Abstract

Alzheimer’s disease (AD) is one of the most complicated progressive neurodegeneration diseases that involve many genes, proteins, and their complex interactions. No effective medicines or treatments are available yet to stop or reverse the progression of the disease due to its polygenic nature. To facilitate discovery of new AD drugs and better understand the AD neurosignaling pathways involved, we have constructed an Alzheimer’s disease domain-specific chemogenomics knowledgebase, AlzPlatform (www.cbligand.org/AD/) with cloud computing and sourcing functions. AlzPlatform is implemented with powerful computational algorithms, including our established TargetHunter, HTDocking, and BBB Predictor for target identification and polypharmacology analysis for AD research. The platform has assembled various AD-related chemogenomics data records, including 928 genes and 320 proteins related to AD, 194 AD drugs approved or in clinical trials, and 405 188 chemicals associated with 1 023 137 records of reported bioactivities from 38 284 corresponding bioassays and 10 050 references. Furthermore, we have demonstrated the application of the AlzPlatform in three case studies for identification of multitargets and polypharmacology analysis of FDA-approved drugs and also for screening and prediction of new AD active small chemical molecules and potential novel AD drug targets by our established TargetHunter and/or HTDocking programs. The predictions were confirmed by reported bioactivity data and our in vitro experimental validation. Overall, AlzPlatform will enrich our knowledge for AD target identification, drug discovery, and polypharmacology analyses and, also, facilitate the chemogenomics data sharing and information exchange/communications in aid of new anti-AD drug discovery and development.
1. Introduction
Alzheimer’s disease (AD) is a progressive neurodegeneration and a complex multifactorial disorder among the elderly.1 The disorder is reaching epidemic proportions with heavy social and economic costs.2 The pathological features of AD are the loss of neurons in conjunction with the presence of oxidative stress, axonal dystrophy, senile plaques, and neurofibrillary tangles.3 Because of its polygenic nature, AD is thought to be caused not by defects in a single gene, but instead by variations in many genes, proteins, and their complex interactions.4 Thus, it is challenging to develop novel effective medications targeting multiple proteins in order to stop or reverse the progression of the disease.
Great efforts have been devoted to carrying out bioscience research with rapid accumulation of a large volume of scientific data relevant to AD. In particular, studies involved in AD neurosignaling pathways and AD-targeted new chemical ligands have been steadily proliferating.5 The quantity and the quality of the AD special class of molecules are expected to grow at a much faster rate in the future, thanks to rapid technology advancement in biochemistry, biophysics, medicinal chemistry, and pharmacology. Unfortunately, the venues to publicize AD-specific research have been limited to archival journals and periodicals. Although SciFinder and other databases have archived most of the documentation, the reported works are scattered. Thus, it is difficult to find, associate, and validate reported AD-related active chemical molecules and reuse the reported research results for AD target research.
Several AD-related databases have been reported to explore the molecular mechanisms, such as AlzGene and Alzpathway. The AlzGene database has been developed for investigating genetic association in the field of AD. It contains almost all genes related to AD and focuses on systematic meta-analyses of information on AD genetic association.6 AlzPathway, a comprehensive map of signaling pathways of AD, was constructed for exploring the AD pathogenesis.7 However, these databases were mainly designed to investigate the pathogenic mechanisms of AD. To our knowledge, there is still no publicly available AD specific chemical genomics (or chemogenomics) database focusing on small molecules that target proteins related to AD for drug research.
Herein, an integrated cloud computing server, AlzPlatform, has been developed in response to the needs. The platform assembles a large repertoire of AD related chemogenomics data, including genes, protein targets, and small chemical molecules with their bioactivity records, bioassays, and references, as well as approved drugs or those in clinical trial for AD treatments. AlzPlatform enables cloud computing and sourcing services and provides powerful computational algorithms and implemented online computing programs/tools, including our established TargetHunter, HTDocking, and BBB predictor for target identification, drug repurposing, and polypharmacology analysis associated with AD (Figure 1). Therefore, AlzPlatform is a valuable platform for investigating and sharing AD targets and small chemical drug molecules at chemogenomics scale for better understanding the mechanisms of system polypharmacology in aid of new anti-AD drug discovery.
Figure 1.
Overview of AlzPlatform database featured with integrated computing and data-mining functions (www.CBLigand.org/AD).
2. Materials and Methods
2.1. Database Infrastructure
AlzPlatform was constructed based on the established molecular database prototype CBID (www.CBLIgand.org/CBID),8,9 with a MySQL (http://www.mysql.com) database and an apache (http://www.apache.org/) web server. Openbabel10 is the search engine for chemical structures. The web interface is written in PHP language (http://www.php.net/).
2.2. Data Collection and Content
The information of protein targets and chemicals associated with AD was gathered according to the approved drugs, clinical trial drugs, and literatures from various databases, including the DrugBank, ClinicalTrials.gov, BindingDB, AlzGene, PubChem, ChEMBL, and SciFinder database. The corresponding information on signaling pathway of these targets was compiled from the KEGG database. All the chemical structures, affinity values, and additional data including pathways, bioassays, and references were archived in relational database structure formats at the backend of the AlzPlatform database.
2.3. Web Interface
AlzPlatform provides a user-friendly interface with a powerful search engine for the detailed information on AD chemicals and targets.
(i) Keywords Search
This includes gene/protein symbol, compound name/ID, and basic pharmacological properties. Searches involving a combination of these keywords are supported without being case sensitive.
(ii) Structure Query
This provides two types of search functions: substructure and similarity. JME is used as the input interface,11 and OpenBabel is the search engine at the backend.12,13 In the structure search window, users can either sketch the structure in the JME interface or upload a file containing a small chemical molecule. After submission, the search is performed by OpenBabel at the server side and the results will be returned to the client side by loading a new page, including ID, structure of compound, target name, and the corresponding links to the literature.
2.4. Chemoinformatics Tools
Target identification and drug design with desired properties are top priorities for medicinal chemists. The data on chemogenomics and cheminformatics collected in AlzPlatform provide a valuable opportunity to explore the underlying targets; absorption, distribution, metabolism, and excretion (ADME) and toxicity prediction; and also calculation of molecular properties and drug-likeness. As such, state-of-art machine learning algorithms and chemoinformatics tools have been deployed on the platforms for facilitating AD drug design and target identification as briefed below.
TargetHunter
TargetHunter was implemented in AlzPlatform to provide online computing algorithm to predict the possible targets or off-targets of compounds. A detailed description of the algorithm has been published.9 The basic principle of the TargetHunter program is based on a known medicinal chemistry concept: structurally similar compounds have similar physical properties that may result in similar biological profiles. This predicts the targets of a query compound by use of the powerful data-mining algorithm (TAMOSIC), which assigns the targets associated with the most similar compounds of a query chemical as the predicted targets. TargetHunter is a powerful cloud computing tool with attractive features: (i) ease of use; (ii) query data retrieval function; (iii) user choices of desired fingerprints and databases; (iv) high accuracy; and (v) Bioassay finder implemented BioassayGeoMap function to find the authors who have published a bioassay for validation. Such a tool will assist researchers to develop bioactive compounds for research on AD target. TargetHunter is available at http://cbligand.org/TargetHunter.
