Abstract

The principal green tea polyphenol, (−)-epigallocatechin-3-O-gallate (EGCg), may provide chemoprotection against conditions ranging from cardiovascular disease to cancer. Binding to plasma proteins stabilizes EGCg during its transport to targeted tissues. This study explored the details EGCg binding to bovine serum albumin. Both fluorescence lifetime and intensity data showed that the hydrophobic pocket between subdomains IIA and IIIA is the binding site for EGCg. Fluorescence and circular dichroism were used to establish the roles of the flavan-3-ol and galloyl moieties of the EGCg in binding and to demonstrate a binding-dependent conformational change in the protein. Competitive binding experiments confirmed the location of binding, and molecular modeling identified protein residues that play key roles in the interaction. This model of EGCg–BSA interactions improves the understanding of the likely physiological fate of this green tea-derived bioactive polyphenol.
Keywords: polyphenol, tea, tannin−protein interaction, molecular modeling, phenylbutazone, tolbutamide
Introduction
Green tea, a beverage that is consumed throughout the world, is believed to reduce the risks of chronic diseases, such as Alzheimer’s disease,1 cardiovascular diseases,2 and cancer.3 These health-promoting effects are attributed to polyphenolic components, especially (−)-epigallocatechin-3-O-gallate (EGCg), the principal green tea polyphenol that has potent chemoprotective activities.4,5 In vitro studies suggest that the health benefits mediated by EGCg can be achieved not only via nonspecific effects including radical scavenging, metal chelating, and protein cross-linking1 but also via specific binding interactions with certain proteins in a wide array of molecular signaling pathways.6
Unlike other polyphenols, EGCg can be absorbed without metabolic modification. Consumption of green tea by humans produces peak plasma EGCg concentrations ranging from 0.04 to 1.0 μM,7 a level effective for the suppression of tumor growth in cell culture studies.8 However, EGCg is chemically highly reactive and requires stabilization during transport to prevent potentially damaging side reactions and to maintain its activity. For example, EGCg is a radical scavenger with a reduction potential lower than α-tocopherol (vitamin E),9 one of the major redox buffers in the plasma. Influx of EGCg could potentially alter the dynamic equilibrium of the blood oxidation–reduction system, possibly harming liver and kidney functions. Furthermore, oxidation of EGCg is accompanied by spontaneous ring opening under physiological conditions, with a chemical half-life of around 45 min at pH 7.4 and 37 °C.10 Recent studies indicate that the presence of serum albumin inhibits EGCg oxidation in vitro.11,12 We propose that binding to plasma protein might not only protect EGCg from degradation but also mask the reactivity of EGCg during transport through the bloodstream, allowing effective delivery of the intact bioactive species to target tissues without collateral damage. The role of EGCg in human health can therefore be understood only in the context of its binding to serum albumin.
Serum albumin (SA) is the principal plasma protein and has an in vivo concentration around 7 × 10–4 M for humans.13 This soft globular protein consists of three homologous domains (I, II, and III) that undergo conformation changes in response to pH change or ligand binding.13,14 As the main carrier protein in plasma, SA is able to bind endogenous and exogenous compounds, controlling their deposition, pharmacokinetics, stability, and toxicity.13 Human serum albumin (HSA) has a large hydrophobic pocket on subdomain IIA, encompassing the Sudlow I binding pocket near the front and two smaller pockets near the back. A second hydrophobic pocket, the Sudlow II binding site, is found on subdomain IIIA. In HSA, the single Trp residue is found near Sudlow I in domain II. Bovine serum albumin (BSA) is 76% homologous to the human protein in its amino sequence, but has three hydrophobic pockets and two Trp residues.15 Trp134 is in a solvent-exposed region of subdomain IB and is highly accessible, whereas Trp213, like Trp214 in HSA, is buried in the hydrophobic pocket near the Sudlow I site in subdomain IIA.15
Extensive experiments have been performed to characterize the interaction between EGCg and SA via circular dichroism (CD),16 electron paramagnetic resonance spectroscopy,17 fluorescence,18 and polyacrylamide gel electrophoresis.12 EGCg quenches the fluorescence of the single Trp residue in HSA, with an apparent association constant of 5 × 104 M–1 for HSA–EGCg binding.19 Binding involves one or both of the hydrophobic pockets on domains IIA and IIIA of the protein, but neither physical methods nor competitive binding studies were able to define a specific binding site.16,19,20
Further study of the interactions between a series of flavan-3-ols with HSA or BSA demonstrated a role for the galloyl ester D ring of the flavonoid (Figure 1a) in the formation of stable complexes.18,20 These complexes could be covalently stabilized by reaction with cysteine or lysine residues.12,17,21 Covalent modification of a specific cysteine residue by EGCg has been demonstrated for glyceraldehyde-3-phosphate dehydrogenase22 but not for SA.
Figure 1.
Structures of (a) epigallocatechin gallate (EGCg), (b) epigallocatechin (EGC), and (c) methyl gallate (MG). The A, B, C, and D rings of the flavan-3-ols are labeled in (a).
Only a few molecular docking studies have been performed for EGCg and SA. A docking study based on the crystal structure of HSA supports the experimental evidence for binding at both hydrophobic sites.19 Docking studies with BSA have been hampered because the structure of the protein has not been available, but one study that used an amino acid sequence-based simulation for the BSA structure suggested preferred binding near Trp213.20 However, there is no strong experimental evidence for site-specific binding of BSA by EGCg.
