this yielded a significantly better fit than a first-order absorption rate constant. The OFV was 45.60 units lower with zero-order absorption. The estimates of the PK parameters are listed in Table 2. Only the BSV for inter-compartment clearance (Q) was included in the final PK model. The T0 (the duration of the zero-order absorption) obtained was 10.47 h. The maximal plasma concentration (Cmax) of DEX was achieved within 8 h and declined bi-exponentially, with a terminal half-life (T1/2) of 2.64 h. The volume of distribution at steady state (Vss, equal to the sum of Vc and Vp) was very large, indicating that DEX has a significant extravascular distribution in rats. The PK profile for DEX was similar to that reported by a previous study31.
Figure 2.
Time course of plasma DEX concentrations after 100 mg/kg ip administration. The symbols represent individual data from rats and the solid line represents the model fit lines. The dashed lines depict 95% prediction interval of the PK model.
Table 2. Pharmacokinetic parameters for dexamethasone.
| Parameters | Definitions | Values (% RSE) | BSV (% RSE) |
|---|---|---|---|
| CL/F (mL·kg−1·h−1) | Clearance | 172.7 (6.70) | – |
| Q/F (mL·kg−1·h−1) | Inter-compartmental clearance | 14.32 (36.52) | 0.655 (36.66) |
| Vc/F (mL/kg) | Central volume | 657.4 (12.55) | – |
| Vp/F (mL/kg) | Peripheral volume | 263.2 (31.95) | – |
| T0 (h) | The duration of the zero-order absorption | 10.47 (6.36) | – |
BSV, between subject variability; RSE, relative standard error.
CYP3A1 and CYP3A2 mRNA dynamics
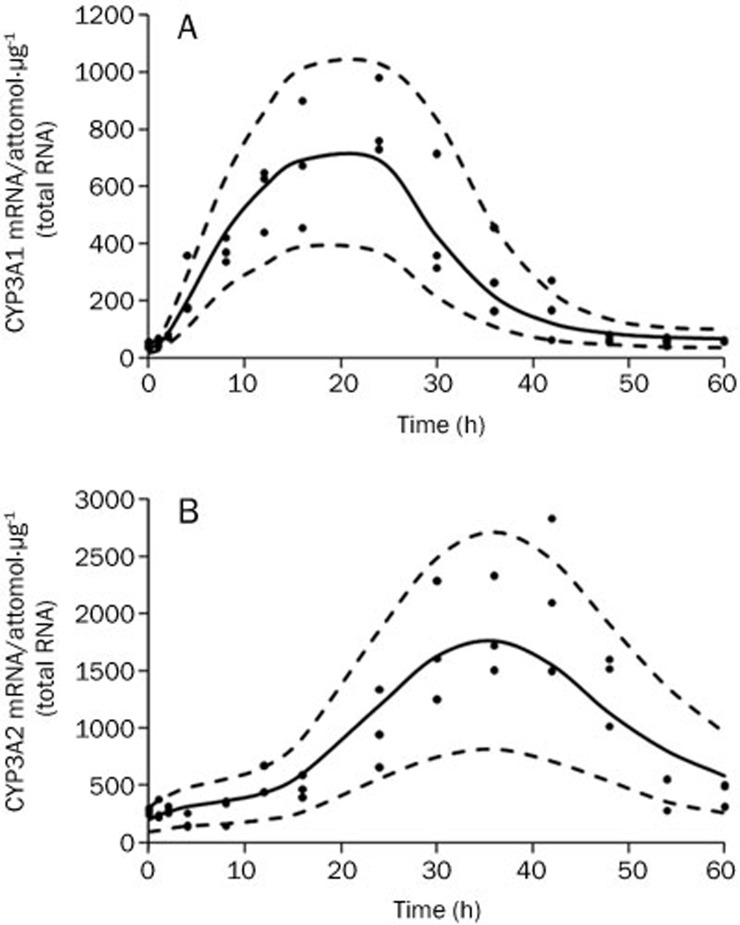
The CYP3A1 and CYP3A2 mRNA levels in vehicle-treated animals remained relatively constant throughout the time course (data not shown), indicating that the baselines were stable. The model corresponding to Equations (3)–(8), with different numbers of transit compartments, was evaluated to describe the profiles of CYP3A1 and CYP3A2 mRNA concentrations versus time. The transit model using one transit compartment provided the lowest OFV and the best fit for the CYP3A1 mRNA induction profile. For CYP3A2 mRNA, an OFV of 1836.70 was obtained for the model with no transit compartment, whereas an OFV of 1794.16 was obtained for a transit compartment model using eight transit compartments. The OFV continued to decrease with the inclusion of additional transit compartments, but there was no further visual improvement in the model predictions. The results corresponding to the VPC, as shown in Figure 3, confirmed that the model fit the data and was appropriate to describe the mean tendency of the data and their dispersion. The estimated CYP3A1 and CYP3A2 mRNA baseline levels were 32.48 attomol/μg (total RNA) and 203.46 attomol/μg (total RNA), respectively. The CYP3A1 and CYP3A2 mRNA values increased gradually