HTDocking
In addition to the ligand-based TargetHunter tool online, we have also established a high-throughput docking (HTDocking, http://www.cbligand.org/AD/docking_search.php) program. It is a web-based computing tool that automates docking procedure to search for protein targets and to explore interactions between compound and protein. In the current version of AlzPlatform database, crystal structures of proteins related to AD have been collected from the Protein Data Bank (PDB) to build an AD domain-specific subset. AutoDock Vina is used as the docking engine at the backend.14 Water molecules and ligands were removed, hydrogen atoms were added, and the active sites of each protein were defined by the residues around the cocrystallized ligands or generated using the AutoDock utility scripts. AutoDock Vina can provide 3–5 predicted binding affinity values (ΔG values) from different docking poses for each compound in a binding pocket of a protein. In our HTDocking program, we only consider the best binding affinity value which is further transformed as docking score. The docking score is calculated as pKi, where pKi = −log(predicted Ki) and the predicted Ki = exp(ΔG×1000/(1.9871917×298.15). The docking score of a queried compound from each protein structure is used to assess and rank the potential protein partners or targets.
Blood–Brain Barrier (BBB) Predictor
The blood–brain barrier (BBB) is the bottleneck in AD drug development and is the single most important factor limiting the future growth of neurotherapeutics.15 Considering this, a BBB predictor was specially designed to classify whether a compound can cross the blood–brain barrier (BBB+) or not (BBB−). This predictor was built by applying the support vector machine (SVM)16 and LiCABEDS17 algorithms on four types of fingerprints of 1593 reported compounds.18 The BBB predictor is available at http://www.cbligand.org/BBB/.
In addition, AlzPlatform provides toxicity prediction with the Toxtree package19 (http://cbligand.org/Tox), an online service for removal of false positive results20 (http://cbligand.org/PAINS), and property calculator for the calculation of molecular properties, such as molecular weight, formula, number of rotatable bonds, hydrogen bond donors and acceptors, polar surface area, xLogP, and Lipinski’s rule of five.21 The properties calculator was implemented with the CDK package22 and is available at http://www.cbligand.org/cbid/Property_Explorer.php. Furthermore, links are provided for quickly accessing chemoinformatics resources, such as actelion’s property explore for ADME prediction and calculation of molecular properties and drug-likeness with Molinspiration. Thus, AlzPlatform acts as a chemoinformatics hub to other tools and databases, which can facilitate researchers in AD drug development and target identification.
2.5. Chemicals and Reagents
Huperzine A was purchased from J&K Scientific Ltd. (Beijing, China). Methyl sandaracopimarate (MS), a known diterpenoid compound, was isolated and identified from the extract of seeds of Platycladus orientalis.
2.6. Caenorhabditis elegans Strains and Maintenance
The Caenorhabditis elegans (C. elegans) strains Cl4176, also the Escherichia coli OP50 strain, were obtained from the Caenorhabditis Genetics Center (CGC; University of Minnesota, Minneapolis, MN). The transgenic nematode CL4176 strain, as an AD model,23 is a temperature-sensitive mutant strain that expresses human Aβ1–42 when it reaches nonpermissive temperatures. The expression of Aβ1–42 in muscle cells causes paralysis in these mutants. The nematodes were maintained and assayed on nematode growth medium (NGM) agar plates with Escherichia coli OP50 at 16 °C. All worms used were raised from eggs obtained after sodium hypochlorite treatment of hermaphrodites.24
2.7. Paralysis Assay
The paralysis assay was measured according to the method described previously,25 with slight modifications. The strain CL4176 maintained at 16 °C was egg-synchronized onto the 35 mm culture plates with or without methyl sandaracopimarate (300 μM). Transgene expression was induced by increasing the temperature from 16 to 26 °C. Induction occurred 36 h after egg laying and lasted until the last worm became paralyzed. For the paralysis assay, the survival of worms was determined by touch-provoked movement.26 Worms were scored as paralyzed when they failed to respond to repeated touching with a platinum wire. Every experiment was conducted three times in a double-blind manner.
2.8. Cell Culture and Transient Transfection
HepG2 cells were plated at a density of 2 × 106 cells on a 48 well plate 24 h before transfection. Plasmids were transfected using Trans-IT LT (Mirus, Madison, WI) according to the manufacturer’s protocol. To evaluate the binding of methyl sandaracopimarate to PPARγ, triplicate transfections were performed using the following plasmids: pCMX-tk-PPRE-LUC (400 ng), pCMX-PPARγ (200 ng), pCMX (200 ng), and (Beta galactosidase) β-gal (50 ng). At 24 h after transfection, the cells were treated with Rosiglizone (10 μM), vehicle, or the methyl sandaracopimarate at increasing concentrations (0.1, 1, 10, 50, 100, and 500 μM). At 24 h after the compound treatment, the cells were lysed and the Luciferase signal was quantified using a standard luminometer (Perkin-Elmer).The Luciferase signal was normalized to β-gal signal.27 All transfections were performed at least three times.
3. Results
AlzPlatform (www.cbligand.org/AD/) archived 928 genes, 320 AD related proteins, 194 AD drugs approved and in clinical trials, and 405 188 chemicals associated with 1 023 137 records of reported AD bioactivities from 38 284 AD corresponding bioassays and 10 050 references.