In this study, we tested the model proposed by Skrt et al.20 that describes a single binding site on BSA for EGCg. We used fluorescence, CD, and competitive binding studies to characterize the interaction and improved the molecular docking studies by using the recently established crystal structure for BSA.15 Our data demonstrate that binding of EGCg induces a conformational change in BSA and requires the synergistic efforts of both the flavan-3-ol and galloyl moiety subunits. The former subunit binds to the entrance of the hydrophobic pocket, perhaps via covalent bonds, whereas the latter binds to the bottom mainly via hydrogen bonds. Our data provide insights into EGCg–SA interactions that are relevant to the human as well as the bovine protein.
Materials and Methods
Materials
Bovine serum albumin (BSA) (fatty acid free, ≥96%), methyl gallate (MG), phenylbutazone (PB), and tolbutamide (TB) were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). EGCg (>95%) was provided by Lipton Tea Co. (Newark, NJ, USA). (−)-Epigallocatechin (EGC) (>95%) was purified from commercial tea (SilvaTeam, San Michele, Italy) by Sephadex LH20 column after tannase treatment and was characterized by HPLC and LC-MS (unpublished data). The chemical structures of the three phenolic compounds (EGCg, MG, EGC) are depicted in Figure 1.
All stock solutions were prepared by initially dissolving the compounds in water so there were no carrier solvent effects on ligand–protein interactions. BSA and EGCg concentrations were determined spectrophotometrically by their extinction coefficients (ε280 (BSA) = 43000 M–1 cm–1 and ε280(EGCg) = 9700 M–1 cm–1). Samples were diluted into 0.2 M sodium phosphate buffers (pH 3.0, 5.0, and 7.4) containing 0.1 M sodium chloride.
Fluorescence Spectroscopy and Quenching Constant Calculation
BSA was mixed with the desired amount of phenolic solution (EGC, EGCg, or MG), diluted to 5.0 mL and a final concentration of 3 μM protein with the appropriate phosphate buffer, vortexed for 10 s, immediately sparged with nitrogen for 20 min, and incubated for 1 h at 22 °C. The concentration of phenolic compound was varied from 0 to 25 μM in 5 μM increments. The stability of each phenolic compound under these conditions was checked by HPLC as described below.
The method of Soares et al.23 was modified to evaluate the BSA fluorescence spectra between 290 and 500 nm with an LS55 luminescence spectrometer (PerkinElmer Inc., USA) with the excitation wavelength set at 280 nm. Slit widths for excitation and emission were 7.5 and 4.5 nm, respectively. Fluorescence intensity was corrected by eq 1 to compensate for the inner filter effect24−26 due to sample absorbance at 280 and 350 nm.
| 1 |
Fcor and Fobs are, respectively, the corrected and observed fluorescence intensity. Abs280 and Abs350 represent the absorbances of samples in a 1 cm path length cell at the excitation and emission wavelengths. In this study, all fluorescence intensities were corrected prior to data analysis.
Fluorescence quenching is usually described by the Stern–Volmer equation (eq 2).27 This classic model assumes that all fluorophores are equally accessible to the quencher.
| 2 |
F0 and F are the fluorescence intensities before and after the addition of the quencher, respectively. KSV is the Stern–Volmer quenching constant, [Q] is the concentration of the quencher, kq is the bimolecular quenching constant, and τ0 is the native lifetime of the fluorophore. KSV can thus be determined by linear regression of a plot of F0/F against [Q].
Negative deviations from the Stern–Volmer equation are frequently found in the systems that have multiple fluorophores, such as BSA. The values of F0/F, instead of increasing linearly with [Q] in the classic model, trend downward toward the x axis at high [Q], signifying differences in the accessibilities of fluorophores to the quencher.27 The modified Stern–Volmer equation (eq 3) includes a factor for fractional accessibility fa, allowing calculation for the modified SV constant Ka for systems with more than one fluorophore.
| 3 |
Time Domain Fluorescence Lifetime
Time-resolved fluorescence lifetime was measured with a TemPro 01 system (Horiba Jobin Yvon, USA), using a FluoroHub-B single-photon counting module with TBX detector. The excitation light (250 nm) was provided by a NanoLED light source, and the number of counts gathered in the channel of maximum intensity was 10000. The data were analyzed with CAS6 V6.6 decay analysis software. As BSA has two fluorophores with different lifetimes, eq 4 was used to describe the time-dependent decay of the biexponential fluorescence system.
| 4 |
τ1 and τ2 are native fluorescence lifetimes for Trp134 and Trp213, respectively, whereas F1 and F2 represent their relative amplitudes. Samples for the lifetime measurement were prepared in the same way as for the intensity measurements.
EGCg Stabilization and Competitive Binding
EGCg–SA interactions can be indirectly assessed by determining the BSA-mediated stabilization of EGCg.12 We used that method to carry out competitive binding experiments between the drug PB or TB and EGCg. BSA (30 μM) was incubated at pH 7.4 with EGCg (25 μM) and either PB or TB (0–900 μM) for 40 min at 37 °C before the amount of EGCg remaining was determined by high-performance liquid chromatography (HPLC) as described below. The amount of EGCg that remained in the samples at the end of the incubation was proportional to the amount of EGCg bound to the serum albumin.
