Abstract
Aim:
Huntingtin protein (Htt) was a neuropathological hallmark in human Huntington's Disease. The study aimed to investigate whether the macroautophagy regulator, Beclin1, was involved in the degradation of Htt.
Methods:
PC12 cells and primary cultured brain neurons of rats were examined. pDC316 adenovirus shuttle plasmid was used to mediate the expression of wild-type Htt-18Q-552 or mutant Htt-100Q-552 in PC12 cells. The expression of the autophagy-related proteins LC3 II and Beclin1, as well as the lysosome-associated enzymes Cathepsin B and L was evaluated using Western blotting. The locations of Beclin1 and Htt were observed with immunofluorescence and confocal microscope.
Results:
Htt552 expression increased the expression of LC3 II, Beclin1, cathepsin B and L in autophagy/lysosomal degradation pathway. Treatment with the autophagy inhibitor 3-MA or the proteasome inhibitors lactacystin and MG-132 increased Htt552 levels in PC12 cells infected with Ad-Htt-18Q-552 or Ad-Htt-100Q-552. The proteasome inhibitor caused a higher accumulation of Htt552-18Q than Htt552-100Q, and the autophagy inhibitor resulted in a higher accumulation of Htt552-100Q than Htt552-18Q. Similar results were observed in primary cultured neurons infected with adenovirus. In Htt552-expressing cells, Beclin1 was redistributed from the nucleus to the cytoplasm. Htt siRNA prevented Beclin1 redistribution in starvation conditions. Blockade of Beclin1 nuclear export by leptomycin B or Beclin1 deficiency caused by RNA interference induced the formation of mHtt552 aggregates.
Conclusion:
Beclin1 regulates the accumulation of Htt via macroautophagy.
Keywords: Huntingtin (Htt), Beclin1, protein degradation, autophagy, RNA interference, ubiquitin-proteasome system, autophagy/lysosome pathway
Introduction
Huntington's disease (HD) is a neurodegenerative disorder caused by the expansion of a trinucleotide (CAG) repeat encoding polyglutamine (polyQ) in the N-terminal region of the Huntingtin (Htt) protein. A neuropathological hallmark in human HD and mouse models is the intracellular accumulation of N-terminal Htt fragments1, suggesting that aberrant Htt proteolysis and/or dysfunctional clearance of Htt fragments may underlie the neuropathology in HD. Several proteases, including caspases, calpains and aspartyl endopeptidases, cleave Htt within the N-terminal region, and in vitro studies have demonstrated that N-terminal Htt fragments with expanded polyglutamine have enhanced cytotoxicity2. Although some evidence shows that wild-type Htt has an essential role in developmental and cellular processes3,4,5,6,7, the physiological role of Htt still needs further investigation. DiFiglia's lab was the first to find that cytoplasmic mHtt aggregates had a distribution similar to that of autophagosomes-lysosomes in postmortem HD brains8 and suggested a possible role for autophagy in HD. Later, with in vitro and invertebrate model systems, other work also indicated that autophagy is an important component of the cellular response to mHtt9,10,11,12,13. Recently, Heng et al employed a novel knock-in HD mouse model and reported an association of mHtt immunoreactive cytoplasmic aggregates with autophagosomes and the early and sustained induction of autophagy-associated proteins, suggesting that autophagy is indeed an important component of the neuronal response to mHtt expression in vivo14.
Htt is the first neuronal protein shown to be a caspase substrate with defined sites for caspase-3 at amino acids 513 and 552, for caspase-2 at amino acid 552, and for caspase-6 at amino acid 586. Using an antibody that only detects caspase-cleaved Htt, Wellington et al demonstrated that Htt was cleaved in vivo specifically at the caspase consensus site at amino acid 552. This form of Htt was also detected in control human brains and in HD brains with early phase neuropathology, as well as in wild-type and HD transgenic mouse brains before the onset of neurodegeneration. These data suggest that caspase cleavage of Htt would be a normal physiological event15. However, in HD, the N-terminal fragments resulting from the cleavage of mutant Htt have the potential to increase cytotoxicity and accumulation because of the presence of the expanded polyglutamine tract. In previous research, various fragments (N-terminal 171 aa or 5′-3 kb) were used13,16, but all of these fragments do not exist in physiological conditions. In this study, the 552 aa fragment was used to produce results which would approach the HD pathophysiological conditions closely.
In HD, mHtt forms aggregates (Htt body) both in the nucleus and the cytoplasm, including in the neuronal synapse17,18. Numerous studies verified that an expanded polyQ tract provoked a dominant gain-of-function neurotoxicity. Treatment with Congo Red or trehalose reduced the accumulation of overexpressed expanded polyQ-positive proteins, increased the rate of their degradation and alleviated neurological symptoms in HD transgenic mouse models12,19,20. Eukaryotic cells have two major protein degradation pathways. One is the ubiquitin-proteasome pathway that is responsible for the selective degradation of most short-lived proteins21,22. Neuronal N-terminal-Htt inclusions are highly ubiquitinated. However, it was reported that mutant Htt impaired synaptic ubiquitin-proteasome system activity in cultured neurons and in HD mouse brains expressing either N-terminal or full-length mutant Htt17. The other protein degradation pathway is the autophagy/lysosomal pathway that consists of the delivery of intracellular and endocytosed proteins to the lysosomes. Autophagosomes sequester the cytoplasmic portions, intracellular organelles fuse with lysosomes and the sequestered materials are then degraded by cathepsins found in the lysosomes23,24. The addition of 3-methyladenine (3-MA), an inhibitor of class III phosphatidylinositol 3-kinase and autophagy, increased Htt aggregate formation in x57 cells, while rapamycin, an inducer of autophagy, reduced them11. The transgenic mice with N-terminal fragment had improved performance in behavioral tests when the Htt aggregates were decreased. These results support a potential role of both the proteasome and autophagy in regulating the turnover of expanded polyQ proteins.