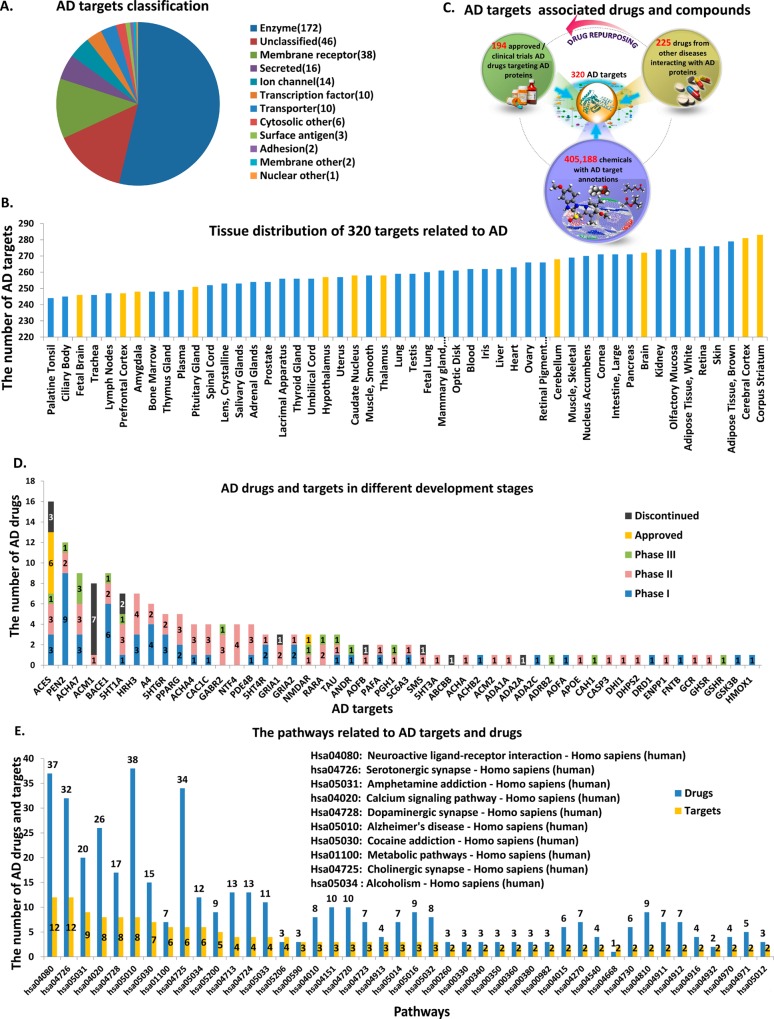
Figure 1 shows an overview of the web-interfaced molecular information database for AD with implemented chemoinformatics computing tools and programs (www.CBLigand.org/AD). The current version of AlzPlatform consists of varieties of AD related proteins (a total of 320): (i) 172 enzymes such as acetylcholinesterase (ACES), monoamine oxidase B, angiotensin-converting enzyme, and cyclooxygenase-2; (ii) 38 membrane receptors, such as serotonin receptors, C–C chemokine receptors, and beta adrenergic receptors; (iii) 14 ion channels, such as glutamate N-methyl-d-aspartate (NMDA) receptors and neuronal acetylcholine receptors; and (iv) 96 other proteins (Figure 2A). For most of these targets, their RNA can be detected in the brain. For example, at least 283 of them are expressed in the corpus striatum and 281 are expressed in the cerebral cortex (Figure 2B). Besides the 194 drugs for AD clinical treatments, AlzPlatform also contains 225 additional drugs that are reported to interact with AD-related proteins but are used for treatment of other diseases. Some of these drugs may have the potential to be repurposed for AD research or treatment. Moreover, small molecules and their bioactivities against these targets could be used for systematic in silico screening of anti-AD lead compounds (Figure 2C). The statistics on these AD drugs, in different development phases, were plotted according to their interacting targets. As shown in Figure 2D, AD drugs approved by the FDA interact with hands of targets, including ACES and NMDA receptors. Among these proteins, muscarinic acetylcholine receptor M1 (ACM1) probably is not an ideal drug target for AD treatment because 7 of 8 drugs targeting ACM1 are discontinued or withdrawn due to undesirable adverse effects,28 and only one is in phase II of clinical trial. In addition, it is well-known that cleavage of amyloid precursor protein (APP) by β-secretase (BACE1) is the rate-limiting step in beta-amyloid production, which suggests a potential target for drug development of AD. Currently, most of BACE1 inhibitors are still in phase I and II clinical trials. MK-8931 developed by Merck is the only BACE1 inhibitor that is currently in phase III clinical trial. However, the research and development pace on BACE1 as the major AD therapeutic target has been slow. Several concerns have been raised about the potential side effects of BACE1-targeted inhibitors,29 because BACE1 also has important roles in myelination, retinal homeostasis, brain circuitry, and synaptic function.30 Therefore, inhibition of the BACE1 enzyme could have toxic consequences. PEN2 could be a good target because it has a gradient number of drugs (9, 2, and 1, respectively) in phases I, II, and III.
Figure 2.
Chemogenomics data archived in AlzPlatform. (A) Summary of AD-related targets. (B) Tissue distribution of targets associated with AD. The yellow lines denote that these tissues are located in the central nervous system. (C) Drugs and compounds associated with AD targets. (D) AD drugs in different development phases and their corresponding targets. These approved and clinical trials AD drugs were classified by different phases with distinct colors. The yellow and black lines indicate the approved and discontinued AD drugs, respectively. The blue, pink, and green lines denote clinical trial drugs in phases I, II, and III, respectively. (E) AD targets and their drugs were plotted according to the pathways the targets involved. The blue and yellow bars indicate drugs and targets, respectively.
The statistics of AD targets and their drugs were plotted according to the pathways the targets involved (Figure 2E). It is not a surprise that Alzheimer’s disease pathway (KEGGID: hsa05010) is among the top of the pathway list. We also notice that pathways related with drug addiction, such as amphetamine addiction (KEGGID: hsa05031), cocaine addiction (KEGGID: hsa05030), and alcoholism (KEGGID: hsa05034), are also enriched on the top list, which could imply that AD shares some common pathways with drug addictions.31 Furthermore, our data show that cannabinoid receptors, the key drug abuse related proteins, are among the top list, which is congruent with the reports that cannabinoid receptors are important in the pathology of AD, and cannabinoids succeed in preventing the neurodegenerative process in AD.32
AlzPlatform also features an integrated cloud computing service with intrinsic scalability and convenient features for further expansion. It provides powerful computing and sourcing services for both computational queried data storage and reretrieval, which can facilitate AD drug research and development. In the next section, three case studies demonstrate the usage of these computational tools on data-mining of the chemogenomics database for AD drug research.
Case Study 1: Prediction and Experimental Validation of Peroxisome Proliferator-Activated Receptor Gamma (PPARγ) as the Neuroprotective Target of a Natural Product Methyl Sandaracopimarate (MS)
Target identification of small chemicals is essential for unraveling the underlying mechanisms of their bioactivities. Often, natural products exhibit significant efficacy, yet their molecular mechanisms remain elusive. We used a natural product to illustrate how the AlzPlatform and high-throughput docking (HTDocking, https://www.cbligand.org/AD/) can be used to identify potential targets and explore the mechanism of action for natural products in which there are often multiple chemical components.
In our previous study, the extract of seeds of Platycladus orientalis can significantly extend lifespan of C. elegans and protect against Aβ toxicity in transgenic C. elegans expressing human Aβ.33 In addition, methyl sandaracopimarate (MS), a known diterpenoid compound, was isolated and identified from the active extract. In order to evaluate the protective effect of the compound against Aβ-induced toxicity, the paralysis assay was conducted in the transgenic C. elegans.CL4176 strain, which expresses human Aβ1–42. As shown in Figure 3A, MS significantly protected against Aβ-induced rapid paralysis at 300 μM in comparison with the untreated control (p < 0.05). Moreover, the mean survival rate of the treated groups was increased by 7.2%.
Figure 3.
Target identification and experimental validation for a bioactive natural product. (A) Time course of Aβ-induced paralysis in the transgenic C. elegans.CL4176 treated with standard nematode growth medium (NGM) and methyl sandaracopimarate (MS). Huperzine A was used as a positive control. (B) The chemical structure query window for AD targets prediction of MS by HTDocking server. (C) Molecular docking study of MS in the active site of PPARγ (PDB: 2OM9). The residues interact with MS through hydrophobic and hydrogen bonds. Yellow dotted lines denote hydrogen bonds and key residues are labeled in black. (D) The predicted target was further validated by in vitro PPARγ responsive luciferase assay. MS activates PPARγ in a concentration dependent manner with EC50 value of 15 μM. The results represent mean ±SD of values. The significance of differences from normal control group is at *p < 0.05.
To explore further the underlying mechanisms of neuroprotection for MS, the AlzPlatform and HTDocking program were used to predict the possible targets. The result shows that the compound is predicted to interact with the peroxisome proliferator-activated receptor gamma (PPARγ), acetylcholinesterase and cGMP-specific 3′, 5′-cyclic phosphodiesterase (PDE5A) (Figure 3B). Among them, PPARγ is listed as one of the top targets with docking score 6.5, suggesting that PPARγ is likely to be a key target for MS. Furthermore, the predicted interactions between the compound and the ligand binding domain of PPARγ were shown in Figure 3C. Residues Phe264, Ser342, Ile341, Phe287, Cys285, Arg288, Gly284, and His266 form a hydrophobic interaction cleft around MS, and the carbonyl group of the compound exhibits the hydrogen-bonding interactions with the backbone of His266 and Lys265, which are vital residues modulating the activation of the receptor,34 suggesting that MS might be a potential PPARγ agonist.