HPLC Analysis
EGC, EGCg, and methyl gallate were quantitated with an Agilent 1100 series HPLC system equipped with a diode array detector (Agilent Technologies, USA). Separation was performed on a Zorbax Eclipse XD8 C8 column (5 μm, 4.6 × 150 mm i.d.) at a flow rate of 0.5 mL/min. Samples were eluted with a gradient of 0.1% trifluoroacetic acid in water (solvent A) and 0.13% trifluoroacetic acid in acetonitrile (solvent B) as follows: 0–10 min, 5% B; 10–20 min, 20% B; 20–30 min, 35% B; 30–34 min, 95% B. The elution profiles were recorded at 280 nm. Samples (10 μL) were injected into HPLC after filtration through 0.22 μm cellulose acetate spin filters. BSA solutions without EGCg were used as the background, and Agilent Chemstation A.09.03 software was used for peak integration.
Circular Dichroism Spectroscopy
The method of Nozaki et al.28 was used in this study. A J-810 spectropolarimeter (Jasco Inc., Japan) was employed to collect CD spectra between 355 and 245 nm with a 1 mm path length cell. The system was purged with dry nitrogen gas during measurement at 298–301 K. CD spectra were built on accumulation of 12 cycles of scans with 0.5 nm data pitch, 1.0 nm bandwidth, 100 nm/min speed, and standard sensitivity.
All samples were prepared in 0.2 M sodium phosphate buffer at pH 7.4. The BSA concentration was 30 μM, whereas the concentrations of EGC, EGCg, and MG ranged from 0 to 200 μM. The spectrum of the buffer was subtracted from all samples to eliminate background noise. Induced CD spectra for BSA–EGCg samples (or EGC or MG) were obtained with eq 5.
| 5 |
CDBSA–EGCg is the spectra of the BSA–EGCg sample, and CDBSA and CDEGCg represent the spectra of samples containing only BSA and EGCg.
Molecular Modeling
The crystal structure of BSA (code: 3 V03)15 was downloaded from the Protein Data Bank. The 3D structure of EGCg (ChEBI:4806) was initially obtained from Chemical Entities of Biological Interest as a MOL file and converted afterward to a PDB file by OpenBabelGUI version 2.3.1. AutoDock Tool 4.2 v. 1.5.6rc329 was used to optimize both BSA and EGCg structures for docking experiments. For BSA, chain B and all water molecules were removed to produce a monomer structure. For EGCg, the structure was prepared by defaults setting with the gasteiger charges computed. Both BSA and EGCg structures had hydrogen atoms added.
The mechanism of the BSA–EGCg interaction was predicted on the molecular level with AutoDock Tool 4.2. The grid box was set up with 126 points in the X, Y, and Z dimensions, respectively. Each grid point had a spacing of 0.65 Å. Docking experiments were performed with default genetic algorithm parameters to generate 10 possible conformations with the lowest Gibbs free energy of binding.
Results
Quenching of BSA Fluorescence Spectra by EGCg
Changes in pH affected the fluorescence spectrum of native BSA. At pH 3.0, the emission spectrum of BSA ranged from 290 to 480 nm with a maximum emission (λem) at 340 nm (Figure 2a). When the pH was increased to 5.0 or 7.4, the spectrum was red-shifted to 280–500 nm with a λem at 350 nm (Figure 2b,c).
Figure 2.
Quenching of 3 μM BSA by EGCg. Changes in the emission spectra of BSA were measured at (a) pH 3.0, (b) pH 5.0, and (c) pH 7.4. (d) The efficiencies of quenching were compared at pH 3.0 (●), 5.0 (■), and 7.4 (▲) by plotting the relative fluorescence intensity (F/F0 × 100) against EGCg concentration. All fluorescence intensities were corrected by eq 1 with a λex = 280 nm and a λem = 350 nm, and the concentrations of EGCg were 0, 5, 10, 15, 20, and 25 μM.
At all pH values tested, BSA fluorescence was affected by EGCg in a dose-dependent manner (Figure 2a–c). In acidic solutions (pH 3.0 and 5.0), addition of EGCg did not change the λem of BSA. However, λem was red-shifted from 350 to 365 nm at pH 7.4 when the EGCg concentration was increased from 0 to 25 μM. The efficacy of EGCg-induced quenching depended largely on the pH (Figure 2d). At pH 3.0, 25 μM EGCg quenched the BSA fluorescence to 68.4% of control value. At pH 5.0, the same concentration of EGCg brought the emission intensity down to 33.4% of control value. When the pH was increased to 7.4, only 20.7% of the control fluorescence remained in the presence of 25 μM EGCg.
The Stern–Volmer plots of EGCg-induced quenching of BSA fluorescence at pH 3.0 and 5.0 were linear, whereas at pH 7.4 the data were nonlinear, consistent with two fluorophores with different accessibilities27 (Figure 3a). The data at pH 7.4 yielded a linear modified Stern–Volmer plot (Figure 3b). The Stern–Volmer constants (Table 1) were consistent with pH-dependent binding that is stronger at higher pH values.
Figure 3.