Class III PI 3-kinase and its product, phosphatidylinositol 3-phosphate (PI 3-P), are involved in the autophagy signaling pathway. The PI 3-kinase inhibitors, wortmannin and 3-MA, can inhibit the formation of autophagosomes. This indicates that PI 3-kinase activity is important in the early phase of autophagic vesicle formation25. Beclin1 is the ortholog of Atg6, a part of the PI 3-kinase complex, and it plays a role in autophagic vesicle formation in yeast. The PI3-kinase complex may also be functional in the mammalian system26. The expression of Beclin1 in MCF7 cells activated the formation of autophagic vesicles and the degradation of long-lived proteins upon amino-acid starvation. In addition, 3-MA is able to restrain autophagy induced by Beclin1 in MCF-7 cells27. The accumulated mutant Htt can recruit Beclin1 and impair the Beclin1-mediated long-lived protein turnover. Thus, sequestration of Beclin1 in the vulnerable neuronal population of HD patients might reduce Beclin1 function and the autophagic degradation of mutant Htt in these neurons28.
In this study, we verified that the autophagy/lysosome pathway is involved in the degradation of both wild-type and mutant Htt552 in PC12 cells and primary neurons. Htt stimulates the nuclear export of Beclin1, thereby facilitating the autophagic process. Additionally, we found that Beclin1 was an important protein involved in Htt552 degradation, particularly degradation of the mutant form of the protein.
Materials and methods
Materials and drugs
The Htt expression plasmids, pcDNA3-Htt-18Q-969aa-552stop and pcDNA3-Htt-100Q-969aa-552stop, were provided by Dr Marian DiFIGLIA (Massachusetts General Hospital and Harvard Medical School, USA). Both of the two constructs encode an N-terminal Htt fragment of 969 aa with a stop codon after 552 aa. The 18Q is a wild-type Htt with 18 repeats of CAG, while 100Q is a mutant Htt with 100 CAG repeats. Rat PC12 cells were purchased from the Shanghai Institute of Cell Biology (Shanghai, China). Leptomycin B was purchased from Merck Chemicals (Germany). The 3-methyladenine (3-MA) was obtained from Sigma-Aldrich Co (St Louis, MO, USA). The MG-132 and lactacystin were from Enzo Life Sciences, Inc (Farmingdale, NY, USA). Concentrations of 10 nmol/L leptomycin B, 10 mmol/L 3-MA, 10 nmol/L MG-132, and 200 nmol/L lactacystin were used in experiments.
Cell culture
The PC12 cells were cultured in 60-mm dishes in DMEM supplemented with 10% FBS, 4.5% glucose, 100 mg/L streptomycin, 100 units/L penicillin, and incubated in a humidified atmosphere of 5% CO2 at 37 °C.
Pregnant rats at embryonic day 16 were used to create primary neuronal cultures. This procedure included the anesthetization of the rat, sterilization of the incision site and removal of the embryos into a dish containing pre-cooled sterile PBS. The brains of the pups were collected, and then the cortical hemispheres were dissected and placed in a new tube with 10 mL PBS on ice. The tissue was rinsed with pre-cooled PBS 2–3 times, incubated with 10 mL of 2.5% trypsin: DMEM medium (1:1) at 37 °C for 15–20 min, centrifuged at 1200 r/min at 4 °C for 3 min and rinsed 2–3 times with DMEM/F12 medium containing 10% FBS. The tissue pellets were resuspended and incubated in 0.125 mg DNase I in 10 mL of DMEM/F12 containing 10% FBS at 37 °C for 5 min. Samples were then centrifuged at 1400 r/min at 4°C for 3 min. Cells were resuspended to the desired concentration with neurobasal medium (NBM) supplemented with B27, 1% pen/strep and 25 μmol/L glutamine. Single cell suspensions were obtained by passing the cell suspension through a 40-μm cell strainer. Cells were plated on poly-D-lysine-coated dishes or plates with a density of 1×105 cells/cm2 and then incubated in a humidified atmosphere of 5% CO2 at 37 °C. After 3–4 d, half of the medium was replaced with fresh DMEM with 10% FBS/Pen-strep and 10 μmol/L AraC. After another 24 h, the medium was replaced with NBM supplemented with B27, pen/strep, and 2 mmol/L L-glutamine. Lastly, half of the medium was replaced every 3–4 d. The neurons matured after 7 d in culture.
Adenovirus mediated Htt expression system
The N-terminus of wild-type (18Q) and mutant (100Q) Htt, with 3 kb cDNAs and a stop codon at 552 aa, were cloned into the pDC316 adenovirus shuttle plasmid. Wild-type and mutant Htt cDNAs were excised from their parental vectors using BamHI and XbaI and then ligated to BamHI/XbaI-digested pUC18, an intermediate vector. Next, these cDNAs were ligated to BglII/SalI-digested pDC316. Two independent adenovirus shuttle plasmids, pDC316-Htt-18Q-552stop and pDC316-Htt-100Q-552stop, were obtained.
Three independent adenoviruses, including Ad-null, Ad-Htt-18Q-552stop, and Ad-Htt-100Q-552stop, were obtained by co-transfection of 293 cells with the backbone plasmid pBHG10 and the shuttle plasmids, pDC316, pDC316-Htt-18Q-552stop, or pDC316-Htt-100Q-552stop. Cytopathic effects happened at 7th day after transfection and both the cells and the supernatant were collected at 10th day. This was the first generation adenovirus. The later proliferation of adenovirus was also manuplated in 293 cells. The fourth generation adenovirus was used in the later experiments.
Protein preparation and Western blot analysis
Cells were harvested and rinsed with ice-cold PBS twice. Each volume of the cell pellet was lysed by five volumes of the Western blot lysing buffer. After being centrifuged at 10 600×g at 4 °C for 10 min, the supernatant was preserved at -70 °C for later use. The protein concentration was determined with a BCA kit (Pierce, USA). The protein samples were denatured by boiling for 5 min in loading buffer, subjected to 10% SDS-PAGE, and then electroblotted onto nitrocellulose membranes (Amersham Biosciences, Piscataway, USA). The membranes were then blocked with 5% non-fat milk for 1 h and first probed with the mouse monoclonal Htt antibody (Chemicon, Temecula, CA) at a dilution of 1:2000 overnight at 4 °C. Next, the membranes were incubated with a horseradish peroxidase-conjugated secondary antibody for 1 h. Lastly, they were visualized by an enhanced chemiluminescence (ECL) kit (Pierce, Rockford, IL, USA).