The prediction was further validated by in vitro PPARγ responsive luciferase assay that allows the quantification of the ligand activated PPARγ on the basis of its specific binding to PPRE sequences.35 The result indicated that the MS activates PPARγ in a concentration dependent manner with EC50 value of 15 μM (Figure 4D). According to accumulating evidence, PPARγ is involved in the regulation of β-secretase1 and neuroinflammatory responses.13,36 PPARγ overexpression decreases β-secretase1 gene transcription and reduces the intracellular and plaque Aβ generation in vivo.37,38 Moreover, neurotoxic activities under inflammatory conditions of microglia and astrocytes are reduced by PPARγ agonists.39 On the basis of our study, enhancing PPARγ expression may be one of the mechanism by which MS protected against Aβ-induced toxicity.
Figure 4.
Overview of the application of the TargetHunter program for AD target prediction of small molecules. (A) Input interface; (B) backend server; (C) output predicted results; and (D) the Bioassay GeoMap function can be used to find potential collaborators for targets validation experimentally.
Case Study 2: Prediction of Endoplasmic Reticulum-Associated Amyloid Beta-Peptide-Binding Protein (ERAB) and Cyclooxygenase-2 (COX-2) as Novel Targets for Acteoside and Search of Potential Collaborators for Experimental Validation by TargetHunter
In addition to the protein-based HTDocking program, we have established the ligand-based TargetHunter tool (http://cbligand.org/TargetHunter) which is an online program for targets identification and drug repurposing. We also used natural product to illustrate the application of TargetHunter for target prediction. Acteoside, isolated from Orobanche minor, can significantly inhibit the aggregation of amyloid-β with IC50 value of 8.9 μM40 and can attenuate the Aβ induced toxicity.41 However, the neuroprotective mechanisms are still not exactly known. To identify further the underlying targets, the structure of acteoside was submitted as a query to the TargetHunter program. As shown in Figure 4A–C, two related compounds (ChEMBL510539 and 455827, with scores of 0.82 and 0.75, respectively) were retrieved. The first compound, ChEMBL510539, targets the endoplasmic reticulum-associated amyloid beta-peptide-binding protein (ERAB, tested in the PubChem bioassay AID: 886 with potency value of 0.1 μM). The protein is an intracellular Aβ-binding protein that contributes to the pathogenesis of AD. The toxic effect of Aβ on neuroblastoma cells is prevented by blocking ERAB and is enhanced by overexpression of ERAB.42,43 Therefore, the inhibition of ERAB is likely to the cause of the protection against Aβ induced toxicity by acteoside. In addition, the Cyclooxygenase-2 (COX-2) targeted by another compound (ChEMBL455827) is an important target associated with inflammatory regulation in the pathogenesis of AD.44 To facilitate the further target validation, BioassayGeoMap (http://cbligand.org/TargetHunter/bioassaygeomap.php) is implemented in the AlzPlatform to locate the nearby potential collaborators who reported their established bioassay. As show in Figure 4D, two research laboratories near the University of Pittsburgh were found by the program, and they could be potential collaborators for further experimental validation of the predicted target COX-2.
Case Study 3: The Prediction of Polypharmacology of Known AD Drugs for Multitarget Drug Discovery
Prediction of polypharmacology of known drugs is highly useful for finding new system polypharmacotherapy. There has been increasing interest in identifying additional targets for known drugs and predicting drug–target associations by in silico and experimental methods.45 Accordingly, we used established chemoinformatics tools to predict potential target and polypharmacology for five FDA-approved AD drugs. Among them, four AD drugs (tacrine, rivastigmine, galantamine, and donepezil) are acetylcholinesterase (AChE) inhibitors and the other one (memantine) is an N-methyl-d-aspartate (NMDA) receptor antagonist.46 The protein targets identified by HTDocking program for each known AD drug were tabulated in an output window and ranked by docking scores. The five drugs and their top candidate targets (docking score higher than 6.0, green and pink nodes) were compiled to build a polypharmacological interacting network with Cytoscape 2.8 (Figure 5). Unsurprisingly, the result shows that the known acetylcholinesterase and NMDA receptors (green nodes) were targeted by four AChE inhibitors and memantine, respectively. Moreover, the comparison of the predicted and experimental pKi values for known AD drugs was visualized in Table 1. The result illustrates that the predicted targets and the binding affinities are correlated with reported experimental data. Indeed, the additional predicted associations or drug/protein networks (green nodes and edges), such as beta-secretase1 (BACE1), glycogen synthase kinase-3 beta (GSK3B), and monoamine oxidase type B (MAO-B), have already been reported in the literature47−55 (Table 2), indicating the reliability of the HTDocking program. Also, the remaining predicted targets (pink nodes) could be the new targets for the known drugs that merit further validation by experiments.
Figure 5.
Illustration of HTDocking server (https://www.cbligand.org/AD/) for polypharmacology analysis of 5 approved AD drugs. The large circles (cyan) represent FDA-approved AD drugs (tacrine, donepezil, rivastigmine, galantamine, and memantine). Each drug is linked to its predicted targets. Among them, the green nodes and edges denote the known targets of drugs. Others pink nodes represent new potential off-targets and their interactions are linked by cyan dotted edges.
Table 1. Comparison of the Experimental pKi and the Predicted pKd Values for the FDA-Approved AD Drugs.
| drug | target | experimental Ki (nM) | experimental (−pKi) | HTDocking score predicted (−pKd) |
|---|---|---|---|---|
| tacrine | acetylcholinesterase | 225a | 6.65 | 6.11 |
| galantamine | acetylcholinesterase | 62b | 7.21 | 7.18 |
| rivastigmine | acetylcholinesterase | 920c | 6.04 | 6.08 |
| donepezil | acetylcholinesterase | 23d | 7.64 | 7.25 |
| glutamate [NMDA] receptor subunit 3a | 700e | 6.15 | 7.17 | |
| glutamate [NMDA] receptor subunit 3b | 540f | 6.27 | 6.33 | |
| memantine | glutamate [NMDA] receptor subunit zeta-1 | 1200g | 5.92 | 6.82 |
| glutamate [NMDA] receptor subunit epsilon 2 | 1020h | 6.00 | 6.33 |
Table 2. Verification of Other Predicted Targets by Experiments for FDA-Approved AD Drugs.