SV plots for the quenching of 3 μM BSA at pH 3.0 (●), pH 5.0 (■), and pH 7.4 (▲): (a) SV plots for EGCg-mediated quenching of BSA fluorescence; (b) modified SV plot for EGCg-mediated quenching of BSA fluorescence at pH 7.4; (c) SV plots for MG-mediated quenching of BSA fluorescence; (d) SV plots for EGC-mediated quenching of BSA fluorescence. All fluorescence intensities were corrected by eq 1 with a λex = 280 nm and a λem = 350 nm, and the concentrations of EGCg were 0, 5, 10, 15, 20, and 25 μM.
Table 1. Stern–Volmer Constants Describing BSA Fluorescence Quenching by EGC, EGCg, or MG at pH 3.0, 5.0, and 7.4.
|
KSV (× 104 M–1) |
|||
|---|---|---|---|
| pH 3.0 | pH 5.0 | pH 7.4 | |
| EGCg | 1.6 | 8.0 | 461a |
| MG | 0.85 | 0.81 | 5.4 |
| EGC | 0.30 | 0.23 | 0.64 |
Estimated as Ka from modified SV plot.
Quenching of BSA Fluorescence Spectra by EGC or MG
MG did not quench as effectively as EGCg, but the pH dependence of quenching was similar to that of EGCg (Supporting Information Figure S1). The linear Stern–Volmer plots (Figure 3c) yielded KSV values consistent with weaker interactions between BSA and MG compared to EGCg, but with a similar pH dependence (Table 1).
EGC was a very weak quencher under experimental conditions. After incubation with EGC, the spectra of BSA fluorescence were only slightly changed and the reduction of fluorescence intensity was small at λem (Supporting Information Figure S2). The linear SV plots (Figure 3d) yielded binding constants consistent with very weak, pH-independent interactions (Table 1).
BSA Fluorescence Lifetime
We used fluorescence lifetime to distinguish the effects of EGCg on Trp134, a long-lifetime species, versus Trp213, a short-lifetime species.30,31 EGCg substantially diminished the fluorescence lifetime of Trp213 in a pH-dependent fashion, reducing the lifetime by about 50% at pH 5.0 or 7.4 (Table 2). At the same pH values, EGCg diminished the fluorescence lifetime of Trp134 about 15–25% (Table 2). Methyl gallate diminished the lifetime of Trp213 only at pH 7.4 (30%), whereas EGC did not change the lifetime of either Trp at any pH (Table 2).
Table 2. Fluorescence Lifetime of Two Trp Residues in Bovine Serum Albumin in the Absence or Presence of 25 μM EGC, EGCg, or MG at pH 3.0, 5.0, and 7.4.
| τ
(ns) |
||||||
|---|---|---|---|---|---|---|
| pH 3.0 |
pH 5.0 |
pH 7.4 |
||||
| Trp134 | Trp213 | Trp134 | Trp213 | Trp134 | Trp213 | |
| BSA | 5.38 | 2.13 | 6.50 | 2.88 | 6.56 | 2.89 |
| BSA + EGCg | 5.24 | 1.86 | 5.49 | 1.43 | 4.92 | 1.34 |
| BSA + MG | 5.38 | 2.11 | 6.49 | 2.85 | 6.29 | 2.06 |
| BSA + EGC | 5.41 | 2.15 | 6.58 | 3.04 | 6.54 | 2.93 |
The bimolecular quenching constant kq was estimated from the KSV and the fluorescence lifetime data (eq 2) to distinguish static from dynamic binding. The upper limit of fluorescence lifetime is about 6 ns, so for the compounds we tested at pH 7.4, kq ranges from about 1011 M–1 s–1 for EGC to about 1014 M–1 s–1 for EGCg.
Impact of Phenolics on the Conformation of BSA
In the presence of EGCg, the CD spectra of BSA were changed in a dose-dependent manner with positive amplitudes centering on 280 nm and negative amplitudes centering on 320 nm (Figure 4a). EGCg at 200 μM induced the largest change in the CD spectrum, giving ∼0.8 mdeg at 280 nm and ∼ –0.4 mdeg at 320 nm. MG or EGC did not significantly alter the conformation of BSA, as the amplitudes of the spectra fluctuated by only ∼0.2 mdeg in the presence of 200 μM of either compound (Figure 4b,c).
Figure 4.
Induced CD spectra of 30 μM BSA in the presence of (a) EGCg, (b) EGC, and (c) MG. All induced CD spectra were obtained via eq 5. The concentrations of EGCg were 20, 75, and 200 μM, and the concentrations of EGC and MG were 200 μM.
EGCg Stabilization and Competitive Binding
In the absence of BSA, 29.2% of the EGCg remained after 40 min at pH 7.4, whereas with the addition of BSA 65.8% remained. We used the BSA-induced stabilization of EGCg as a surrogate for measuring binding of EGCg to BSA and did competitive binding assays with the drugs PB and TB. In competitive binding assays with 25 μM EGCg, the IC50 for PB was 450 μM, whereas that for TB was >900 μM (Supporting Information Figure S3).