Immunofluorescence
PC12 cells were plated in 24-well plates containing poly-D-lysine-coated coverslips, and the transfection was performed when cells reached 80% confluence, approximately 24 h after plating. According to the MOI 20, mixed adenovirus in 0.25 mL medium with 5% FBS was transferred to planted cells. After a 2-h incubation, the medium was removed and the cells were cultured in 0.5 mL of fresh, antibiotic-free DMEM. Every 24 h, the medium was changed with fresh, antibiotic-free complete medium. After 24, 48, or 72 h, cells were washed with PBS and fixed in pre-cooled ethanol for 15 min at room temperature. After another wash with PBS, they were incubated in PBS with 0.1% Triton X-100 for 10 min. The cells were then incubated for 1 h in a blocking solution of PBS supplemented with 2% non-fat milk (Guangming Milk, Shanghai, China) at room temperature. Next, the cells were incubated overnight at 4 °C in a blocking solution containing the primary antibody. The cells were then incubated for 2 h at room temperature with secondary antibodies coupled with fluorophores. The expression of Htt was detected with the mouse monoclonal Htt antibody at a dilution of 1:2000 as the first antibody and the FITC-conjugated donkey anti-mouse antibody with a dilution of 1:800 as the second antibody. The expression of Beclin1 was detected with the rabbit polyclonal BECN antibody (Santa Cruz Biotechnologies, Santa Cruz, CA, USA) with a dilution of 1:400 as the first antibody and cy3-conjugated donkey anti-rabbit antibody with a dilution of 1:800 as the second antibody. Lastly, cells were incubated with 300 nmol/L 4',6-diamidino-2-phenylindole (DAPI) for 15 min. The immunostained cells were examined with laser confocal microscopy (C1Sl, Nikon, Tokyo, Japan).
Quantitative analysis of cells
To quantify the location of Htt and Beclin1, PC12 cells were examined with a 60×objective. Confocal images were obtained from ten fields each for the control and treatment conditions. Each field contained 15–20 cells for analysis. Images were coded before evaluation. Up to 100 cells per coverslip were selected at random for the analysis of the signal intensity using Sigma Scan Pro 5. The boundaries of the cell membrane and the nucleus were outlined, and the average intensity was recorded. The immunoreactivity of Htt and Beclin1 was determined with Sigma Scan Pro 5 using a calculation of the ratio of the stain intensity in the nucleus and in the whole cell. At least 50 cells were used for the quantitative analysis. All data are presented as the mean±SEM. Statistical analysis was carried out using Dunnett's test, considering P<0.05 as significant.
RNA interference
The double-stranded Beclin1 and Htt siRNA sequences were designed according to the cDNAs sequences of rat Beclin1 and human Htt. We chose the most effective pair among three pairs tested for their efficiency in the knockdown of target genes. The sequences used for the experiments included 5′-UGAGGAUGACAGUGAACAGTT-3′ and 5′-CUGUUCACUGUCAUCCUCATT-3′ for Beclin1 and 5′-GCUUCGGAGUGACAAGGAATT-3′ and 5′-UUCCUUGUCACUCCGAAGCTG-3′ for Htt. The siRNA was synthesized by the Genepharma company in China.
PC12 cells were plated in 24-well plates and incubated in complete medium without antibiotics for 24 h. The knockdown experiment was then performed. First, 1 μL Lipofectamine 2000 was diluted with 50 μL Opti-MEMI reduced serum medium and then incubated at room temperature for 5 min. A volume of 2 μL siRNA was also diluted with 50 μL Opti-MEMI reduced serum medium. The two solutions were gently mixed and incubated for another 20 min at room temperature. The mixture solution was added to cells containing approximately 400 μL normal medium. The cells were then incubated in a humidified atmosphere of 5% CO2 at 37 °C. After the cells were transfected for 24 h, the efficiency of the siRNAs was determined by Western blot.
Results
The high efficiency expression of Htt552 in PC12 cells
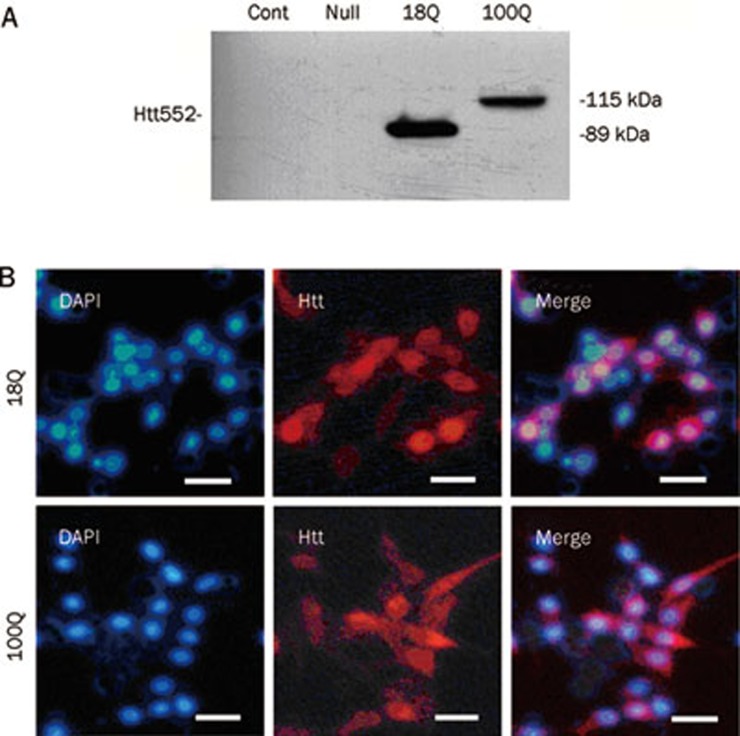
The expression of Htt552 was achieved by insertion of a stop codon after amino acid residue 552 in the 3-kb Htt cDNA. As expected, the expressed wild-type Htt with 1–552 aa (Htt552-18Q) migrated at 89 kDa, while mutant Htt (Htt552-100Q) migrated at 115 kDa (Figure 1A). Our immunofluorescent results showed that the transfection efficiency was approximately 60%–70% at an MOI 30 (Figure 1B) and approximately 90% at an MOI 100 (data not shown). Both wild-type and mutant Htt552 were primarily distributed in the cytoplasm with little or no aggregation.