| drug | target | experimental potency | ref |
|---|---|---|---|
| galantamine | beta-secretase1 (BACE1) | 44% decrease in BACE1 level/0.3 μM | (74) |
| donepezil | beta-secretase1 (BACE1) | IC50 = 3.2 μM | (75) |
| donepezil | nitric oxide synthase, brain (NOS1) | increase expression of NOS1/5 mg/kg in vivo | (76) |
| donepezil | glycogen synthase kinase-3 beta (GSK3B) | decrease 77% in vivo/(1 mg/kg) | (77) |
| memantine | monoamine oxidase type B (MAO-B) | inhibition of 64%/1 mM | (78) |
| memantine | Adenosine receptor A2a (AA2AR) | increase 43% in vivo (25 mg/kg) | (79) |
| memantine | nitric oxide synthase, brain (NOS1) | active in vivo (10 mg/kg) | (80) |
| memantine | metabotropic glutamate receptor 2 (GRM2) | active/100 μM | (81) |
| memantine | glycogen synthase kinase-3 beta (GSK3B) | inhibit GSK-3/100 μM | (82) |
Another finding in the network is the polypharmacological effects for two known drugs, galantamine and memantine (Figure 5). The network shows that besides binding to AChE, galantamine is predicted to interact with BACE1, mitogen-activated protein kinase 14 (MAPK14), and adenosine A2a receptor (AA2AR). Inhibitions of these proteins have effects on decreasing the Aβ production and Aβ-induced toxicity47 and increasing the expression of nicotinic receptors.56 Similarly, memantine is predicted to interact with GSK3B, BACE1, MAO-B, and nitric oxide synthase 1 (NOS1) besides binding to NMDA receptors. Inhibition of these proteins can prevent the accumulation of the misfolded proteins (Tau and Aβ) and enhance neuronal function.51,55,57 Such in silico analysis of polypharmacological effects may explain why the combined use of memantine and galantamine can produce greater memory improvement than either treatment alone in clinical trials,58 which will guide to design and discover new drug-like leads with the multitarget synergetic therapeutics for AD.
4. Discussion
Alzheimer’s disease is a complex multifactorial disorder.1 With the extensive accumulation of molecular biological elucidations of AD signaling pathway at genes and proteins levels, several AD-related databases have been developed, such as AlzGene6and Alzpathway.7 These databases together with other disease specific databases, such as HLungDB (human lung cancer database)59 and CVDHD (Cardiovascular Disease Herbal Database),60 provide alternative avenues to explore the molecular mechanisms and signaling pathways of diseases. However, there is no comprehensive AD specific chemical genomics knowledgebase available for polypharmacology targets identification to facilitate novel AD drug discovery. Comparing with other general in silico docking platforms, such as DOCK Blaster,61 our AlzPlatform also offers an AD domain-specific chemogenomics database with user-friendly query functions and polytarget identification algorithms implemented with ligand-based TargetHunter and protein structure-based HTDocking. AlzPlatform provides a promising alternative to bridge the knowledge gap between biology and chemistry related to AD, enhancing AD target research, polypharmacology analysis, and new drug discovery.
Our pilot studies demonstrated that the protein-based HTDocking program has been successfully used to identify the AD-related targets for small molecules, such as drugs, lead compounds, and natural product. HTDocking program provides a list of predicted targets and corresponding computational docking-based binding affinity (scores). The reliability of the HTDocking program has been confirmed by comparison of the predicted with the experimental pKi values reported for known AD drugs, also by our in vitro experimental validation for an active natural product. Of course, HTDocking has certain limitations on availability of high-quality protein structures. As a complementary partner, the ligand-based TargetHunter tool is designed to predict the potential targets and off-targets of chemicals using our established chemogenomics database.9 Our established programs also can be useful in the application of drug repurposing, and in the investigation of potential side effects related to AD drugs.62 TargetHunter is a powerful cloud-computing tool with attractive features: usability, flexibility, and veracity. Furthermore, it embeds an important query function, i.e., the geographical bioassay locator can assist users to find nearby collaborators who have reported suitable bioassays in order to validate the target prediction, which will enhance the productivity of collaborative researchers and facilitate the chemogenomics data sharing and information communications.
In addition, our chemoinformatics tools can be used in mapping the drug–target network for polypharmacology investigation. Understanding drug–target associations can benefit the discovery of novel therapeutic applications and also reveal the possible side effects of drugs. It will transform the one-target-one-drug development process to a new multitarget–multidrug paradigm, thereby expanding the opportunity for system multitarget drug discovery.63−65 By assembling many AD related drugs and small molecules with target annotations, AlzPlatform provides specific data and tools to help researchers conduct in-depth analysis for AD related targets and drugs and will also enable the chemists to design multitarget small molecules and to perform bioactivity test with the collaborators, which will boost the more effective system pharmacotherapy and drug design discovery.
5. Conclusion
AlzPlatform, a one-stop integrated cloud computing server, has been specifically developed as a public repository http://www.cbligand.org/AD/ for AD drug and targets research. The cloud computing server will augment our capacity to benefit the AD research community and will help break to the knowledge barrier, enhance the productivity of chemogenomics researchers, and accelerate advances in system biology computer-aided drug design by consolidating existing data and computational technology.
Acknowledgments
Authors would like to acknowledge the financial support for the laboratory at the University of Pittsburgh from the NIH DA025612 and HL109654 (Xie), and Science and Technological Program for Dongguan’s Higher Education, Science and Research, and Health Care Institutions, Guangdong Province, China (Grant No. 2012105102002). We are also grateful to Ph.D. students Chibueze A. Ihunnah and Bingfang Hu in Prof. Wen Xie’s laboratory at School of Pharmacy, University of Pittsburgh (Pittsburgh, PA, U.S.A), for their assistance in biological experiments.
Author Contributions
⊥ H.L. and L.W.: These authors contributed equally to this work.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Querfurth H. W.; LaFerla F. M. Alzheimer’s disease. New Engl. J. Med. 2010, 362, 329–44. [DOI] [PubMed] [Google Scholar]
- Wimo A.; Winblad B.; Jonsson L. The worldwide societal costs of dementia: Estimates for 2009. Alzheimer’s Dementia: J. Alzheimer’s Assoc. 2010, 6, 98–103. [DOI] [PubMed] [Google Scholar]
- Mattson M. P. Pathways towards and away from Alzheimer’s disease. Nature 2004, 430, 631–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray M.; Ruan J.; Zhang W. Variations in the transcriptome of Alzheimer’s disease reveal molecular networks involved in cardiovascular diseases. Genome Biol. 2008, 9, R148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S.; Duan Y.; Hu Y.; Zhao Z. Advances in the pathogenesis of Alzheimer’s disease: a re-evaluation of amyloid cascade hypothesis. Translational Neurodegeneration 2012, 1, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L.; McQueen M. B.; Mullin K.; Blacker D.; Tanzi R. E. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 2007, 39, 17–23. [DOI] [PubMed] [Google Scholar]
- Mizuno S.; Iijima R.; Ogishima S.; Kikuchi M.; Matsuoka Y.; Ghosh S.; Miyamoto T.; Miyashita A.; Kuwano R.; Tanaka H. AlzPathway: a comprehensive map of signaling pathways of Alzheimer’s disease. BMC Syst. Biol. 2012, 6, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Wang J.; Xie Z.-J.; Ma C.; Myint K. Z.; Mu Y.; Xie X.-Q. Cloud Computing Webserver for Cannabinoid Research. In 22nd International Cannabis Research Society (ICRS) Meeting, Freiburg, Germany, July 22–27, 2012.