to their peak levels (21.29- and 8.67-fold increase, respectively, vs baseline) within 24 h and 36 h, respectively, and then returned to their baseline levels 60 h after drug administration. The estimated parameters for CYP3A1 and CYP3A2 mRNA dynamics are listed in Table 3. The estimated Smax,i values for CYP3A1 and CYP3A2 mRNA synthesis were 22.42- and 8.55-fold of the kin,i, respectively. The high Hill coefficient estimates (8.01 and 5.00 for CYP3A1 and CYP3A2 mRNA, respectively) could be explained by one of the following: 1) the CYP3A1/2 DNA responsive elements were initially saturated with the DEX-PXR complex because the plasma concentration was much higher than the SC50,i values (2.39 μg/mL for CYP3A1 mRNA and 2.82 μg/mL for CYP3A2 mRNA) within 18 h after DEX administration, or 2) the affinity of the DEX-PXR complex binding to CYP3A1/2 DNA responsive elements could be increased dramatically by heterodimerization with the nuclear hormone receptor RXR32. Subsequently, the two chains of transit compartments with different numbers of compartments and mean transit times resolved the delay of the mRNA transcription relative to the concentration of DEX. The first chain for CYP3A1 mRNA had one compartment and a τ1 of 4.59 h, whereas the second chain for CYP3A2 mRNA had eight compartments and a τ2 of 2.58 h.
Figure 3.
Time course of CYP3A1 (A) and CYP3A2 (B) mRNA expression. The symbols represent individual data from rats and the solid line represents the model fit lines. The dashed lines depict 95% prediction interval of the mRNA PD models.
Table 3. Dynamic parameters for CYP3A1 and CYP3A2 mRNA after DEX administration.
| Parameters | Definitions | Values (% RSE) |
|---|---|---|
| kin,1 (attomol·h−1·μg−1 total RNA) | Transcription rate for CYP3A1 mRNA | 7.47 (4.65) |
| kout,1 (h−1) | Degradation rate for CYP3A1 mRNA | 0.23 (6.45) |
| Smax,1 | Maximum stimulation of the transcription rate of CYP3A1 mRNA | 22.42 (10.90) |
| SC50,1 (μg/mL) | DEX concentration required to reach 50% Smax,1 | 2.39 (15.89) |
| γ1 | Hill-type parameter for CYP3A1 mRNA | 8.01 (6.92) |
| τ1 (h) | Mean transit time of the stimulation of kin,1 | 4.59 (4.71) |
| n1 | Number of the transit compartments for CYP3A1 mRNA | 1 (Fixed) |
| kin,2 (attomol·h−1·μg−1 total RNA) | Transcription rate for CYP3A2 mRNA | 40.00 (19.56) |
| kout,2 (h−1) | Degradation rate for CYP3A2 mRNA | 0.197 (21.00) |
| Smax,2 | Maximum stimulation of the transcription rate of CYP3A2 mRNA | 8.55 (11.72) |
| SC50,2 (μg/mL) | DEX concentration required to reach 50% Smax,2 | 2.82 (38.50) |
| γ2 | Hill-type parameter for CYP3A2 mRNA | 5.00 (13.16) |
| τ2 (h) | Mean transit time of the stimulation of kin,2 | 2.58 (12.79) |
| n2 | Number of the transit compartments for CYP3A2 mRNA | 8 (Fixed) |
RSE, relative standard error.