Figure 1.
The expression of Htt552 in PC12 cells. PC12 cells were infected with Ad-Htt-18Q-552, Ad-Htt-100Q-552, or Ad-null at MOI 30 and harvested 24 h later for Western blot analysis. Both wild-type and mutant Htt552 were expressed at the expected molecular mass (A). PC12 cells were infected with Ad-Htt-18Q-552, Ad-Htt-100Q-552 at MOI 30 for 24 h and then processed for double immunofluorescence of Htt (red) and the nuclear marker DAPI (blue). Cells were examined with fluorescent microscopy (400×, scale bar=10 μm). Both wild-type and mutant Htt552 had a high expression of Htt552, and the rate of transfection reached approximately 60%–70%.
Htt552 expression increased autophagy activity
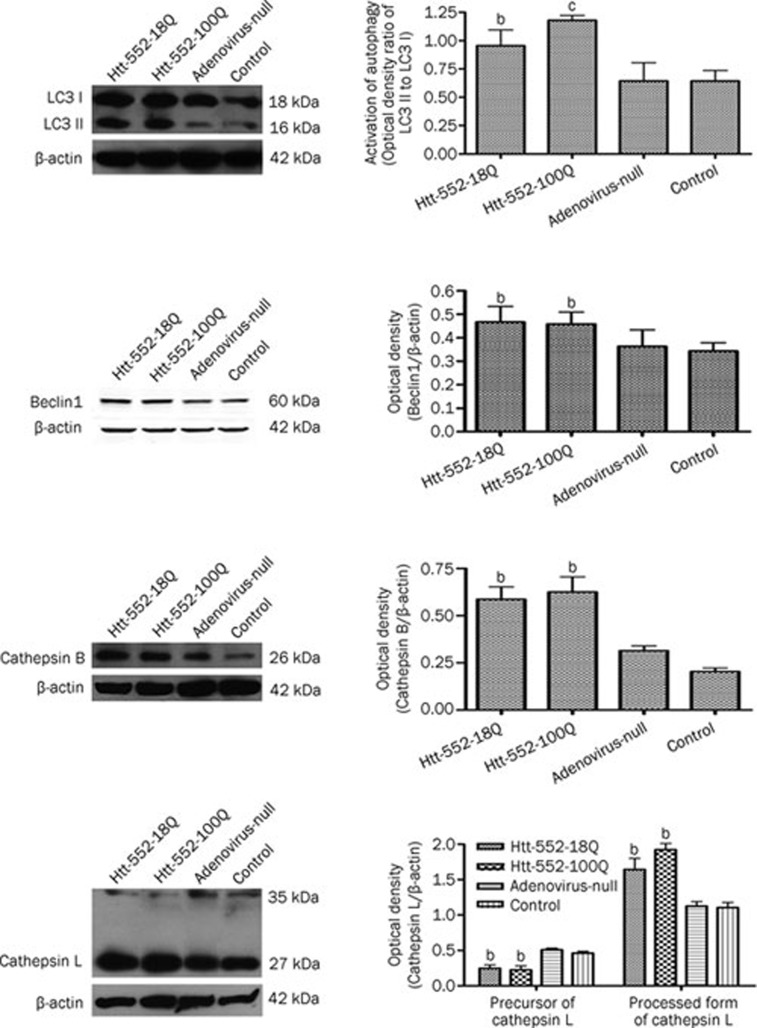
LC3 II is an important protein involved in the formation of autophagosomes. Western blot analysis showed that the level of LC3 II was lower in both normal cells and adenovirus-null infected cells, but it was significantly higher in the cells expressing wild-type or mutant Htt552. Beclin1 is essential for the formation of autophagosomes and cytosol-to-vacuole vesicles29. Beclin1 was also prominently increased after Htt552 expression. In addition, the levels of cathepsin B and L were increased in Htt-expressing cells. Both the expression of autophagy-related proteins and the enzymes found in lysosomes increased, suggesting that autophagy was activated in Htt-expressing cells (Figure 2).
Figure 2.
Autophagy was activated by Htt552 expression. PC12 cells were infected with Ad-Htt-18Q-552, Ad-Htt-100Q-552, or Ad-null at MOI 30 and harvested 24 h later for Western blot analysis. The protein levels of LC3 I and LC3 II increased after Htt552 expression. The data are expressed as a ratio of LC3 II/LC3 I. Beclin1, cathepsin B and the processed form of cathepsin L (bottom panel) were also upregulated after Htt552 expression. The densities of protein bands were analyzed with an image analyzer (Sigma Scan Pro 5) and normalized to the loading control (β-actin). The data are represented as the mean±SEM. Statistical comparisons were carried out with an ANOVA followed by a Dunnett's test. bP<0.05, cP<0.01 vs control.
Both autophagy and proteasome prevent the accumulation of Htt552
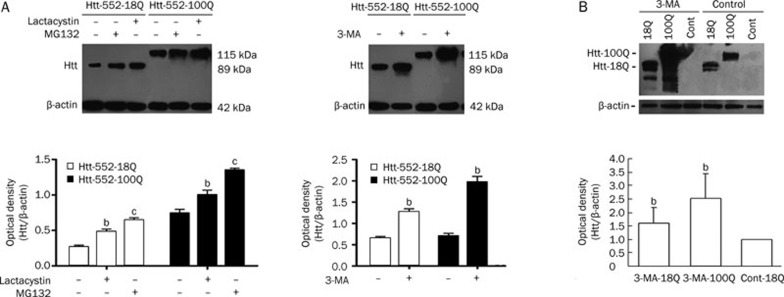
The ubiquitin-proteasome system and autophagy/lysosome pathway are two primary protein degradation pathways that can be activated by different substrates. Treatment with the autophagy inhibitor 3-MA or the proteasome inhibitors lactacystin and MG-132 increased Htt552 levels in PC12 cells infected with Ad-Htt-18Q-552 or Ad-Htt-100Q-552 (Figure 3A). The proteasome inhibitor caused a higher accumulation of Htt552-18Q than Htt552-100Q, and the autophagy inhibitor resulted in a higher accumulation of Htt552-100Q than Htt552-18Q. This suggests that the proteasome plays a key role in the metabolism of Htt552-18Q, whereas autophagy plays a major role in the metabolism of Htt552-100Q. Similar results were observed for our primary cultured neurons infected with adenovirus (Figure 3B). These results support that both the ubiquitin-proteasome system and autophage-lysosomal pathway play a role in the clearance of Htt552.