- Wang L.; Ma C.; Wipf P.; Liu H.; Su W.; Xie X.-Q. TargetHunter: an in silico target identification tool for predicting therapeutic potential of small organic molecules based on chemogenomic database. AAPS J. 2013, 15, 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N. M.; Banck M.; James C. A.; Morley C.; Vandermeersch T.; Hutchison G. R. Open Babel: An open chemical toolbox. J. Cheminf. 2011, 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertl P. Molecular structure input on the web. J. Cheminf. 2010, 2, 10.1186/1758-2946-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N. M.; Banck M.; James C. A.; Morley C.; Vandermeersch T.; Hutchison G. R. Open Babel: An open chemical toolbox. J. Cheminf. 2011, 3, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daynes R. A.; Jones D. C. Emerging roles of PPARs in inflammation and immunity. Nat. Rev. Immunol. 2002, 2, 748–59. [DOI] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardridge W. M. The blood-brain barrier: bottleneck in brain drug development. NeuroRx: J. Am. Soc. Exp. NeuroTherapeutics 2005, 2, 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik V. N.The nature of statistical learning theory; Springer: New York, 1995. [Google Scholar]
- Xie X. Q.; Chen J. Z.; Billings E. M. 3D structural model of the G-protein-coupled cannabinoid CB2 receptor. Proteins 2003, 53, 307–19. [DOI] [PubMed] [Google Scholar]
- Zhao Y. H.; Abraham M. H.; Ibrahim A.; Fish P. V.; Cole S.; Lewis M. L.; de Groot M. J.; Reynolds D. P. Predicting penetration across the blood-brain barrier from simple descriptors and fragmentation schemes. J. Chem. Inf. Model 2007, 47, 170–175. [DOI] [PubMed] [Google Scholar]
- Patlewicz G.; Jeliazkova N.; Safford R. J.; Worth A. P.; Aleksiev B. An evaluation of the implementation of the Cramer classification scheme in the Toxtree software. SAR QSAR Environ. Res. 2008, 19, 495–524. [DOI] [PubMed] [Google Scholar]
- Baell J. B.; Holloway G. A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010, 53, 2719–40. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 2001, 46, 3–26. [DOI] [PubMed] [Google Scholar]
- Steinbeck C.; Han Y.; Kuhn S.; Horlacher O.; Luttmann E.; Willighagen E. The Chemistry Development Kit (CDK): an open-source Java library for Chemo- and Bioinformatics. J. Chem. Inf. Comput. Sci. 2003, 43, 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald C. Y.; Li C. Understanding the molecular basis of Alzheimer’s disease using a Caenorhabditis elegans model system. Brain Struct. Funct. 2010, 214, 263–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. A.; Fleming J. T.. Basic culture methods. In Caenorhabditis elegans: Modern biologicalanalysis of an organism; Epstein H. F., Shakes D. C. Eds.; 1995; p 4–30. [Google Scholar]
- Sutphin G. L.; Kaeberlein M. Measuring Caenorhabditis elegans life span on solid media. J. Vis. Exp.: JoVE 2009, 10.3791/1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow G. J.; White T. M.; Melov S.; Johnson T. E. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc. Natl. Acad. Sci. U.S.A. 1995, 92, 7540–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Z.; Wada T.; Gramignoli R.; Li S.; Strom S. C.; Huang M.; Xie W. MicroRNA hsa-miR-613 targets the human LXRalpha gene and mediates a feedback loop of LXRalpha autoregulation. Mol. Endocrinol. 2011, 25, 584–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich J. N.; Butera J. A.; Carrick T.; Kramer A.; Kowal D.; Lock T.; Marquis K. L.; Pausch M. H.; Popiolek M.; Sun S. C.; Tseng E.; Uveges A. J.; Mayer S. C. Pharmacological comparison of muscarinic ligands: historical versus more recent muscarinic M1-preferring receptor agonists. Eur. J. Pharmacol. 2009, 605, 53–6. [DOI] [PubMed] [Google Scholar]
- Klaver D. W.; Wilce M. C.; Cui H.; Hung A. C.; Gasperini R.; Foa L.; Small D. H. Is BACE1 a suitable therapeutic target for the treatment of Alzheimer’s disease? Current strategies and future directions. Biol. Chem. 2010, 391, 849–59. [DOI] [PubMed] [Google Scholar]
- Evin G.; Hince C. BACE1 as a therapeutic target in Alzheimer’s disease: rationale and current status. Drugs Aging 2013, 30, 755–64. [DOI] [PubMed] [Google Scholar]
- Cai Z.; Ratka A. Opioid system and Alzheimer’s disease. Neuromol. Med. 2012, 14, 91–111. [DOI] [PubMed] [Google Scholar]
- Ramírez B. G.; Blázquez C.; del Pulgar T. G.; Guzmán M.; de Ceballos M. L. Prevention of Alzheimer’s disease pathology by cannabinoids: neuroprotection mediated by blockade of microglial activation. J. Neurosci. 2005, 25, 1904–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Liang F.; Su W.; Wang N.; Lv M.; Li P.; Pei Z.; Zhang Y.; Xie X. Q.; Wang L.; Wang Y. Lifespan extension by n-butanol extract from seed of Platycladus orientalis in Caenorhabditis elegans. J. Ethnopharmacol. 2013, 147, 366–72. [DOI] [PubMed] [Google Scholar]
- Ambrosio A. L.; Dias S. M.; Polikarpov I.; Zurier R. B.; Burstein S. H.; Garratt R. C. Ajulemic acid, a synthetic nonpsychoactive cannabinoid acid, bound to the ligand binding domain of the human peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 2007, 282, 18625–33. [DOI] [PubMed] [Google Scholar]
- Lee J. H.; Gong H.; Khadem S.; Lu Y.; Gao X.; Li S.; Zhang J.; Xie W. Androgen deprivation by activating the liver X receptor. Endocrinol. 2008, 149, 3778–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastre M.; Dewachter I.; Landreth G. E.; Willson T. M.; Klockgether T.; van Leuven F.; Heneka M. T. Nonsteroidal anti-inflammatory drugs and peroxisome proliferator-activated receptor-gamma agonists modulate immunostimulated processing of amyloid precursor protein through regulation of beta-secretase. J. Neurosci.: Off. J. Soc. Neurosci. 2003, 23, 9796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneka M. T.; Sastre M.; Dumitrescu-Ozimek L.; Hanke A.; Dewachter I.; Kuiperi C.; O’Banion K.; Klockgether T.; Van Leuven F.; Landreth G. E. Acute treatment with the PPARgamma agonist pioglitazone and ibuprofen reduces glial inflammation and Abeta1–42 levels in APPV717I transgenic mice. Brain: J. Neurol. 2005, 128, 1442–53. [DOI] [PubMed] [Google Scholar]
