Abstract
The potassium channel KcsA offers a unique opportunity to explicitly study the dynamics of the moving parts of ion channels, yet our understanding of the extent and dynamic behavior of the physiologically relevant structural changes at the inner gate in KcsA remains incomplete. Here, we use electron paramagnetic resonance, nuclear magnetic resonance, and molecular dynamics simulations to characterize the extent of pH-dependent conformational changes of the inner gate in lipid bilayers or detergent micelles. Our results show that under physiological conditions the inner gate experiences a maximal diagonal opening of ∼24 Å with the largest degree of dynamics near the pKa of activation (pH ∼3.9). These results extend the observation that the C-terminus is necessary to limit the extent of opening and imply that the inner gate regulates the extent of conformational change at the zone of allosteric coupling and at the selectivity filter.
Despite its apparent simplicity, the prokaryotic potassium channel KcsA displays a complex gating cycle characterized by the coupled action of a H+-activated gate at the inner bundle crossing and a H+-independent gate at the selectivity filter.1 Previous studies have shown that low pH promotes interhelical separation at the inner gate, seen from the central axis of symmetry.2,3 Several studies have suggested that these changes are allosterically coupled to the selectivity filter and promote the transition from a conductive (activated) to a nonconductive (inactivated) channel.4,5 The role of the C-terminus as the regulator of this process has been suggested by the effects of its deletion on the rate of C-type inactivation and exaggerated opening of the inner gate during pH gating.4,6,7 Conversely, mechanisms that tend to stabilize the conductive conformation of the selectivity filter (KcsA-E71A, permeation by Rb+ or other long dwell-time ions) lower the proton concentration required to open the inner gate.4 These results strongly support the idea of a bidirectional coupling between the two gates.
Despite extensive static structural and functional studies of KcsA, understanding the extent of conformational dynamics at the inner gate during activation gating remains incomplete. One question is how far the inner gate opens to trigger the conformational wave leading to inactivation. Crystallographic studies suggest that critical reorientations in the selectivity filter that ultimately lead to a loss of ion occupancy take place beyond inner gate openings of 17–23 Å.5
Here, we have conducted a systematic measurement of inner gate fluctuations in KcsA using electron paramagnetic resonance (EPR), nuclear magnetic resonance (NMR), and molecular dynamics (MD) simulations to evaluate the conformational space of the inner helical bundle during pH-induced gating. The extent of opening at the inner gate was measured in full-length KcsA using both continuous wave (CW) and double-electron–electron resonance (DEER) EPR spectroscopy in lipids and detergent micelles at residue G116. These observations have been extended using solution NMR to monitor inner gate dynamics. We demonstrate a correlation between the sigmoidal pH dependence in the chemical shift at residue G116 and changes in local backbone dynamics. In both cases, local dynamics are maximal at or near the pKa of activation, suggesting an increase in the total number of conformers at the midpoint of activation. Finally, we suggest that the extent of opening in full-length KcsA at position G116 is ∼24 Å under physiological conditions.
Dynamics of Inner Gate Opening
In wild-type (WT) KcsA, singly spin-labeled monomers generate two sets of interspin interactions: adjacent (α) and diagonally related (δ), with a distance relation of δ = √2α. The use of tandem dimer constructs allows the placement of a cysteine residue on diagonally related subunits, simplifying the distance distribution determination by EPR spectroscopy to a single peak.2 With this approach, we determined intersubunit distances and distance distributions at sequential pHs from a spin-label placed at the bottom of TM2 (residue G116). We observed an abrupt increase in the diagonal intersubunit distance at G116 near pH 4.0 (Figure 1A), which is in good agreement with previously measured values (functional pKa of 4.21). Intersubunit distances range from 10 to 23.7 Å over the pH range, a gate opening that is narrower than the largest diameter determined from crystal structures of truncated KcsA (∼32 Å). This result furthers the observation that deletion of the C-terminus promotes inner gate opening beyond its physiological range.5,6 In Figure 1A, each pH measurement is associated with a distance distribution (Figures 2 and 3 of the Supporting Information), where the width of these distributions (i.e., error) reflects the total number of interconverting conformations of the KcsA inner bundle gate. A key observation from this data set is that distance distributions broaden as the pH reaches the pKa (i.e., pH 3.9), a minimum for the energetic costs to interconvert. As expected, these distributions narrow at extreme pHs when either the open or closed conformations are most stable and must overcome larger energy barriers to interconvert. Further, the degree of opening in the full-length channel appears to be more restrained than in the open mutant KcsA structural studies.5 Notably, residue F103, which has been implicated in allosteric coupling of the inner gate to the selectivity filter,4 and residue F114, located near the inner gate (facing outward), display a similar abrupt change in distance and orientation of the side chain (Figure 1B).