Figure 3.
Both the UPS and ALP play a role in the clearance of Htt552. PC12 cells were infected with Ad-Htt-18Q-552, Ad-Htt-100Q-552, or Ad-null at MOI 30, and primary neurons were infected at MOI 100. After 24 h, cells were treated with the proteasome inhibitors, MG132 and lactacystin, or the autophagy inhibitor, 3-MA, for 18 h. Cells were then lysed for Western blot analysis. Treatment with MG-132, lactacystin (left panel in A) and 3-MA (right panel in A) increased the levels of Htt552. Treatment with 3-MA also increased the levels of Htt552 in rat primary cultured neurons (B). The densities of protein bands were analyzed with an image analyzer (Sigma Scan Pro 5) and normalized to the loading control (β-actin). The data are represented as the mean±SEM. Statistical comparisons were carried out with an ANOVA followed by a Dunnett's test. bP<0.05, cP<0.01 vs control.
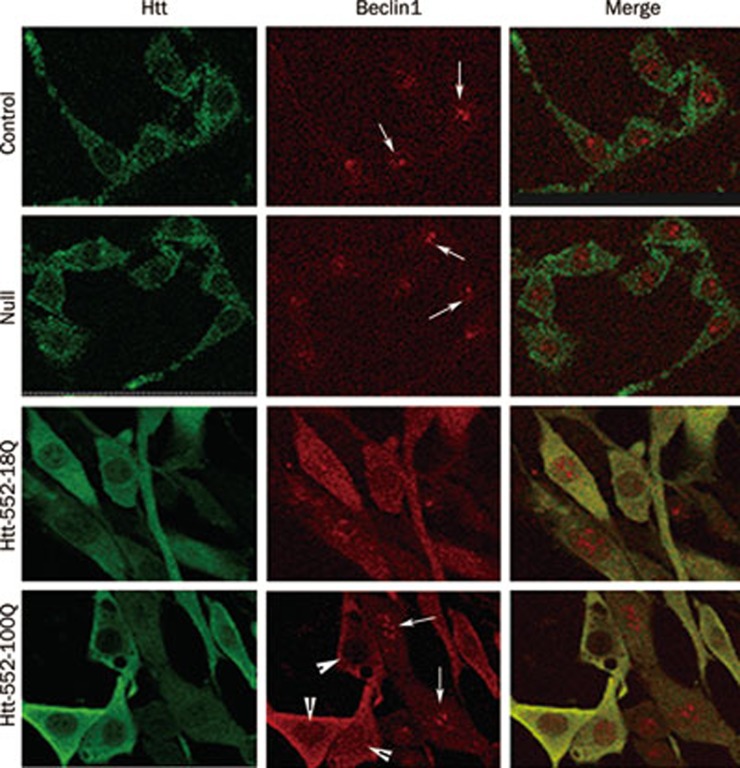
Beclin1 redistributed from the nucleus in Htt552-expressing cells
The export of Beclin1 from the nucleus is the first step in the process of autophagy30. Beclin1 deficiency leads to impaired autophagy function in mammalian cells26,31. Our immunofluorescence results showed that the basal level of Beclin1 was low and distributed mainly in the nucleus of control cells. After expression of Htt552, the levels of Beclin1 greatly increased in the cytosol. Furthermore, the expression of Beclin1 in the cytosol increased as the expression of Htt552 increased (Figure 4).
Figure 4.
Beclin1 levels increased after Htt552 expression in PC12 cells. PC12 cells expressing Htt552 were fixed with pre-cooled EtOH and processed for double immunofluorescence of Htt552 (green) and Beclin1 (red). Cells were analyzed with a confocal microscope (600×). After Htt552 expression, Beclin1 was upregulated in the cytoplasm but reduced in the nucleus, indicating the activation of Beclin1 and the presence of autophagy. The thin arrow refers to Beclin1 in the nucleus, and the arrowhead refers to the export of Beclin1 to the cytoplasm.