- Sastre M.; Dewachter I.; Rossner S.; Bogdanovic N.; Rosen E.; Borghgraef P.; Evert B. O.; Dumitrescu-Ozimek L.; Thal D. R.; Landreth G.; Walter J.; Klockgether T.; van Leuven F.; Heneka M. T. Nonsteroidal anti-inflammatory drugs repress beta-secretase gene promoter activity by the activation of PPARgamma. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunard R.; Ricote M.; DiCampli D.; Archer D. C.; Kahn D. A.; Glass C. K.; Kelly C. J. Regulation of cytokine expression by ligands of peroxisome proliferator activated receptors. J. Immunol. 2002, 168, 2795–802. [DOI] [PubMed] [Google Scholar]
- Kurisu M.; Miyamae Y.; Murakami K.; Han J.; Isoda H.; Irie K.; Shigemori H. Inhibition of amyloid beta aggregation by acteoside, a phenylethanoid glycoside. Biosci., Biotechnol., Biochem. 2013, 77, 1329–32. [DOI] [PubMed] [Google Scholar]
- Wang H.; Xu Y.; Yan J.; Zhao X.; Sun X.; Zhang Y.; Guo J.; Zhu C. Acteoside protects human neuroblastoma SH-SY5Y cells against beta-amyloid-induced cell injury. Brain Res. 2009, 1283, 139–47. [DOI] [PubMed] [Google Scholar]
- Yan S. D.; Fu J.; Soto C.; Chen X.; Zhu H.; Al-Mohanna F.; Collison K.; Zhu A.; Stern E.; Saido T.; Tohyama M.; Ogawa S.; Roher A.; Stern D. An intracellular protein that binds amyloid-beta peptide and mediates neurotoxicity in Alzheimer’s disease. Nature 1997, 389, 689–95. [DOI] [PubMed] [Google Scholar]
- Yan S. D.; Shi Y.; Zhu A.; Fu J.; Zhu H.; Zhu Y.; Gibson L.; Stern E.; Collison K.; Al-Mohanna F.; Ogawa S.; Roher A.; Clarke S. G.; Stern D. M. Role of ERAB/L-3-hydroxyacyl-coenzyme A dehydrogenase type II activity in Abeta-induced cytotoxicity. J. Biol. Chem. 1999, 274, 2145–56. [DOI] [PubMed] [Google Scholar]
- Samy A. S.; Igwe O. J. Regulation of IL-1beta-induced cyclooxygenase-2 expression by interactions of Abeta peptide, apolipoprotein E and nitric oxide in human neuroglioma. J. Mol. Neurosci.: MN 2012, 47, 533–45. [DOI] [PubMed] [Google Scholar]
- Keiser M. J.; Setola V.; Irwin J. J.; Laggner C.; Abbas A. I.; Hufeisen S. J.; Jensen N. H.; Kuijer M. B.; Matos R. C.; Tran T. B.; Whaley R.; Glennon R. A.; Hert J.; Thomas K. L.; Edwards D. D.; Shoichet B. K.; Roth B. L. Predicting new molecular targets for known drugs. Nature 2009, 462, 175–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohanka M. Cholinesterases, a target of pharmacology and toxicology. Biomed. Pap. Med. Faculty Univ. Palacky, Olomouc, Czechoslovakia 2011, 155, 219–29. [DOI] [PubMed] [Google Scholar]
- Li Q.; Wu D.; Zhang L.; Zhang Y. Effects of galantamine on beta-amyloid release and beta-site cleaving enzyme 1 expression in differentiated human neuroblastoma SH-SY5Y cells. Exp. Gerontol. 2010, 45, 842–7. [DOI] [PubMed] [Google Scholar]
- Mohamed T.; Yeung J. C.; Vasefi M. S.; Beazely M. A.; Rao P. P. Development and evaluation of multifunctional agents for potential treatment of Alzheimer’s disease: application to a pyrimidine-2,4-diamine template. Bioorg. Med. Chem. Lett. 2012, 22, 4707–12. [DOI] [PubMed] [Google Scholar]
- Kamat P. K.; Tota S.; Rai S.; Swarnkar S.; Shukla R.; Nath C. A study on neuroinflammatory marker in brain areas of okadaic acid (ICV) induced memory impaired rats. Life Sci. 2012, 90, 713–20. [DOI] [PubMed] [Google Scholar]
- Song X. Y.; Hu J. F.; Chu S. F.; Zhang Z.; Xu S.; Yuan Y. H.; Han N.; Liu Y.; Niu F.; He X.; Chen N. H. Ginsenoside Rg1 attenuates okadaic acid induced spatial memory impairment by the GSK3beta/tau signaling pathway and the Abeta formation prevention in rats. Eur. J. Pharmacol. 2013, 710, 29–38. [DOI] [PubMed] [Google Scholar]
- Onogi H.; Ishigaki S.; Nakagawasai O.; Arai-Kato Y.; Arai Y.; Watanabe H.; Miyamoto A.; Tan-no K.; Tadano T. Influence of memantine on brain monoaminergic neurotransmission parameters in mice: neurochemical and behavioral study. Biol. Pharmaceut. Bull. 2009, 32, 850–5. [DOI] [PubMed] [Google Scholar]
- Marvanova M.; Wong G. Adenosine A2A receptor mRNA expression is increased in rat striatum and nucleus accumbens after memantine administration. Brain Res. Mol. Brain Res. 2004, 120, 193–6. [DOI] [PubMed] [Google Scholar]
- Rai S.; Kamat P. K.; Nath C.; Shukla R. A study on neuroinflammation and NMDA receptor function in STZ (ICV) induced memory impaired rats. J. Neuroimmunol. 2013, 254, 1–9. [DOI] [PubMed] [Google Scholar]
- Wegner F.; Kraft R.; Busse K.; Schaarschmidt G.; Hartig W.; Schwarz S. C.; Schwarz J. Glutamate receptor properties of human mesencephalic neural progenitor cells: NMDA enhances dopaminergic neurogenesis in vitro. J. Neurochem. 2009, 111, 204–16. [DOI] [PubMed] [Google Scholar]
- Goni-Oliver P.; Avila J.; Hernandez F. Memantine inhibits calpain-mediated truncation of GSK-3 induced by NMDA: implications in Alzheimer’s disease. J. Alzheimer’s Disease: JAD 2009, 18, 843–8. [DOI] [PubMed] [Google Scholar]
- Nordberg A. Mechanisms behind the neuroprotective actions of cholinesterase inhibitors in Alzheimer disease. Alzheimer Disease Assoc. Disorders 2006, 20, S12–8. [DOI] [PubMed] [Google Scholar]
- Wu H. M.; Tzeng N. S.; Qian L.; Wei S. J.; Hu X.; Chen S. H.; Rawls S. M.; Flood P.; Hong J. S.; Lu R. B. Novel neuroprotective mechanisms of memantine: increase in neurotrophic factor release from astroglia and anti-inflammation by preventing microglial activation. Neuropsychopharmacol.: Off. Publication Am. College Neuropsychopharmacol. 2009, 34, 2344–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano O.; Ito H.; Takazawa T.; Kawase Y.; Murata K.; Iwamoto K.; Nagaoka T.; Hirayama T.; Miura K.; Nagata R.; Kiyozuka T.; Aoyagi J.; Sato R.; Eguchi T.; Ikeda K.; Iwasaki Y. Clinically meaningful treatment responses after switching to galantamine and with addition of memantine in patients with Alzheimer’s disease receiving donepezil. Neuropsychiatric Disease Treatment 2013, 9, 259–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Xiong Y.; Sun Y.; Fang Z.; Li L.; Ji H.; Shi T. HLungDB: an integrated database of human lung cancer research. Nucleic Acids Res. 2010, 38, D665–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J.; Gui Y.; Chen L.; Yuan G.; Xu X. CVDHD: a cardiovascular disease herbal database for drug discovery and network pharmacology. J. Cheminf. 2013, 5, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin J. J.; Shoichet B. K.; Mysinger M. M.; Huang N.; Colizzi F.; Wassam P.; Cao Y. Automated docking screens: a feasibility study. J. Med. Chem. 2009, 52, 5712–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X.-Q.; Wang L.; Liu H.; Ouyang Q.; Fang C.; Su W., Chemogenomics Knowledgebased Polypharmacology Analyses of Drug Abuse Related G-Protein Coupled Receptors and Their Ligands. Frontiers Pharmacol. 2014, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins A. L. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–90. [DOI] [PubMed] [Google Scholar]