Molecular Simulation of the Interaction between BSA and EGCg
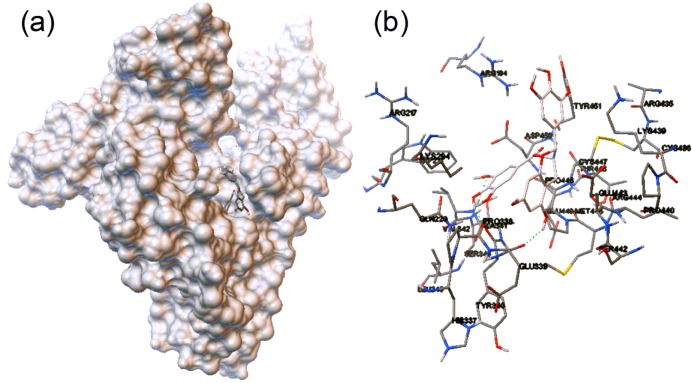
The two docking models with the lowest free energies (−20.8 and −20.3 kJ mol–1) put the Trp residues out of the effective quenching distance of EGCg. The molecular simulation with the next most favorable free energy (−17.9 kJ mol–1) indicated that EGCg binds to the pocket between subdomains IIA and IIIA in the vicinity of Sudlow’s site I. The galloyl moiety is projected toward the bottom, and the flavan-3-ol subunit is positioned in the entrance to the pocket (Figure 5a). The electrostatic interactions were weak between the protein–ligand pair, adding −0.46 kJ mol–1 to the binding energy, whereas van der Waals interactions, hydrogen bonds, and desolvation effects contributed −32.4 kJ mol–1.
Figure 5.
Docking simulation of EGCg for the hydrophobic pocket between subdomains IIA and IIIA of BSA: (a) complexation between BSA and EGCg; (b) amino residues involved in binding EGCg. The stick model indicates the involved amino acid residues and EGCg.
In this simulation model, the binding between EGCg and BSA consists of three hydrogen bonds and various hydrophobic interactions (Figure 5b). The hydrogen atom of the 3-hydroxyl group and the oxygen atom of the 4-hydroxyl group in the galloyl moiety (ring D, Figure 1a) and the hydrogen atom of the 5-hydroxyl group in ring A of the flavan-3-ol subunit of EGCg form hydrogen bonds with the atoms in the peptide backbone of Glu339, Tyr340, and Val342. The simulation results suggest bond lengths of 2.2, 1.8, and 2.0 Å, respectively. EGCg interactions with other nearby amino acid residues, such as Trp134 and Pro446, are dominated by van der Waals interactions and the hydrophobic effect. Electrostatic effects also contribute to EGCg binding, for example, by interactions between the electron-dense hydroxyl groups of the B and D rings with positively charged Arg194, Lys294, and Lys439. The nitrogen atom of the Lys439 side chain is especially close to the oxygen atom of a B ring hydroxyl group (3.9 Å). In addition, the distance from the Trp213 residue to the galloyl moiety (6–10 Å) is shorter than the distance to the flavan-3-ol subunit (12–14 Å).
Discussion
Our fluorescence studies demonstrated that EGCg interacts with BSA, with an apparent binding constant of about 4.6 × 10–5 M–1 at pH 7.4, similar to that obtained in other studies of EGCG–BSA interactions20 and about 10-fold higher than that reported for EGCg–HSA.18,19 However, the Stern–Volmer plot at pH 7.4 exhibited downward curvature, concave toward the x axis, at high EGCg concentrations. This phenomenon is often observed in systems with fluorophores that are unequally quenched due to their accessibility in the protein. Trp134, which is responsible for 85% of the intrinsic fluorescence of BSA, is on the surface of the protein and is generally more accessible than Trp213, which is buried in the hydrophobic pocket.15,32 It has been proposed that EGCg binds near the buried Trp213 residue rather than at the surface-exposed Trp134,20 and our data confirm that idea.
We used measurements of fluorescent lifetimes to more closely examine the binding site of EGCg. The two Trp residues can be distinguished by their fluorescent lifetimes, with lifetime of the surface Trp134 (around 5–6 ns) much longer than that of the buried Trp213 (around 2–3 ns) (Table 2). Fluorescence lifetime decay studies revealed large changes in the fluorescent lifetime of Trp213 induced by EGCg at pH 5 and 7 and demonstrate that EGCg does bind near this residue, deep in the hydrophobic pocket between domains IIA and IIIA. Our data support the binding site on BSA proposed by Skrt et al.20 and demonstrate strong homology to binding with human serum albumin, which clearly involves the single, buried Trp214 on this protein.18,19
The pH dependence of the interaction between EGCg and BSA revealed by the magnitude of the fluorescence quenching is in accord with the importance of the hydrophobic pocket in the BSA–EGCg interaction. At pH 3.0, native BSA exhibits an expanded conformation by unfolding domains I and III,14,33 disassembling the hydrophobic pocket and exposing Trp213 to a surface environment similar to that of Trp134. EGCg-induced quenching effect was weak under this condition with small changes in the fluorescence lifetimes for both Trp residues, suggesting weak nonspecific binding. In addition, λem of the BSA fluorescence spectrum was neither red-shifted nor blue-shifted in the presence of EGCg, indicating a stable molecular environment around two Trp residues. When the pH is increased to 5.0 or 7.4, native BSA achieves its physiological conformation (or heart-shaped structure) with formation of the hydrophobic pocket,14,33 and the affinity for EGCg increases, along with increased specific interaction at Trp213. The λem of the BSA fluorescence spectrum was also gradually red-shifted with increasing EGCg concentration at pH 7.4 (Figure 2c), indicating possible changes in BSA conformation. We propose that EGCg specifically binds to BSA in the hydrophobic pocket between domains IIA and IIIA at physiological pH.