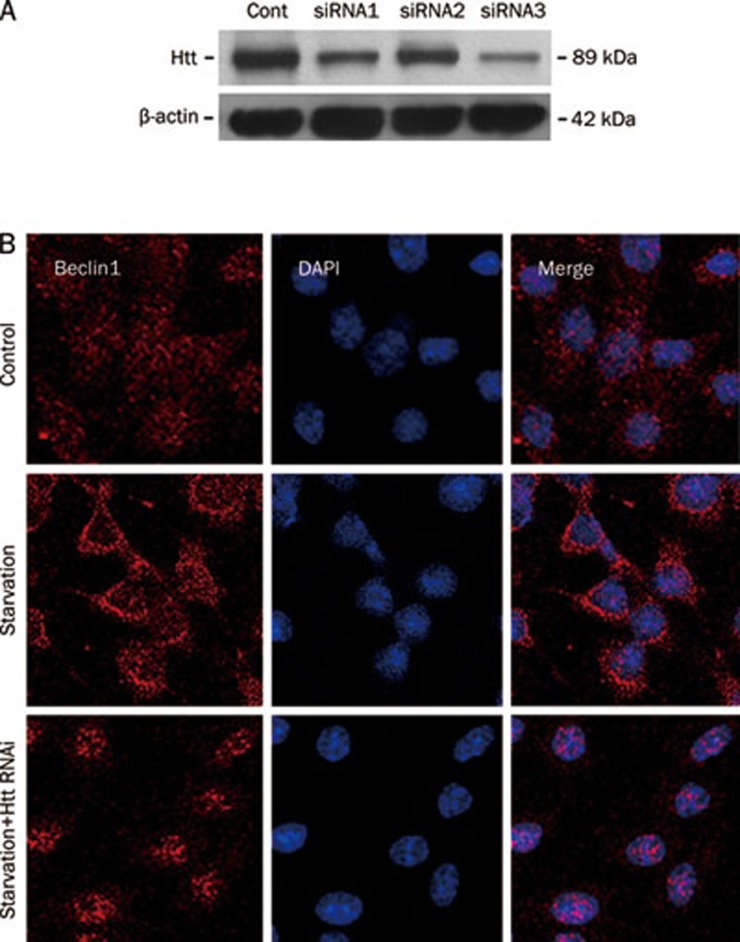
Htt siRNA prevented the redistribution of Beclin1 in starvation conditions
It is known that starvation or amino acid deprivation stimulates the nuclear export of Beclin1 and activates autophagy. Thus, we evaluated the role of Htt in the redistribution of Beclin1 during starvation conditions. The knockdown of endogenous Htt was achieved with the transfection of PC12 cells with Htt siRNA. The Htt siRNA was selected from three siRNAs that were designed based on Htt cDNA. The endogenous Htt in PC12 cells was difficult to detect by normal Western blot; therefore, PC12 cells were transfected with Htt siRNAs 6 h before they were infected with Ad-Htt-18Q-552. After 24 h, Western blot analysis was used to detect the levels of Htt552. siRNA3 was the most efficient in reducing Htt552 expression and was used for the subsequent studies. PC12 cells were transfected with Htt siRNAs for 24 h in normal culture medium with 10% FBS and then in fresh medium without FBS for another 24 h. The cells were harvested for immunofluorescence procedure. The results showed that the redistribution of Beclin1 during starvation was inhibited by the knockdown of Htt552, suggesting that Htt may be involved in Beclin1 activation through the enhancement of Beclin1 exportation from the nucleus to the cytoplasm (Figure 5).
Figure 5.
The effect of Htt on the activation of Beclin1. PC12 cells were transfected with Htt siRNAs for 6 h. They were then infected with Ad-Htt-18Q-552 at MOI 30 and harvested 24 h later for Western blot analysis. siRNA3 was the most efficient of the samples tested (A). Next, PC12 cells were transfected with Htt siRNA in normal culture medium with 10% FBS for 24 h and then changed to fresh medium without FBS and cultured for another 24 h. Lastly, cells were harvested for immunofluorescence. Knockdown of Htt retained Beclin1 in the nucleus during starvation conditions (B). This result suggests that the activation of Beclin1 was inhibited.
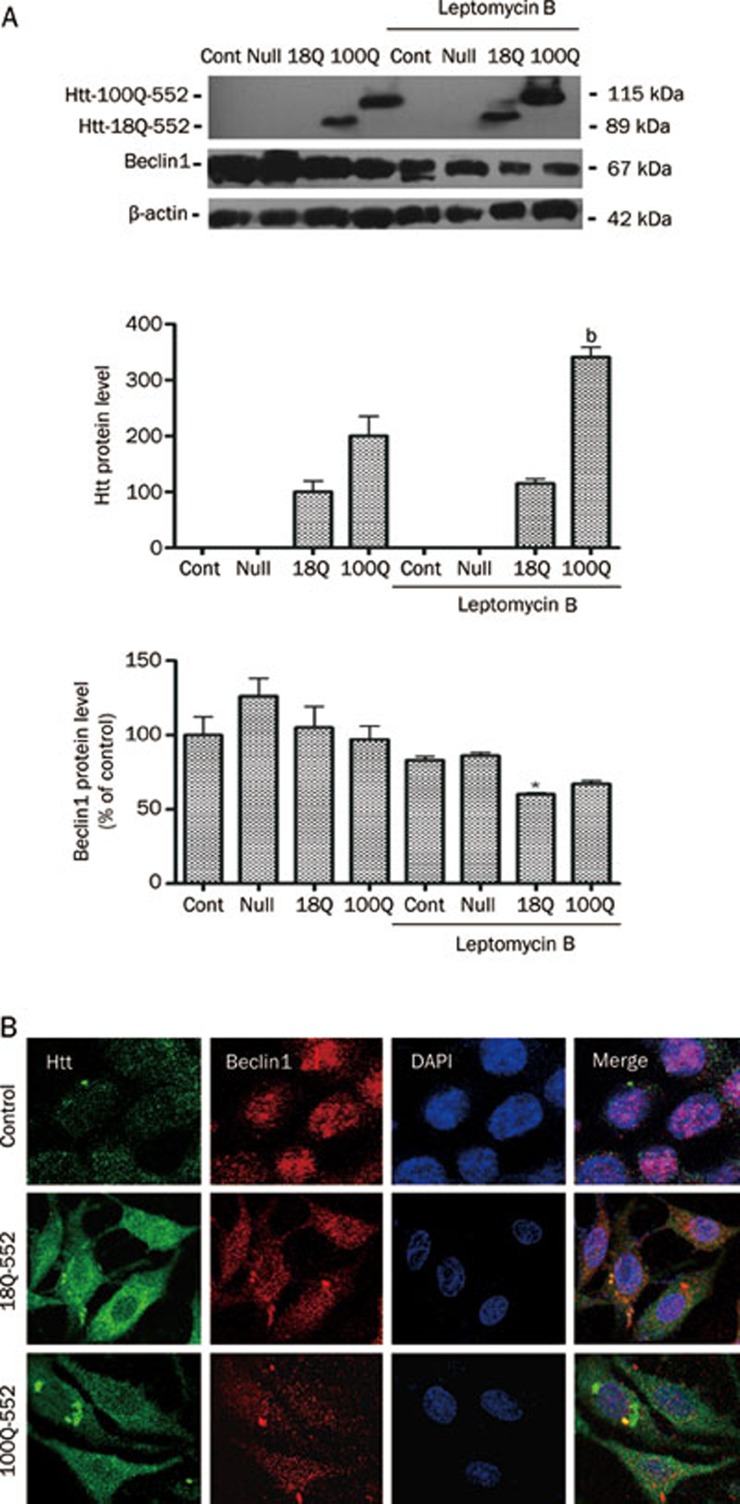
Blockade of Beclin1 nuclear export or Beclin1 deficiency induced the formation of mHtt552 aggregates
The Rev-type NES forms a complex with the nuclear export receptor, CRM1, and Ran GTP. Leptomycin B can bind to CRM1 and block the formation of this complex, thereby blocking the nuclear exportation of the Rev-type nuclear export signal (NES)-containing proteins. Beclin1 contains a leptomycin B-sensitive leucine-rich nuclear export signal that is responsible for its efficient nuclear export. This export is required for autophagy and tumor suppressor function30. Therefore, 10 nmol/L leptomycin B was added to PC12 cells after Htt552 expression. Leptomycin B was added 18 h after adenovirus infection to prevent its influence on the viral infection capacity. After 24 h of treatment with leptomycin B, the cells were fixed and immunofluorescence was performed. The results showed that leptomycin B restrained Beclin1 in the nucleus, and mHtt formed aggregates in the cytosol in some Htt-expressing cells (Figure 6B). In addition, Western blot analysis showed that the expression of Htt552 increased after the treatment with leptomycin B (Figure 6A).