Figure 1.
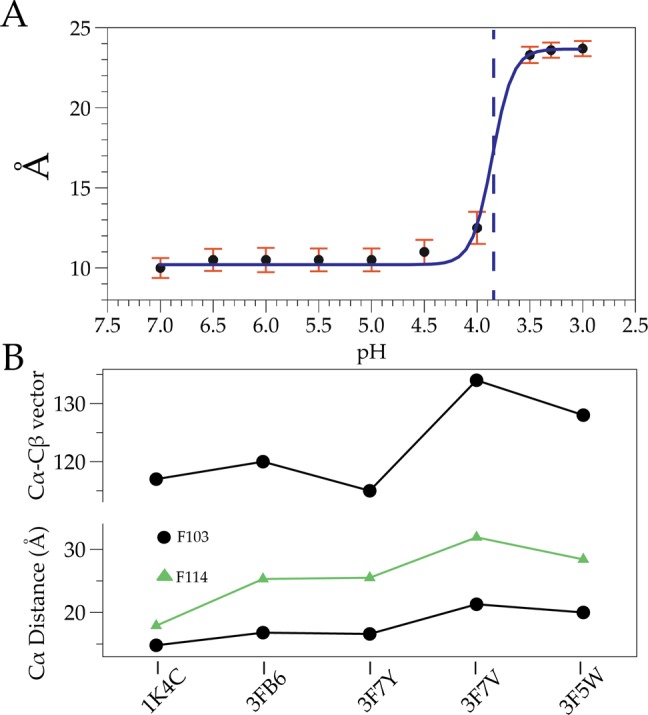
Conformational changes at the inner gate. (A) EPR measurements of G116. Error bars are half-maximal population widths normalized to pH 4 and the pKa indicated with a vertical dashed line. (B) Conformational changes at residues F103 and F114.2,5,12 The top trace shows the Cα–Cβ vector relative to the α-helix at F103. The bottom trace shows the Cα–Cα diagonal distance of F103 and F114 in angstroms.
NMR of the Inner Gate
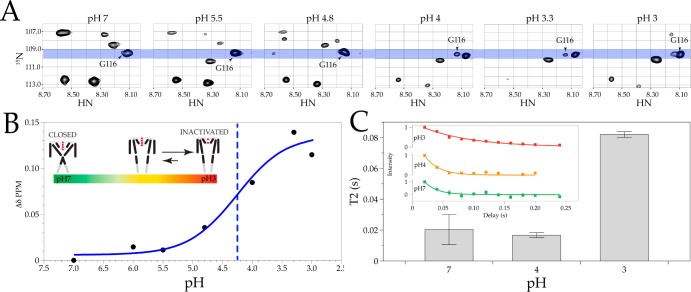
In solution NMR, inherent conformational dynamics can be estimated through three primary observables: chemical shift, signal intensity (relative population), and line width.8 A C-terminally truncated version of KcsA (Δ125, 2H–15N) in detergent micelles was evaluated at sequential pHs by monitoring chemical shift changes using transverse relaxation-optimized spectroscopy (15N–1H TROSY). The cross-peak corresponding to G116 (a key reporter in previous EPR analyses) was tracked at several titration points (Figure 2A). In agreement with the EPR data in Figure 1A, the pH dependence of the chemical shift is sigmoidal and can be fit with a Boltzmann function [pKa = 4.27 ± 0.24; ρ = 0.97 (Figure 2B)]. The shift in the observed pKa reflects the C-terminal truncation also seen by EPR and agrees well with the functional data.1,9 The shape of the cross-peaks during pH-dependent gating suggests a change in dynamics and underscores the need for further analysis.8 Finally, the T2 times of residue G116 differ with respect to pH, with an increase seen at pH 3 (Figure 2C). These observations correlate well with previous EPR studies showing mobility increases (and, thus, dynamic changes) at G116 as the pH decreases.3
Figure 2.