CYP3A1 and CYP3A2 protein dynamics
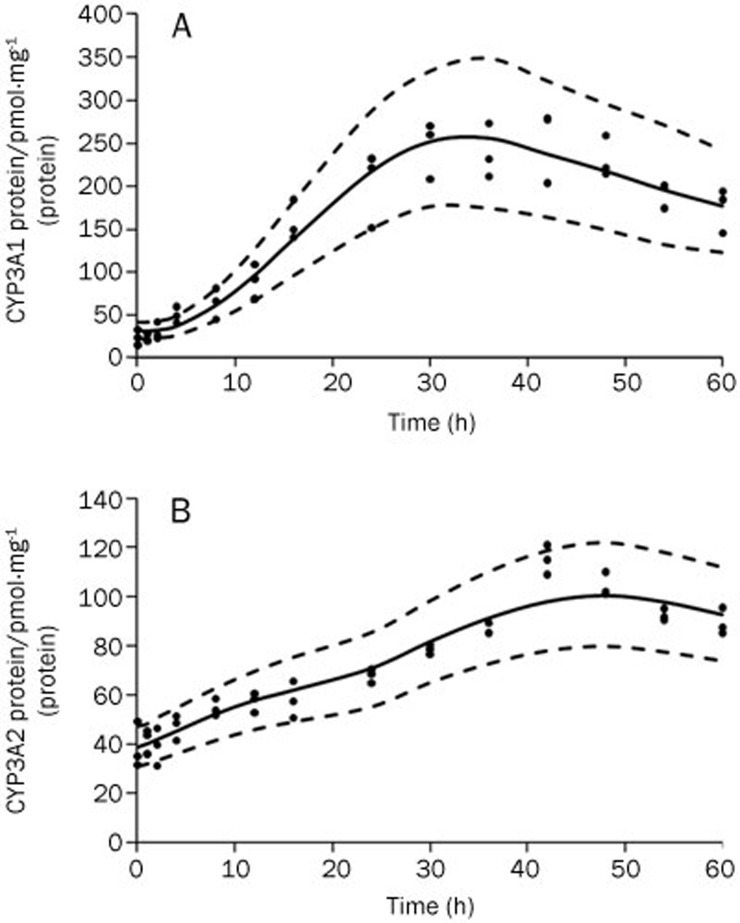
The time courses of CYP3A1 and CYP3A2 protein expression were simulated by the model (Figure 4), and the lag time between mRNA and protein expression changes was approximately 12 h. The CYP3A1 protein exhibited an inductive potency in DEX-treated rats (from 31.92 to 256.03 pmol·mg−1 protein, an 8.02-fold increase), which was greater than that of the CYP3A2 protein (from 39.39 to 97.99 pmol·mg−1 protein, a 2.49-fold increase). The estimated parameters for Equation (9) are listed in Table 4. A visual predictive check indicated that the time courses of CYP3A1 and CYP3A2 protein dynamics were well described by the model.
Figure 4.
Time courses of CYP3A1 protein (A) and CYP3A2 protein (B) after DEX administration. The symbols represent individual data from rats and the solid line represents the model fit lines. The dashed lines depict 95% prediction interval of the protein PD models.
Table 4. Pharmacodynamic parameters for CYP3A1 and CYP3A2 protein after DEX administration.
| Parameters | Definitions | Values (% RSE) |
|---|---|---|
| ksyn,1 (pmol·h−1·mg−1 protein)/(attomol mRNA·μg−1 total RNA) | Synthesis rate for CYP3A1 protein | 0.0359 (18.68) |
| kdeg,1 (h−1) | Degradation rate for CYP3A1 protein | 0.0268 (15.99) |
| m1 | Amplification factor for CYP3A1 protein | 0.911 (3.99) |
| ksyn,2 (pmol·h−1·mg−1 protein)/(attomol mRNA·μg−1 total RNA) | Synthesis rate for CYP3A2 protein | 0.486 (11.77) |
| kdeg,2 (h−1) | Degradation rate for CYP3A2 protein | 0.0567 (11.79) |
| m2 | Amplification factor for CYP3A2 protein | 0.287 (9.68) |
RSE, relative standard error.
CYP3A1/2 enzyme activity dynamics
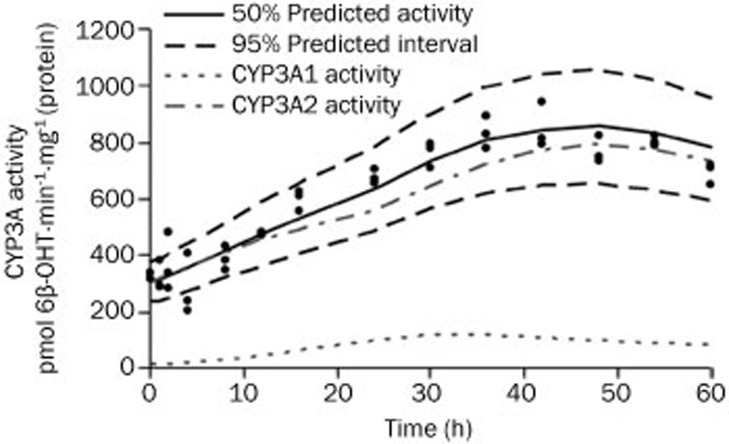
The maximum induction of testosterone 6β-hydroxylation activity was observed after DEX dosing, as depicted in Figure 5. Because both CYP3A1 and CYP3A2 proteins are capable of catalyzing 6β-hydroxylation of testosterone, the enzyme activities were composed of two parts, as described by Equation (12). The gray dashed line and the dotted line in Figure 5 show the contributions of CYP3A1 and CYP3A2, respectively, to 6β-OHT formation, which were generally parallel to their protein levels. The rates of formation of 6β-OHT for CYP3A1 and CYP3A2, as shown in Table 5, were estimated to be 0.463 pmol·min−1·pmol−1 CYP3A1 and 7.49 pmol·min−1·pmol−1 CYP3A2, respectively.