Figure 6.
Leptomycin B induced the formation of Htt552 aggregates. PC12 cells were infected with Ad-Htt-18Q-552, Ad-Htt-100Q-552, or Ad-null at MOI 30 and were then treated with leptomycin B for 18 h. Western blot results showed that leptomycin B inhibited the increase of Beclin1 after Htt552 expression (A). The densities of protein bands were analyzed with an image analyzer (Sigma Scan Pro 5) and normalized to the loading control (β-actin). The data are represented as the mean±SEM. Statistical comparisons were carried out using an ANOVA followed by Dunnett's test. bP<0.05 vs cells without leptomycin B treatment. Immunofluorescence results showed that leptomycin B restrained Beclin1 (red) in the nucleus (B). Protein aggregates of mutant Htt552 (green) were observed in some cells. Cells were detected with a confocal microscope (×600).
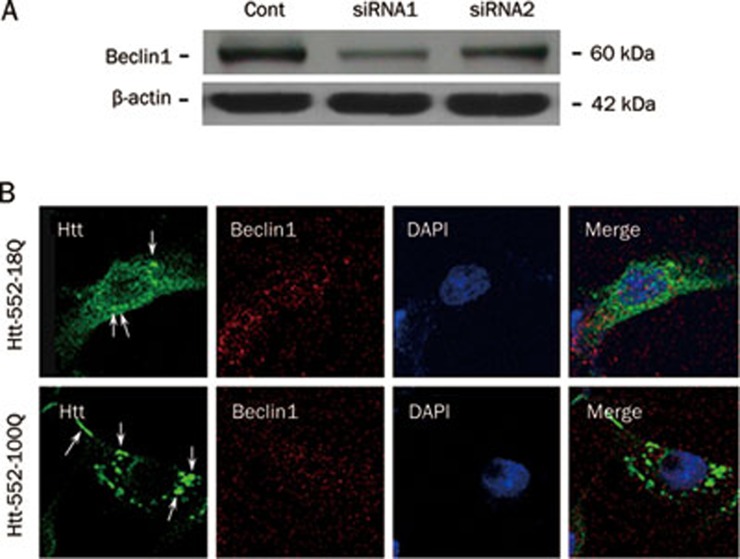
To verify the important role of Beclin1 in Htt degradation, two Beclin1 siRNAs were designed and tested for their efficiency in reducing Beclin1 levels. siRNA1 was more efficient at reducing the levels of Beclin1 and was used in subsequent experiments. The RNA interference of Beclin1 resulted in the formation of mutant Htt552 aggregates (Figure 7), suggesting that Beclin1 is involved in the clearance of Htt552.
Figure 7.
Beclin1 RNAi induced the formation of Htt552 aggregates. To detect the efficiency of Beclin1 RNAi, PC12 cells were transfected with Beclin1 siRNAs and harvested 24 h later for Western blot analysis. siRNA1 was the most efficient of the samples tested (A). PC12 cells were transfected with Beclin1 siRNA1 for 6 h and were then infected with Ad-Htts. After 48 h, cells were fixed and processed for double immunofluorescence of Htt (green) and the nuclear marker DAPI (blue). Cells were analyzed with a confocal microscope (×600). Mutant Htt552 clearly formed aggregates after Beclin1 levels were reduced (B).
Discussion
The accumulation of misfolded proteins in the affected neurons is a common characteristic of several neurological disorders. Macroautophagy (commonly referred to as autophagy) is a crucial cellular process for the bulk degradation of organelles and long-lived proteins. Cytoplasmic substrates are delivered via autophagosomes to lysosomes and degraded. Several studies have suggested an altered autophagy function in HD. Our present results show that both proteasome and autophagy participate in the degradation of wild-type and mutant Htt552. Previous data suggest that proteasome inhibition causes a greater accumulation of mutant Htt than inhibition of autophagy32. According to our experiments, we believe that the different model systems and different concentrations of lactacystin or 3-MA can produce conflicting results. To verify the roles of these pathways, additional work should be done to determine the concentration and the specificity of stimulators or inhibitors of autophagy or the proteasome and utilize the knockdown of specific proteins. However, our results are consistent with previous work which shows that both the proteasome and autophagy affect the degradation of Htt. Far less is known about the molecular events controlling Htt degradation by autophagy in mammalian cells.
Beclin1 is a mammalian ortholog of Atg6/Vps30, a component of the Class III phosphatidylinositol 3-kinase complex in yeast33. The Class III PI 3-kinase and its product, PI 3-P, are involved in the autophagy signaling pathway. It has been verified that Beclin1 contains a core of closely spaced leucine residues that conform to the consensus motif of the NES27,30. Subcellular redistribution and compartmental sequestration of proteins have emerged as important mechanisms in the regulation of cellular responses. Beclin1 plays an important role in macroautophagy by stimulating the formation of autophagosomes. In our study, the levels of Beclin1 increased in the cytoplasm. We also observed that the high expression of mHtt552 resulted in a greater effect on reducing the nuclear level of Beclin1 when compared to lower levels of mHtt552. Moreover, the downregulation of Beclin1 by siRNA and the inhibition of Beclin1 nuclear export by leptomycin B led to the accumulation of Htt552 and the formation of mHtt552 oligomers, even aggregates. These results indicate that Beclin1 is a necessary protein in the autophagic degradation of Htt552.