- Bolognesi M. L.; Matera R.; Minarini A.; Rosini M.; Melchiorre C. Alzheimer’s disease: new approaches to drug discovery. Curr. Opin. Chem. Biol. 2009, 13, 303–8. [DOI] [PubMed] [Google Scholar]
- Ouyang Q.; Tong Q.; Feng R.; Myint K.-Z.; Yang P.; Xie X.-Q. Trisubstituted Sulfonamides: A New Chemotype for Development of Potent and Selective CB2 Receptor Inverse Agonists. ACS Med. Chem. Lett. 2013, 4, 387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamulakova S.; Janovec L.; Hrabinova M.; Kristian P.; Kuca K.; Banasova M.; Imrich J. Synthesis, design and biological evaluation of novel highly potent tacrine congeners for the treatment of Alzheimer's disease. Eur. J. Med. Chem. 2012, 55, 23–31. [DOI] [PubMed] [Google Scholar]
- Peng D. Y.; Sun Q.; Zhu X. L.; Lin H. Y.; Chen Q.; Yu N. X.; Yang W. C.; Yang G. F. Design, synthesis, and bioevaluation of benzamides: novel acetylcholinesterase inhibitors with multi-functions on butylcholinesterase, Abeta aggregation, and beta-secretase. Bioorg. Med. Chem. 2012, 20, 6739–6750. [DOI] [PubMed] [Google Scholar]
- Sterling J.; Herzig Y.; Goren T.; Finkelstein N.; Lerner D.; Goldenberg W.; Miskolczi I.; Molnar S.; Rantal F.; Tamas T.; Toth G.; Zagyva A.; Zekany A.; Finberg J.; Lavian G.; Gross A.; Friedman R.; Razin M.; Huang W.; Krais B.; Chorev M.; Youdim M. B.; Weinstock M. Novel dual inhibitors of AChE and MAO derived from hydroxy aminoindan and phenethylamine as potential treatment for Alzheimer's disease. J. Med. Chem. 2002, 45, 5260–5279. [DOI] [PubMed] [Google Scholar]
- Bolognesi M. L.; Banzi R.; Bartolini M.; Cavalli A.; Tarozzi A.; Andrisano V.; Minarini A.; Rosini M.; Tumiatti V.; Bergamini C.; Fato R.; Lenaz G.; Hrelia P.; Cattaneo A.; Recanatini M.; Melchiorre C. Novel class of quinone-bearing polyamines as multi-target-directed ligands to combat Alzheimer's disease. J. Med. Chem. 2007, 50, 4882–4897. [DOI] [PubMed] [Google Scholar]
- Williams M.; Kowaluk E. A.; Arneric S. P. Emerging molecular approaches to pain therapy. J. Med. Chem. 1999, 42, 1481–1500. [DOI] [PubMed] [Google Scholar]
- Kroemer R. T.; Koutsilieri E.; Hecht P.; Liedl K. R.; Riederer P.; Kornhuber J. Quantitative analysis of the structural requirements for blockade of the N-methyl-D-aspartate receptor at the phencyclidine binding site. J. Med. Chem. 1998, 41, 393–400. [DOI] [PubMed] [Google Scholar]
- Banerjee A.; Schepmann D.; Kohler J.; Wurthwein E. U.; Wunsch B. Synthesis and SAR studies of chiral non-racemic dexoxadrol analogues as uncompetitive NMDA receptor antagonists. Bioorg. Med. Chem. 2010, 18, 7855–7867. [DOI] [PubMed] [Google Scholar]
- Takayama H.; Yaegashi Y.; Kitajima M.; Han X.; Nishimura K.; Okuyama S.; Igarashi K. Design, synthesis, and biological evaluation of tricyclic heterocycle-tetraamine conjugates as potent NMDA channel blockers. Bioorg. Med. Chem. Lett. 2007, 17, 4729–4732. [DOI] [PubMed] [Google Scholar]
- Li Q.; Wu D.; Zhang L.; Zhang Y. Effects of galantamine on beta-amyloid release and beta-site cleaving enzyme 1 expression in differentiated human neuroblastoma SH-SY5Y cells. Exp. Gerontol. 2010, 45, 842–847. [DOI] [PubMed] [Google Scholar]
- Mohamed T.; Yeung J. C.; Vasefi M. S.; Beazely M. A.; Rao P. P. Development and evaluation of multifunctional agents for potential treatment of Alzheimer's disease: application to a pyrimidine-2,4-diamine template. Bioorg. Med. Chem. Lett. 2012, 22, 4707–4712. [DOI] [PubMed] [Google Scholar]
- Kamat P. K.; Tota S.; Rai S.; Swarnkar S.; Shukla R.; Nath C. A study on neuroinflammatory marker in brain areas of okadaic acid (ICV) induced memory impaired rats. Life Sci. 2012, 90, 713–720. [DOI] [PubMed] [Google Scholar]
- Song X. Y.; Hu J. F.; Chu S. F.; Zhang Z.; Xu S.; Yuan Y. H.; Han N.; Liu Y.; Niu F.; He X.; Chen N. H. Ginsenoside Rg1 attenuates okadaic acid induced spatial memory impairment by the GSK3beta/tau signaling pathway and the Abeta formation prevention in rats. Eur. J. Pharmacol. 2013, 710, 29–38. [DOI] [PubMed] [Google Scholar]
- Onogi H.; Ishigaki S.; Nakagawasai O.; Arai-Kato Y.; Arai Y.; Watanabe H.; Miyamoto A.; Tan-no K.; Tadano T. Influence of memantine on brain monoaminergic neurotransmission parameters in mice: neurochemical and behavioral study. Biol. Pharma. Bull. 2009, 32, 850–855. [DOI] [PubMed] [Google Scholar]
- Marvanova M.; Wong G. Adenosine A2A receptor mRNA expression is increased in rat striatum and nucleus accumbens after memantine administration. Brain Res. Mol. Brain Res. 2004, 120, 193–196. [DOI] [PubMed] [Google Scholar]
- Rai S.; Kamat P. K.; Nath C.; Shukla R. A study on neuroinflammation and NMDA receptor function in STZ (ICV) induced memory impaired rats. J. Neuroimmun. 2013, 254, 1–9. [DOI] [PubMed] [Google Scholar]
- Wegner F.; Kraft R.; Busse K.; Schaarschmidt G.; Hartig W.; Schwarz S. C.; Schwarz J. Glutamate receptor properties of human mesencephalic neural progenitor cells: NMDA enhances dopaminergic neurogenesis in vitro. J. Neurochem. 2009, 111, 204–216. [DOI] [PubMed] [Google Scholar]
- Goni-Oliver P.; Avila J.; Hernandez F. Memantine inhibits calpain-mediated truncation of GSK-3 induced by NMDA: implications in Alzheimer's disease. J. Alzheimer's Dis.: JAD 2009, 18, 843–848. [DOI] [PubMed] [Google Scholar]