NMR TROSY spectra of KcsA Δ125 residue G116. (A) Cross-peaks of associated chemical shifts for G116 for 15N and 1H. See the Supporting Information for methods for δ calculations. (B) Line graph of chemical shifts with a Boltzmann fit (blue) and pKa (blue dashed line). (C) T2 times for residue G116 at pH 7 (green), pH 4 (orange), and pH 3 (red) (inset shows two or three experiments and fits).
Inner Gate and Allosteric Coupling
Multiple lines of spectroscopic, crystallographic, and functional evidence point to the C-terminus as an important modulator of inner gate opening and dynamics.4,6,10,11 Additionally, inspection of the Cα–Cα distances at F103 (implicated in allosteric coupling of the inner gate to the selectivity filter) and F114 (located near the inner gate) in several structures, together with correlation of the extent of opening at the inner gate by CW and DEER EPR, suggests that the maximal distance achieved during inner gate opening is roughly 23 Å5,12 (Figure 1B, bottom trace). The Cα–Cβ vector relative to the α-helix also shows a maximal degree of change (Figure 1B, top trace) at the same opening. To elucidate the membrane-bound conformation of open KcsA, MD simulations were performed starting from a series of open structures of KcsA:5 the crystallographic fully open conformation (∼32 Å) and two narrower open states (at 23 and 17 Å openings). Of the three structures tested, only the 23 Å opening (Protein Data Bank entry 3F7V) maintains a stable conformation in the membrane (Figure 3). We suggest that while the inner gate can open to a greater extent, steric hindrance from the C-terminal bundle and allosteric coupling to the selectivity filter (and thus C-type inactivation) likely set the energy optimal for open KcsA at ∼24 Å. Interestingly, the abrupt changes observed via NMR and EPR at the midpoint of activation correlate well with the set of inner bundle distances associated with the largest conformational changes at the selectivity filter (17–23 Å).5 This observation will lead to future evaluations of the inner gate conformational dynamics and the establishment of the minimal set of conformational excursions in the inner gate required to allosterically induce selectivity filter reorientations that lead to C-type inactivation. We propose that the nature of this energetic coupling will improve our understanding of the structural underpinnings of subconductance states by defining such thresholds.5,13,14
Figure 3.
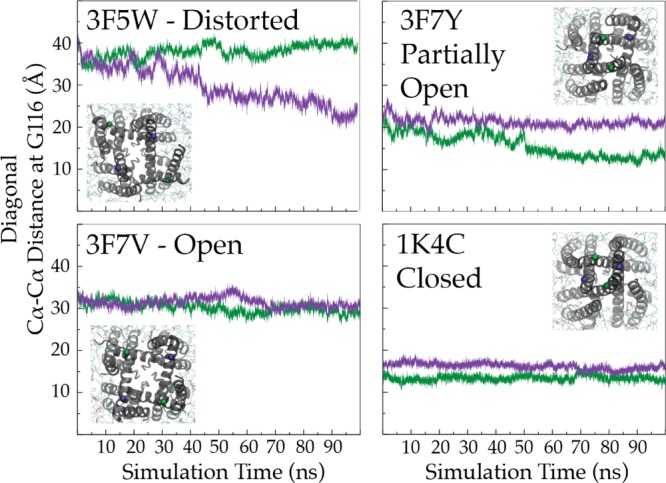
Dynamics of the inner gates of different structures in the membrane observed via MD simulations. Distances between Cα atoms of G116 of diagonally related subunits in different KcsA simulations. The inset shows the cytoplasmic view of the inner gate at the end of the simulation (t = 100 ns). Time series of distances are displayed (purple and green) at diagonal G116.