Our study suggests a possible role of Beclin1 in the autophagic degradation of mutant Htt552. Thus, Beclin1 may be a useful target for the treatment of HD.
Author contribution
Fang LIN and Zheng-hong QIN designed the study. Jun-chao WU, Fang LIN, Lin QI, and Yan WANG performed the research. Kimberly B KEGEL and Jennifer YODER contributed the plasmids. Jun-chao WU and Fang LIN analyzed the data and wrote the manuscript. Zheng-hong QIN and Marian DIFIGILA revised the paper.
Abbreviations
HD, Huntington's disease; Htt, Huntingtin; polyQ, polyglutamine; siRNA, small interference RNA; WT, wild type; LC3, light chain 3.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No 30600197) and the Natural Science Foundation of Jiangsu Province for College (No 09KJB310014).
References
- Kim YJ, Yi Y, Sapp E, Wang Y, Cuiffo B, Kegel KB, et al. Caspase 3-cleaved N-terminal fragments of wild-type and mutant Htt are present in normal and Huntington's disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc Natl Acad Sci USA. 2001;98:12784–9. doi: 10.1073/pnas.221451398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin ZH, Gu ZL, Lin F. The advancement of molecular pathology of Huntington's disease. Chin Pham Bull. 2004;20:378–82. [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pages M, Dompierre JP, Rangone H, Cordelieres FP, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–38. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Rigamonti D, Bauer JH, De-Fraja C, Conti L, Sipione S, Sciorati C, et al. Wild-type huntingtin protects from apoptosis upstream of caspase-3. J Neurosci. 2000;20:3705–13. doi: 10.1523/JNEUROSCI.20-10-03705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir J, Floresco SB, O'Kusky JR, Diewert VM, Richman JM, Zeisler J, et al. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–23. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- Duyao MP, Auerbach AB, Ryan A, Persichetti F, Barnes GT, McNeil SM, et al. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science. 1995;269:407–10. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat Genet. 1995;11:155–63. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- Sapp E, Schwarz C, Chase K, Bhide PG, Young AB, Penney J, et al. Huntingtin localization in brains of normal and Huntington's disease patients. Ann Neurol. 1997;42:604–12. doi: 10.1002/ana.410420411. [DOI] [PubMed] [Google Scholar]
- Kegel KB, Kim M, Sapp E, McIntyre C, Castano JG, Aronin N, DiFiglia M. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20:7268–78. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-proneproteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–17. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–52. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- Qin ZH, Wang Y, Kegel KB, Kazantsev A, Apostol BL, Thompson LM, et al. Autophagy regulates the processing of amino terminal huntingtin fragments. Hum Mol Genet. 2003;12:3231–44. doi: 10.1093/hmg/ddg346. [DOI] [PubMed] [Google Scholar]
- Heng MY, Duong DK, Albin RL, Tallaksen-Greene SJ, Hunter JM, Lesort MJ, et al. Early autophagic response in a novel knock-in model of Huntington disease. Hum Mol Genet. 2010;19:3702–20. doi: 10.1093/hmg/ddq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington CL, Ellerby LM, Gutekunst CA, Rogers D, Warby S, Graham RK, et al. Caspase cleavage of mutant Htt precedes neurodegeneration in Huntington's disease. J Neurosci. 2002;22:7862–72. doi: 10.1523/JNEUROSCI.22-18-07862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W, Jeong H, Cui LB, Krainc D, Tjian R. In vitro analysis of Htt-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–53. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang CE, Orr A, Tydlacka S, Li SH, Li XJ. Impaired ubiquitin-proteasome system activity in the synapses of Huntington's disease mice. J Cell Biol. 2008;180:1177–89. doi: 10.1083/jcb.200709080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamini E, Cavallini G, Donati A, Gori Z. The role of macroautophagy in the ageing process, anti-ageing intervention and age-associated diseases. Int J Biochem Cell B. 2004;36:2392–404. doi: 10.1016/j.biocel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Heiser V, Scherzinger E, Boeddrich A, Nordhoff E, Lurz R, Schugardt N, et al. Inhibition of huntingtin fibrillogenesis by specific antibodies and small molecules: implications for Huntington's disease therapy. Proc Natl Acad Sci U S A. 2000;97:6739–44. doi: 10.1073/pnas.110138997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Rubinsztein DC. Huntington's disease: degradation of mutant huntingtin by autophagy. FEBS J. 2008;275:4263–70. doi: 10.1111/j.1742-4658.2008.06562.x. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Lin F, Qin ZH. The roles of the proteasome pathway in signal transduction and neurodegenerative diseases. Neurosci Bull. 2008;24:183–94. doi: 10.1007/s12264-008-0183-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RJ, Wilson SM. Neurological disease: UPS stops delivering! Trends Pharmacol Sci. 2003;24:18–23. doi: 10.1016/s0165-6147(02)00011-1. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassa A, Roux MP, Attaix D, Bechet DM. Class III phosphoinositide 3-kinase-Beclin1 complex mediates the amino acid-dependent regulation of autophagy in C2C12 myotubes. Biochem J. 2003;376:577–86. doi: 10.1042/BJ20030826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by Beclin 1. Nature (London) 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–85. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- Saeki K, Yuo A, Okuma E, Yazaki Y, Susin SA, Kroemer G, et al. Bcl-2 down-regulation causes autophagy in a caspase-independent manner in human leukemic HL-60 cells. Cell Death Differ. 2000;7:1263–9. doi: 10.1038/sj.cdd.4400759. [DOI] [PubMed] [Google Scholar]
- Liang XH, Yu J, Brown K, Levine B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61:3443–9. [PubMed] [Google Scholar]
- Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of Htt aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–31. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang CE, Huang S, Xu X, Li XJ, Li H, et al. Inhibiting the ubiquitin-proteasome system leads to preferential accumulation of toxic N-terminal mutant huntingtin fragments. Hum Mol Genet. 2010;19:2445–55. doi: 10.1093/hmg/ddq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–5. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]