Figure 5.
Time course of CYP3A1/2 enzyme activity after DEX administration. The symbols represents individual data from rats and the solid line represents the model fit lines. The black dashed lines depict 95% prediction interval of the enzyme activity dynamic model. The gray dashed line and the dotted line represent the simulated contributions of CYP3A1 and CYP3A2 to 6β-OHT formation, respectively.
Table 5. Pharmacodynamic parameters for CYP3A1/2 enzyme activity after DEX administration.
| Parameters | Definitions | Values (% RSE) |
|---|---|---|
| α (pmol·min−1·pmol−1 CYP3A1) | Rate of formation of 6β-OHT for CYP3A1 | 0.463 (22.73) |
| β (pmol·min−1·pmol−1 CYP3A2) | Rate of formation of 6β-OHT for CYP3A2 | 7.49 (5.51) |
RSE, relative standard error.
Discussion
In the present study, a mechanism-based PK/PD model was successfully developed to characterize the complex relationships between DEX PK and the time courses of CYP3A1/2 mRNA, protein and enzyme activity in male Sprague-Dawley rats.
We found that a conventional two-compartment model with a zero-order absorption process best described the pharmacokinetics of DEX in our study. Both one-compartment33, 34 and two-compartment models35 have been found to best describe DEX plasma concentration profiles in other research studies. Our ability to differentiate the bi-exponential elimination of DEX may have been facilitated by the relatively long sampling duration. The CL and Vc estimates for DEX obtained in our study were comparable to the values reported by Varma34. The first-order absorption of DEX has been reported previously36. However, first-order absorption was not an appropriate fit for our data because of the saturation of absorption from the peritoneal cavity when a high dosage of DEX was administered.
The PD data from this study demonstrated that a single dose of DEX could generate markedly elevated levels of CYP3A1 and CYP3A2 mRNA, protein and enzyme activities. The total plasma concentration of DEX was used as a forcing function in the PK/PD system, with the assumptions of rapid diffusion into hepatocytes, binding with PXR and translocation to the nucleus. Our mechanism-based PD model was developed based on the model introduced by Sarangapani25, which described the time course of CYP2B1/2 proteins, their enzyme activities in the liver and increases in liver weight induced by octamethylcyclotetrasiloxane (D4). In this previous study, the authors did not investigate the relationship between CYP2B1/2 mRNA and the inducer. The resultant mRNA regulation serves as a key component of the signaling pathway downstream of CYP induction, and it has been widely used as an important indicator of CYP induction during drug development3. Thus, the CYP3A1/2 mRNA levels were incorporated into our mechanism-based PK/PD model to enhance the integrity and stability of the models and to provide a better understanding of the sequence of molecular events that cause CYP3A1/2 induction. The time course of CYP3A1/2 mRNA was described using an indirect response model, where the impact of DEX on the synthesis rate of CYP3A1/2 mRNA was introduced by a receptor-based sigmoidal induction model with Smax, SC50, and γ parameters. Several functions with an increase in complexity were also tested to select the optimal model for the effect of DEX on the rates of CYP3A1 and CYP3A2 mRNA synthesis. The sigmoid Smax model in Equations (3)–(4) was shown to better fit the data than the linear model (ΔOFV=-31.47, P<0.0001 for CYP3A1 mRNA; ΔOFV=-37.18, P<0.0001 for CYP3A2 mRNA) or the Smax model (ΔOFV=-41.54, P<0.0001 for CYP3A1 mRNA; ΔOFV=-23.28, P<0.0001 for CYP3A2 mRNA).
The indirect response model was found to adequately describe CYP2B1/2 protein profiles after D4 administration. However, the indirect response model was not appropriate for our study because significant time delays between DEX exposure and CYP3A1/2 mRNA induction (approximately 16 h for CYP3A1 mRNA and 28 h for CYP3A2 mRNA) were observed. These delayed onsets of induction are likely due to the complex series of events that are required to trigger an induction in transcription37. In our study, we applied a series of transit compartments, which explained the time spent by mRNA maturation, to account for this observed temporal delay between the DEX plasma concentration and the increase in mRNA levels. Our indirect response models, with a series of one transit compartment and eight transit compartments, adequately characterized the general trends of CYP3A1 and CYP3A2 mRNA dynamics, respectively.