Supporting Information Available
Methods, supplemental Table 1, and supplemental Figures S1–S4. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by National Institutes of Health Grants U54-GM087519, R01-GM57846, and P41-GM104601. MD simulations were performed on XSEDE (Grant MCA06N060).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Chakrapani S.; Cordero-Morales J. F.; Perozo E. (2007) A quantitative description of KcsA gating I: Macroscopic currents. J. Gen. Physiol. 130, 465–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-S.; Sompornpisut P.; Perozo E. (2001) Structure of the KcsA channel intracellular gate in the open state. Nat. Struct. Biol. 8, 883–887. [DOI] [PubMed] [Google Scholar]
- Perozo E.; Cortes D. M.; Cuello L. G. (1999) Structural rearrangements underlying K+-channel activation gating. Science 285, 73–78. [DOI] [PubMed] [Google Scholar]
- Cuello L. G.; Jogini V.; Cortes D. M.; Pan A. C.; Gagnon D. G.; Dalmas O.; Cordero-Morales J. F.; Chakrapani S.; Roux B.; Perozo E. (2010) Structural basis for the coupling between activation and inactivation gates in K+ channels. Nature 466, 272–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello L. G.; Jogini V.; Cortes D. M.; Perozo E. (2010) Structural mechanism of C-type inactivation in K+ channels. Nature 466, 203–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmas O.; Hyde H. C.; Hulse R. E.; Perozo E. (2012) Symmetry-Constrained Analysis of Pulsed Double Electron-Electron Resonance (DEER) Spectroscopy Reveals the Dynamic Nature of the KcsA Activation Gate. J. Am. Chem. Soc. 134, 16360–16369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M.; Onishi Y.; Yanagida T.; Ide T. (2011) Role of the KcsA Channel Cytoplasmic Domain in pH-Dependent Gating. Biophys. J. 101, 2157–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner I. R.; Foster M. P. (2011) An introduction to NMR-based approaches for measuring protein dynamics. Biochim. Biophys. Acta 1814, 942–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello L. G.; Cortes D. M.; Jogini V.; Sompornpisut A.; Perozo E. (2010) A molecular mechanism for proton-dependent gating in KcsA. FEBS Lett. 584, 1126–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uysal S.; Cuello L. G.; Cortes M.; Koide S.; Kossiakoff A.; Perozo E. (2011) Mechanism of activation gating in the full-length KcsA K+ channel. Proc. Natl. Acad. Sci. U.S.A. 108, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M.; Takeuchi Y.; Aoki T.; Yanagida T.; Ide T. (2010) Rearrangements in the KcsA Cytoplasmic Domain Underlie Its Gating. J. Biol. Chem. 285, 3777–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.; Morais-Cabral J. H.; Kaufman A.; Mackinnon R. (2001) Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature 414, 43–48. [DOI] [PubMed] [Google Scholar]
- Chakrapani S.; Cordero-Morales J. F.; Perozo E. (2007) A quantitative description of KcsA gating II: Single-channel currents. J. Gen. Physiol. 130, 479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrapani S.; Cordero-Morales J. F.; Jogini V.; Pan A. C.; Cortes D. M.; Roux B.; Perozo E. (2011) On the structural basis of modal gating behavior in K+ channels. Nat. Struct. Mol. Biol. 18, 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.