Our data demonstrate that both the flavan-3-ol and the galloyl group of EGCg are essential for site-specific, high-affinity binding. The apparent affinity of the flavan-3-ol EGC for BSA is only about 103 M–1, slightly smaller than the value reported for several other flavan-3-ols in an earlier study.23 The pH independence of EGC binding is consistent with nonspecific interactions. MG has a somewhat higher affinity for BSA, and the pH dependence of the binding and the fluorescence lifetimes suggest that MG binds in the hydrophobic pocket near Trp213. However, it binds an order of magnitude more weakly than EGCg. The CD spectra provide further evidence that EGCg binding requires both the flavan-3-ol and galloyl group. EGCg alters BSA conformation in a dose-dependent fashion, but neither MG nor EGC has this effect.
The molecular simulation model is consistent with our experimental work, demonstrating the favorable interaction of EGCg with the hydrophobic pocket of BSA. At pH 7.4, a positively charged microenvironment comprising Arg194, Arg435, Lys187, Lys294, and His438 is found at the entrance to the hydrophobic pocket, similar to that in human serum albumin.19 These residues may be involved in the initial step of BSA–EGCg complexation by binding to the electron-dense B ring of the flavan-3-ol subunit.22,34 In addition, the nitrogen atom of the Lys439 side chain is 3.9 Å from the oxygen atom in the B-ring hydroxyl group. Considering that the van der Waals radii of both nitrogen and oxygen atoms are about 1.6 Å,35 it is plausible to assume that the distance between the surface of two atoms is <1 Å. Covalent binding to lysine has been invoked to explain the BSA-induced stabilization of EGCg,12 and our data now suggest that the reaction might occur specifically between the B ring and Lys439 (Figure 5b). Covalent bonding in the hydrophobic pocket is supported by the values for the bimolecular quenching constant for the BSA–EGCg interaction, which were estimated to be 1014 m–1s–1 at pH 7.4, indicating static quenching typical of highly stable interaction such as covalent bonding. The B ring of EGCg is more reactive than the D ring,17 providing additional indirect support for covalent reaction as one component that stabilizes EGCg binding to BSA.
Although our model predicts that EGC binds in the neck of the pocket in a location similar to EGCg binding, our data showed that EGC binding was very weak and nonspecific. We speculate that in EGCg the galloyl ester is essential to position the flavan-3-ol for tight binding by its interaction at the base of the pocket. The model suggests that the critical role played by the galloyl moiety involves hydrogen bonds with the peptide backbone of two amino acids (Figure 5b), similar to the hydrogen bonds proposed by others.18,20 Furthermore, the galloyl group appeared to rest between Arg217 and Asp450 in the bottom of the hydrophobic pocket, where it might disrupt a salt bridge and thus change the potential along the indole ring of Trp213, leading to the red shift observed in the fluorescence spectra.36
Consistent with previous research,11,12 our study showed that EGCg was stabilized by BSA at pH 7.4. We used that stabilization to monitor BSA–EGCg binding in competitive binding experiments with two drugs, PB and TB. The former binds Sudlow site I with an association constant of 6.2 × 105 M–1, whereas the latter interacts more weakly and less specifically with BSA, binding to both Sudlow site I (8.7 × 104 M–1) and Sudlow site II (0.81 × 104 M–1).37,38 PB effectively inhibited EGCg binding to BSA, consistent with our model in which EGCg binds to the hydrophobic pocket on BSA that overlaps with Sudlow site I. TB did not inhibit EGCg binding even at a 50-fold excess, supporting our model in which EGCg binds specifically and tightly in the hydrophobic pocket, not at other binding sites on BSA such as Sudlow II.
Our study indicated that EGCg binds selectively to the hydrophobic pocket on subdomains IIA and IIIA of BSA, overlapping with the Sudlow I binding site but also involving residues near Trp213 deep in the interior of the protein. The protein undergoes a slight conformational change upon polyphenol binding. The pH dependence of the binding is a consequence of the unfolding of BSA at low pH, destroying the hydrophobic pocket that is the EGCg binding site. The interaction requires both the flavan-3-ol and the galloyl moieties of the bioactive polyphenol and may involve site-specific covalent interaction at Lys439. Lys195 occupies a position in the hydrophobic pocket of human SA that is similar to the position of Lys439 in BSA, leading us to suggest that it plays a similar role in stabilizing the HSA–EGCg interactions. Previous studies suggested a role for electrostatic binding at Lys195 in the interaction between human SA and EGCg,19 but our model suggests covalent bond formation at this site. Formation of carbonyl groups on HSA during reaction with EGCg suggests that the reaction may involve Schiff base formation,12 but direct evidence for that modification is not available. Further studies are needed to establish whether EGCg does covalently bind to a specific lysine residue in serum albumin and to further explore how SA–EGCg complexation controls both EGCg stability and redox reactivity in the bloodstream.
Acknowledgments
We acknowledge Neil D. Danielson for the use of the LS55 luminescence spectrometer and C. Scott Hartley for the use of the TemPro 01 system.