Differential induction of CYP3A1 and CYP3A2 mRNA expression by DEX was observed; this disparity was likely a result of a difference in the rate of CYP3A1 and CYP3A2 transcription. The estimated maximum change in synthesis rates for CYP3A1 and CYP3A2 mRNA due to induction were 22.42- and 8.55-fold, respectively. As reported, the CYP3A1 and CYP3A2 promoters both contain HNF4-binding sites and two DEX response elements, referred to as DexRE-1 and DexRE-238. The DR-3 sequence in DexRE-2, which can be bound by the heterodimer of PXR and RXR, plays a key role in CYP3A1/2 mRNA induction4, 39. The DexRE-1 of CYP3A2 is mainly occupied by COUP-TF, whereas the DexRE-1 of CYP3A1 is preferentially bound by protein complex B, which can stimulate the binding of PXR/RXR to DexRE-239. Therefore, although the CYP3A1 and CYP3A2 genes share highly comparable promoter structures, small sequence differences in DexRE-1 may contribute to the apparent discrepancies in the transcription rates of CYP3A1 and CYP3A2 mRNA.
The baseline levels of CYP3A1 and CYP3A2 protein in liver microsomes were 28.95 pmol/mg and 48.60 pmol/mg, respectively, and this result was consistent with the values previously reported in the literature40. Experiments from previous studies showed significant induction of CYP3A1 and CYP3A2 proteins in rats treated with the same dose of DEX41, 42, which is similar to the results obtained in the present study. Although CYP3A2 protein was less inducible than CYP3A1, the contribution of CYP3A2 protein to 6β-OHT formation was greater than that of CYP3A1 during the time course of induction (Figure 5 and Table 5). This phenomenon was likely observed because CYP3A2 has higher basal protein expression levels than CYP3A1.
Mechanism-based PK/PD models differ from conventional PK/PD models in that they contain specific expressions to characterize, in a quantitative manner, processes in the causal path between drug administration and effect14. In addition, mechanism-based PK/PD models can resolve drug-specific and biological system-specific parameters. Because the system-specific parameters (ie, kin, kout, τ, ksyn, kdeg, α, and β) characterize the physiological functioning of CYP3A1/2 mRNA, protein and enzyme activity in the rat, they should remain generally consistent within the Sprague-Dawley rat population. These values of system-specific parameters can only be estimated by in vivo analysis14. In contrast, drug-specific parameters (ie, Emax and EC50) can often be predicted based on in vitro bioassays43, 44. As a result, this mechanism-based PK/PD approach may serve as a tool for predicting the extent of CYP3A1/2 induction by potential inducers and the magnitude of changes in the PK of a probe substrate, based on drug-specific parameters and the PK of the inducers.
In summary, the present study sought to establish a mechanism-based PK/PD model to characterize the effect of DEX on CYP3A1/2 induction as a function of time. The results of this study illustrate the relationships between the molecular events involved in CYP3A1/2 induction by a single dose of DEX. In addition, the system-specific parameters of this mechanism-based PK/PD model could help to design and conduct DDI studies with the aim of regulating the induction of CYP3A1/2.
Author contribution
Wei LU, Tian-yan ZHOU, and Liang LI designed the research; Liang LI, Zai-quan LI, Miao-ran NING, and Han-qing LI performed the research; Liang LI, Wei LU, Chen-hui DENG, and Shan-shan BI analyzed the data; and Liang LI, Wei LU, and Tian-yan ZHOU wrote the paper.
Acknowledgments
The authors would like to express their appreciation to Dr Ma-gang SHOU for his valuable and constructive suggestions during the planning and development of this research work. The authors would like to thank Dr Feng GUO for his advice on model construction and validation.