Glossary
Abbreviations Used
- BSA
bovine serum albumin
- CD
circular dichroism
- EGC
(−)-epigallocatechin
- EGCg
(−)-epigallocatechin-3-O-gallate
- HPLC
high-performance liquid chromatography
- MG
methyl gallate
- PB
phenylbutazone
- SV
Stern–Volmer
- TB
tolbutamide
Supporting Information Available
Figures depicting additional fluorescence quenching data and competitive binding data. This material is available free of charge via the Internet at http://pubs.acs.org.
Financial support was provided by USDA Specific Cooperative Agreement 58-1932-5-534 and by NIDDK R15DK069285 to A.E.H.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Weinreb O.; Mandel S.; Amit T.; Youdim M. B. H. Neurological mechanisms of green tea polyphenols in Alzheimer’s and Parkinson’s diseases. J. Nutr. Biochem. 2004, 15, 506–516. [DOI] [PubMed] [Google Scholar]
- Velayutham P.; Babu A.; Liu D. M. Green tea catechins and cardiovascular health: an update. Curr. Med. Chem. 2008, 15, 1840–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiawan V. W.; Zhang Z. F.; Yu G. P.; Lu Q. Y.; Li Y. L.; Lu M. L.; Wang M. R.; Guo C. H.; Yu S. Z.; Kurtz R. C.; Hsieh C. C. Protective effect of green tea on the risks of chronic gastritis and stomach cancer. Int. J. Cancer 2001, 92, 600–604. [DOI] [PubMed] [Google Scholar]
- Jagtap S.; Meganathan K.; Wagh V.; Winkler J.; Hescheler J.; Sachinidis A. Chemoprotective mechanism of the natural compounds, epigallocatechin-3-O-gallate, quercetin and curcumin against cancer and cardiovascular diseases. Curr. Med. Chem. 2009, 16, 1451–1462. [DOI] [PubMed] [Google Scholar]
- Yang C. S.; Hong J. G. Prevention of chronic diseases by tea: Possible mechanisms and human relevance. Annu. Rev. Nutr. 2013, 33, 161–181. [DOI] [PubMed] [Google Scholar]
- Singh B. N.; Shankar S.; Srivastava R. K. Green tea catechin, epigallocatechin-3-gallate (EGCg): mechanisms, perspectives and clinical applications. Biochem. Pharmacol. 2011, 82, 1807–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J. D.; Hong J.; Lu H.; Meng X.; Lee M.-J.; Yang C. S.. Bioavailabilities of tea polyphenols in humans and rodents. In Protective Effects of Tea on Human Health; Jain N., Siddiqi M., Weisburger J., Eds.; Cabi Publishing: Wallingford, UK, 2006; pp 25–33. [Google Scholar]
- Tachibana H.; Koga K.; Fujimura Y.; Yamada K. A receptor for green tea polyphenol EGCg. Nat. Struct. Mol. Biol. 2004, 11, 380–381. [DOI] [PubMed] [Google Scholar]
- Jovanovic S. V.; Hara Y.; Steenken S.; Simic M. G. Antioxidant potential of gallocatechins. A pulse radiolysis and laser photolysis study. J. Am. Chem. Soc. 1995, 117, 9881–9888. [Google Scholar]
- Zimeri J.; Tong C. Degradation kinetics of (−)-epigallocatechin gallate as a function of pH and dissolved oxygen in a liquid model system. J. Food Sci. 1999, 64, 753–758. [Google Scholar]
- Bae M. J.; Ishii T.; Minoda K.; Kawada Y.; Ichikawa T.; Mori T.; Kamihira M.; Nakayama T. Albumin stabilizes (−)-epigallocatechin gallate in human serum: binding capacity and antioxidant property. Mol. Nutr. Food Res. 2009, 53, 709–715. [DOI] [PubMed] [Google Scholar]
- Ishii T.; Ichikawa T.; Minoda K.; Kusaka K.; Ito S.; Suzuki Y.; Akagawa M.; Mochizuki K.; Goda T.; Nakayama T. Human serum albumin as an antioxidant in the oxidation of (−)-epigallocatechin gallate: Participation of reversible covalent binding for interaction and stabilization. Biosci., Biotechnol., Biochem. 2011, 75, 100–106. [DOI] [PubMed] [Google Scholar]
- Fanali G.; di Masi A.; Trezza V.; Marino M.; Fasano M.; Ascenzi P. Human serum albumin: from bench to bedside. Mol. Asp. Med. 2012, 33, 209–290. [DOI] [PubMed] [Google Scholar]
- Kun R.; Szekeres M.; Dekany I. Isothermal titration calorimetric studies of the pH induced conformational changes of bovine serum albumin. J. Therm. Anal. Calorim. 2009, 96, 1009–1017. [Google Scholar]
- Majorek K. A.; Porebski P. J.; Dayal A.; Zimmerman M. D.; Jablonska K.; Stewart A. J.; Chruszcz M.; Minor W. Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol. Immunol. 2012, 52, 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki A.; Hori M.; Kimura T.; Ito H.; Hatano T. Interaction of polyphenols with proteins: binding of (−)-epigallocatechin gallate to serum albumin, estimated by induced circular dichroism. Chem. Pharm. Bull. (Tokyo) 2009, 57, 224–228. [DOI] [PubMed] [Google Scholar]
- Hagerman A. E.; Dean R. T.; Davies M. J. Radical chemistry of epigallocatechin gallate and its relevance to protein damage. Arch. Biochem. Biophys. 2003, 414, 115–120. [DOI] [PubMed] [Google Scholar]