References
- Shou M, Hayashi M, Pan Y, Xu Y, Morrissey K, Xu L, et al. Modeling, prediction, and in vitroin vivo correlation of CYP3A4 induction. Drug Metab Dispos. 2008;36:2355–70. doi: 10.1124/dmd.108.020602. [DOI] [PubMed] [Google Scholar]
- Bjornsson TD, Callaghan JT, Einolf HJ, Fischer V, Gan L, Grimm S, et al. The conduct of in vitro and in vivo drug-drug interaction studies: a pharmaceutical research and manufacturers of America (PhRMA) perspective. Drug Metab Dispos. 2003;31:815–32. doi: 10.1124/dmd.31.7.815. [DOI] [PubMed] [Google Scholar]
- Dickins M. Induction of cytochromes P450. Curr Top Med Chem. 2004;4:1745–66. doi: 10.2174/1568026043387115. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- van Schaik IN, Eftimov F, van Doorn PA, Brusse E, van den Berg LH, van der Pol WL, et al. Pulsed high-dose dexamethasone versus standard prednisolone treatment for chronic inflammatory demyelinating polyradiculoneuropathy (PREDICT study): a double-blind, randomised, controlled trial. Lancet Neurol. 2010;9:245–53. doi: 10.1016/S1474-4422(10)70021-1. [DOI] [PubMed] [Google Scholar]
- Leggas M, Kuo KL, Robert F, Cloud G, deShazo M, Zhang R, et al. Intensive anti-inflammatory therapy with dexamethasone in patients with non-small cell lung cancer: effect on chemotherapy toxicity and efficacy. Cancer Chemother Pharmacol. 2009;63:731–43. doi: 10.1007/s00280-008-0767-x. [DOI] [PubMed] [Google Scholar]
- Rossignol J, Michallet AS, Oberic L, Picard M, Garon A, Willekens C, et al. Rituximab-cyclophosphamide-dexamethasone combination in the management of autoimmune cytopenias associated with chronic lymphocytic leukemia. Leukemia. 2011;25:473–8. doi: 10.1038/leu.2010.278. [DOI] [PubMed] [Google Scholar]
- Pascussi JM, Drocourt L, Fabre JM, Maurel P, Vilarem MJ. Dexamethasone induces pregnane X receptor and retinoid X receptor-alpha expression in human hepatocytes: synergistic increase of CYP3A4 induction by pregnane X receptor activators. Mol Pharmacol. 2000;58:361–72. doi: 10.1124/mol.58.2.361. [DOI] [PubMed] [Google Scholar]
- Ejiri N, Katayama K, Doi K. Induction of CYP3A1 by dexamethasone and pregnenolone-16alpha-carbonitrile in pregnant rat and fetal livers and placenta. Exp Toxicol Pathol. 2003;54:273–9. doi: 10.1078/0940-2993-00263. [DOI] [PubMed] [Google Scholar]
- Kostrubsky VE, Lewis LD, Wood SG, Sinclair PR, Wrighton SA, Sinclair JF. Effect of Taxol on cytochrome P450 3A and acetaminophen toxicity in cultured rat hepatocytes: comparison to dexamethasone. Toxicol Appl Pharmacol. 1997;142:79–86. doi: 10.1006/taap.1996.8023. [DOI] [PubMed] [Google Scholar]
- Schmiedlinren P, Benedict PE, Dobbins WO, Ghosh M, Kolars JC, Watkins PB. Cultured adult-rat jejunal explants as a model for studying regulation of Cyp3a. Biochem Pharmacol. 1993;46:905–18. doi: 10.1016/0006-2952(93)90501-m. [DOI] [PubMed] [Google Scholar]
- Hosoe T, Nakahama T, Inouye Y. Divergent modes of induction of rat hepatic and pulmonary CYP3A1 by dexamethasone and pregnenolone 16 alpha-carbonitrile. J Health Sci. 2005;51:75–9. [Google Scholar]
- Ronis MJJ, Chen Y, Liu XL, Blackburn ML, Shankar K, Landes RD, et al. Enhanced expression and glucocorticoid-inducibility of hepatic cytochrome P450 3A involve recruitment of the pregnane-X-receptor to promoter elements in rats fed soy protein isolate. J Nutr. 2011;141:10–6. doi: 10.3945/jn.110.127423. [DOI] [PubMed] [Google Scholar]
- Danhof M, de Jongh J, De Lange EC, Della Pasqua O, Ploeger BA, Voskuyl RA. Mechanism-based pharmacokinetic-pharmacodynamic modeling: biophase distribution, receptor theory, and dynamical systems analysis. Annu Rev Pharmacol Toxicol. 2007;47:357–400. doi: 10.1146/annurev.pharmtox.47.120505.105154. [DOI] [PubMed] [Google Scholar]
- Kumar V, Mostafa S, Kayo MW, Goldberg EP, Derendorf H. HPLC determination of dexamethasone in human plasma and its application to an in vitro release study from endovascular stents. Pharmazie. 2006;61:908–11. [PubMed] [Google Scholar]
- Khan AA, Chow EC, van Loenen-Weemaes AM, Porte RJ, Pang KS, Groothuis GM. Comparison of effects of VDR versus PXR, FXR and GR ligands on the regulation of CYP3A isozymes in rat and human intestine and liver. Eur J Pharm Sci. 2009;37:115–25. doi: 10.1016/j.ejps.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B. A comprehensive collection of experimentally validated primers for polymerase chain reaction quantitation of murine transcript abundance. BMC Genomics. 2008;9:633. doi: 10.1186/1471-2164-9-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao HT, Chang YW, Lan SJ, Yeh TK. The inhibitory effect of tannic acid on cytochrome P450 enzymes and NADPH-CYP reductase in rat and human liver microsomes. Food Chem Toxicol. 2008;46:645–53. doi: 10.1016/j.fct.2007.09.073. [DOI] [PubMed] [Google Scholar]
- Roe AL, Warren G, Hou GQ, Howard G, Shedlofsky SI, Blouin RA. The effect of high dose endotoxin on CYP3A2 expression in the rat. Pharm Res. 1998;15:1603–8. doi: 10.1023/a:1011915402914. [DOI] [PubMed] [Google Scholar]
- Omura T, Sato R. Carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–8. [PubMed] [Google Scholar]
- Chovan JP, Ring SC, Yu E, Baldino JP. Cytochrome P450 probe substrate metabolism kinetics in sprague dawley rats. Xenobiotica. 2007;37:459–73. doi: 10.1080/00498250701245250. [DOI] [PubMed] [Google Scholar]
- Mills JB, Rose KA, Sadagopan N, Sahi J, de Morais SMF. Induction of drug metabolism enzymes and MDR1 using a novel human hepatocyte cell line. J Pharmacol Exp Ther. 2004;309:303–9. doi: 10.1124/jpet.103.061713. [DOI] [PubMed] [Google Scholar]
- Hamberg AK, Dahl ML, Barban M, Scordo MG, Wadelius M, Pengo V, et al. A PK–PD model for predicting the impact of age, CYP2C9, and VKORC1 genotype on individualization of warfarin therapy. Clin Pharmacol Ther. 2007;81:529–38. doi: 10.1038/sj.clpt.6100084. [DOI] [PubMed] [Google Scholar]
- Savic RM, Jonker DM, Kerbusch T, Karlsson MO. Implementation of a transit compartment model for describing drug absorption in pharmacokinetic studies. J Pharmacokinet Pharmacodyn. 2007;34:711–26. doi: 10.1007/s10928-007-9066-0. [DOI] [PubMed] [Google Scholar]
- Sarangapani R, Teeguarden J, Plotzke KP, McKim JM, Jr, Andersen ME. Dose-response modeling of cytochrome p450 induction in rats by octamethylcyclotetrasiloxane. Toxicol Sci. 2002;67:159–72. doi: 10.1093/toxsci/67.2.159. [DOI] [PubMed] [Google Scholar]
- Florian JA, Tornoe CW, Brundage R, Parekh A, Garnett CE. Population pharmacokinetic and concentration-QTc models for moxifloxacin: pooled analysis of 20 thorough QT studies. J Clin Pharmacol. 2011;51:1152–62. doi: 10.1177/0091270010381498. [DOI] [PubMed] [Google Scholar]
- Masui K, Kira M, Kazama T, Hagihira S, Mortier EP, Struys MM. Early phase pharmacokinetics but not pharmacodynamics are influenced by propofol infusion rate. Anesthesiology. 2009;111:805–17. doi: 10.1097/ALN.0b013e3181b799c1. [DOI] [PubMed] [Google Scholar]
- Lu SK, Callahan SA, Jin RY, Brunner LJ. Cyclosporine and bromocriptine-induced suppressions of CYP3A1/2 and CYP2C11 are not mediated by prolactin. Eur J Pharmacol. 2004;501:215–24. doi: 10.1016/j.ejphar.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Zhang LP, Beal SL, Sheiner LB. Simultaneous vs sequential analysis for population PK/PD data I: best-case performance. J Pharmacokinet Phar. 2003;30:387–404. doi: 10.1023/b:jopa.0000012998.04442.1f. [DOI] [PubMed] [Google Scholar]
- Zhang LP, Beal SL, Sheiner LB. Simultaneous vs sequential analysis for population PK/PD data II: robustness of methods. J Pharmacokinet Phar. 2003;30:405–16. doi: 10.1023/b:jopa.0000012999.36063.4e. [DOI] [PubMed] [Google Scholar]