- Trnkova L.; Bousova I.; Stankova V.; Drsata J. Study on the interaction of catechins with human serum albumin using spectroscopic and electrophoretic techniques. J. Mol. Struct. 2011, 985, 243–250. [Google Scholar]
- Maiti T. K.; Ghosh K. S.; Dasgupta S. Interaction of (−)-epigallocatechin-3-gallate with human serum albumin: Fluorescence, fourier transform infrared, circular dichroism, and docking studies. Proteins 2006, 64, 355–362. [DOI] [PubMed] [Google Scholar]
- Skrt M.; Benedik E.; Podlipnik C.; Ulrih N. P. Interactions of different polyphenols with bovine serum albumin using fluorescence quenching and molecular docking. Food Chem. 2012, 135, 2418–2424. [DOI] [PubMed] [Google Scholar]
- Trombley J. D.; Loegel T. N.; Danielson N. D.; Hagerman A. E. Capillary electrophoresis methods for the determination of covalent polyphenol-protein complexes. Anal. Bioanal. Chem. 2011, 401, 1523–1529. [DOI] [PubMed] [Google Scholar]
- Ishii T.; Mori T.; Tanaka T.; Mizuno D.; Yamaji R.; Kumazawa S.; Nakayama T.; Akagawa M. Covalent modification of proteins by green tea polyphenol (−)-epigallocatechin-3-gallate through autoxidation. Free Radical Biol. Med. 2008, 45, 1384–1394. [DOI] [PubMed] [Google Scholar]
- Soares S.; Mateus N.; De Freitas V. Interaction of different polyphenols with bovine serum albumin (BSA) and human salivary α-amylase (HSA) by fluorescence quenching. J. Agric. Food Chem. 2007, 55, 6726–6735. [DOI] [PubMed] [Google Scholar]
- Kubista M.; Sjoback R.; Eriksson S.; Albinsson B. Experimental correction for the inner-filter effect in fluorescence-spectra. Analyst 1994, 119, 417–419. [Google Scholar]
- Ding F.; Diao J. X.; Sun Y. Bioevaluation of human serum albumin-hesperidin bioconjugate: insight into protein vector function and conformation. J. Agric. Food Chem. 2012, 60, 7218–7228. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Cui L. J.; Wang J. M.; Huo K.; Chen C.; Zhan W. H.; Wang Y. L. Interaction of aconitine with bovine serum albumin and effect of atropine sulphate and glycyrrhizic acid on the binding. J. Lumin. 2012, 132, 357–361. [Google Scholar]
- Lakowicz J. R.Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, 2006. [Google Scholar]
- Nozaki A.; Kimura T.; Ito H.; Hatano T. Interaction of polyphenolic metabolites with human serum albumin: a circular dichroism study. Chem. Pharm. Bull. (Tokyo) 2009, 57, 1019–1023. [DOI] [PubMed] [Google Scholar]
- Morris G. M.; Huey R.; Lindstrom W.; Sanner M. F.; Belew R. K.; Goodsell D. S.; Olson A. J. Autodock4 and autodocktools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A. B.; Srivastava S.; Phadke R. S. Interaction of 7-hydroxy-8-(phenylazo) 1,3-naphthalenedisulfonate with bovine plasma albumin – spectroscopic studies. J. Biol. Chem. 1999, 274, 21755–21762. [DOI] [PubMed] [Google Scholar]
- Tian J. N. A.; Zhao Y. C.; Liu X. H.; Zhao S. L. A steady-state and time-resolved fluorescence, circular dichroism study on the binding of myricetin to bovine serum albumin. Luminescence 2009, 24, 386–393. [DOI] [PubMed] [Google Scholar]
- Militello V.; Vetri V.; Leone M. Conformational changes involved in thermal aggregation processes of bovine serum albumin. Biophys. Chem. 2003, 105, 133–141. [DOI] [PubMed] [Google Scholar]
- Peters T.All about Albumin: Biochemistry, Genetics and Medical Applications; Academic Press: San Diego, CA, USA, 1996. [Google Scholar]
- Ishii T.; Minoda K.; Bae M. J.; Mori T.; Uekusa Y.; Ichikawa T.; Aihara Y.; Furuta T.; Wakimoto T.; Kan T.; Nakayama T. Binding affinity of tea catechins for HSA: characterization by high-performance affinity chromatography with immobilized albumin column. Mol. Nutr. Food Res. 2010, 54, 816–822. [DOI] [PubMed] [Google Scholar]
- Batsanov S. S. Van der Waals radii of elements. Inorg. Mater. 2001, 37, 871–885. [Google Scholar]
- Vivian J. T.; Callis P. R. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 2001, 80, 2093–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascenzi P.; Bocedi A.; Notari S.; Menegatti E.; Fasano M. Heme impairs allosterically drug binding to human serum albumin sudlow’s site I. Biochem. Biophys. Res. Commun. 2005, 334, 481–486. [DOI] [PubMed] [Google Scholar]
- Joseph K. S.; Anguizola J.; Hage D. S. Binding of tolbutamide to glycated human serum albumin. J. Pharm. Biomed. Anal. 2011, 54, 426–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.