- Earp JC, Pyszczynski NA, Molano DS, Jusko WJ. Pharmacokinetics of dexamethasone in a rat model of rheumatoid arthritis. Biopharm Drug Dispos. 2008;29:366–72. doi: 10.1002/bdd.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ourlin JC, Lasserre F, Pineau T, Fabre JM, Sa-Cunha A, Maurel P, et al. The small heterodimer partner interacts with the pregnane X receptor and represses its transcriptional activity. Mol Endocrinol. 2003;17:1693–703. doi: 10.1210/me.2002-0383. [DOI] [PubMed] [Google Scholar]
- Samtani MN, Jusko WJ. Comparison of dexamethasone pharmacokinetics in female rats after intravenous and intramuscular administration. Biopharm Drug Dispos. 2005;26:85–91. doi: 10.1002/bdd.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma DR, Yue TL. Influence of protein-calorie malnutrition on the pharmacokinetics, placental transfer and tissue localization of dexamethasone in rats. Br J Pharmacol. 1984;83:131–7. doi: 10.1111/j.1476-5381.1984.tb10127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DK, LaBorde JB, Wall KS, Holson RR, Young JF. Pharmacokinetic considerations of dexamethasone-induced developmental toxicity in rats. Toxicol Sci. 1999;48:230–9. doi: 10.1093/toxsci/48.2.230. [DOI] [PubMed] [Google Scholar]
- Yang L, Panetta JC, Cai X, Yang W, Pei D, Cheng C, et al. Asparaginase may influence dexamethasone pharmacokinetics in acute lymphoblastic leukemia. J Clin Oncol. 2008;26:1932–9. doi: 10.1200/JCO.2007.13.8404. [DOI] [PubMed] [Google Scholar]
- Raybon JJ, Pray D, Morgan DG, Zoeckler M, Zheng M, Sinz M, et al. Pharmacokinetic-pharmacodynamic modeling of rifampicin-mediated cyp3a11 induction in steroid and xenobiotic X receptor humanized mice. J Pharmacol Exp Ther. 2011;337:75–82. doi: 10.1124/jpet.110.176677. [DOI] [PubMed] [Google Scholar]
- Hoen PA, Commandeur JN, Vermeulen NP, Van Berkel TJ, Bijsterbosch MK. Selective induction of cytochrome P450 3A1 by dexamethasone in cultured rat hepatocytes: analysis with a novel reverse transcriptase-polymerase chain reaction assay section sign. Biochem Pharmacol. 2000;60:1509–18. doi: 10.1016/s0006-2952(00)00454-8. [DOI] [PubMed] [Google Scholar]
- Huss JM, Wang SI, Kasper CB. Differential glucocorticoid responses of CYP3A23 and CYP3A2 are mediated by selective binding of orphan nuclear receptors. Arch Biochem Biophys. 1999;372:321–32. doi: 10.1006/abbi.1999.1496. [DOI] [PubMed] [Google Scholar]
- Aiba T, Yoshinaga M, Ishida K, Takehara Y, Hashimoto Y. Intestinal expression and metabolic activity of the CYP3A subfamily in female rats. Biol Pharm Bull. 2005;28:311–5. doi: 10.1248/bpb.28.311. [DOI] [PubMed] [Google Scholar]
- Eeckhoudt SL, Horsmans Y, Verbeeck RK. Differential induction of midazolam metabolism in the small intestine and liver by oral and intravenous dexamethasone pretreatment in rat. Xenobiotica. 2002;32:975–84. doi: 10.1080/0049825021000012655. [DOI] [PubMed] [Google Scholar]
- Yu LS, Lu SJ, Zhao NP, Ni SQ, Yao TW, Zeng S. Male-specific induction of CYP3A2 in rats by zolmitriptan. J Pharm Pharmacol. 2008;60:1601–7. doi: 10.1211/jpp/60.11.0005. [DOI] [PubMed] [Google Scholar]
- Van Der Graaf PH, Van Schaick EA, Math-ot RA, Ijzerman AP, Danhof M. Mechanism-based pharmacokinetic–pharmacodynamic modeling of the effects of N6-cyclopentyladenosine analogs on heart rate in rat: estimation of in vivo operational affinity and efficacy at adenosine A1 receptors. J Pharmacol Exp Ther. 1997;283:809–16. [PubMed] [Google Scholar]
- Visser SA, Wolters FL, Gubbens-Stibbe JM, Tukker E, Van Der Graaf PH, Peletier LA, et al. Mechanism-based pharmacokinetic/pharmacodynamic modeling of the electroencephalogram effects of GABAA receptor modulators: in vitro-in vivo correlations. J Pharmacol Exp Ther. 2003;304:88–101. doi: 10.1124/jpet.102.042341. [DOI] [PubMed] [Google Scholar]