Abstract
Objective
We aimed to study the prognostic value of BCAR1 expression and its associations with clinical-demographical characteristics in multiple centers of non-small cell lung cancer (NSCLC) patients.
Methods
Gene expression microarray (mRNA) of 77 adenocarcinomas from Mayo Clinic, RNA-sequencing of 508 NSCLC from The Cancer Genome Atlas (TCGA), and immunohistochemistry(IHC) stain of BCAR1-protein expression in 150 cases from Daping Hospital were included in the study. The association of mRNA or protein expression with patient clinical characteristics and overall survival was assessed in each dataset. We also predicted microRNAs (miRNA) that target BCAR1- using bioinformatics prediction tools and evaluated miRNA expression patterns with BCAR1 expression in miRNA-sequencing data of 74 lung cancer cases from TCGA dataset.
Results
In the Mayo Clinic dataset, a higher BCAR1-mRNA level was significantly correlated with more advanced tumor-stage and lymphatic metastasis. Similar changes were observed in the TCGA RNA-seq dataset. Additionally, higher BCAR1-mRNA levels predicted poorer survival in adenocarcinoma and squamous carcinoma from the TCGA dataset. The protein levels in the adenocarcinoma cases with lymphatic metastasis were significantly higher than of those without metastasis. Tumor tissues demonstrated remarkably higher levels of protein compared with matched normal tissues although there was no significant difference in BCAR1-mRNA expression between tumor and matched normal tissues was detected. In miRNAs that were down-regulated in the tumors, Let-7f-2 and miR-22 differed the most (P<0.001 and P=0.007, respectively).
Conclusion
We confirmed that increased BCAR1 expression predicts poorer prognosis in NSCLC. We postulate mRNA-protein decoupling of BCAR1 may be a result of reduced inhibition of specific miRNAs in tumor tissues, which warrants further study.
Keywords: BCAR1, NSCLC, prognosis, mRNA, miRNA
1. Introduction
Breast cancer anti-estrogen resistance 1(BCAR1) also named p130cas, is one of the CAS protein (Crk-associated substrate) family members. The name “Crk” is from "CT10 regulator of kinase" where CT10 is the avian virus from which a protein was isolated, lacking kinase domains, but capable of stimulating phosphorylation of tyrosines in cells. It was originally identified as a cellular protein migrating at 130 kDa of molecular weight, and was hyperphosphorylated in v-Crk and v-Src transformed cells.1, 2 As an adapter protein, BCAR1 localizes to chromosome 16q22-q237 and mainly consists of four functional portions, including an amino-terminal Src homology 3 domain, a Src-binding domain, a large substrate domain, and a helix-loophelix domain. 3, 4 BCAR1 has important roles in a variety of cellular processes, e.g., apoptosis, cell cycle, migration, chemotaxis and differentiation, and is involved in a variety of carcinogenic behavior of cancer cells. 5–8 Our studies 8, 9 show its etiological and clinical outcome involvement in non-small cell lung cancer (NSCLC). Serum BCAR1 levels are significantly higher in NSCLC cases than in the control individuals, increasing with the progression of tumor staging and decreasing after the removal of the malignant lesions.9 Elevated BCAR1 protein expressions in tumor tissues predict poor prognosis in a cohort of NSCLC patients (n=151).8 In addition, BCAR1 knockdown caused cell growth arrest, cell migration inhibition, and cell cycle arrest of A549 lung adenocarcinoma cells.8
However, the correlations between BCAR1-expression and clinical-demographical characteristic of NSCLC, e.g., smoking, cell type and tumor staging, are still unclear. The prognostic value of BCAR1 expression should be confirmed in a large sample of NSCLC, based on multiple datasets. The roles of BCAR1-miRNA in tumor and matched normal tissues are still unclear. Herein, we investigated the associations between the levels of BCAR1-mRNA or -protein and clinical-demographical characteristics of NSCLC. We analyzed the prognostic value of BCAR1-mRNA in NSCLC cases. Additionally, we found the decoupling of BCAR1-mRNA and –protein in tumor and normal matched tissues, which is likely regulated by miRNA.
2. Methods
2.1 Datasets and Patients
2.1.1 mRNA
Mayo Clinic Dataset
The gene expression microarray of Mayo Clinic (Rochester, MN, USA) included 77 adenocarcinoma cases from never-smokers. All the cases had matched normal tissues. The clinical-demographical characteristics of the patients are presented as Table 1. The microarray data was generated by whole genome DASL (cDNA-mediated Annealing Selection extension and Ligation) microarray according to manufacturer's instructions as described. 10 All the study protocols were approved by Mayo Clinic’s Institutional Review Board.
Table 1.
Clinical-demographical characteristics of NSCLC dataset
| Adenocarcinoma-mRNA (complete dataset) |
Adenocarcinoma-mRNA (For survival analysis) |
Squamous Carcinoma-mRNA (complete dataset) |
Squamous Carcinoma-mRNA (For survival analysis) |
Adenocarcinoma -protein (complete dataset) |
Squamous Carcinoma-protein (complete dataset) |
|||
|---|---|---|---|---|---|---|---|---|
| Mayo Clinic dataset (DASL, N=77) |
TCGA dataset (RNA-seq V2, N=288) |
Mayo Clinic dataset (DASL, N=77) |
TCGA dataset (RNA-seq V2, N=188) |
TCGA dataset (RNA-seq V2, N=220) |
TCGA dataset (RNA-seq V2, N=169) |
Daping Hospital (IHC, N=75) |
Daping Hospital (IHC, N=75) |
|
| Age | ||||||||
| N | 77 | 269 | 77 | 188 | 212 | 169 | 74 | 75 |
| Mean (SD) | 67.8(12.9) | 65.6(9.7) | 67.8(12.9) | 64.8(10.1) | 67.5(8.4) | 67.8(8.2) | 60.9 | 64.4(8.1) |
| Gender | ||||||||
| Male | 14(18.2%) | 133(46.2%) | 14(18.2%) | 94(50.0%) | 161(73.2%) | 124(73.4%) | 41(54.7%) | 71(94.7%) |
| Female | 63(81.8%) | 155(53.8%) | 63(81.8%) | 94(50.0%) | 59(26.8%) | 45(26.6%) | 34(45.3%) | 4(5.3%) |
| Tumor stage | ||||||||
| I | 51(66.2%) | 147(51.0%) | 51(66.2%) | 95(50.5%) | 114(51.8%) | 91(53.8%) | 27(36.0%) | 27(36.0%) |
| II | 9(11.7%) | 61(21.2%) | 9(11.7%) | 39(20.7%) | 53(24.1%) | 37(21.9%) | 16(21.3%) | 28(37.3%) |
| III | 17(22.1%) | 58(20.1%) | 17(22.1%) | 41(21.8%) | 46(20.9%) | 36(21.3%) | 27(36.0%) | 18(24.0%) |
| IV | 0 (0.0%) | 16(5.6%) | 0 (0.0%) | 10(5.3%) | 4(1.8%) | 3(1.8%) | 5(6.7%) | 2(2.7%) |
| Unknown | 0(0.0%) | 6(2.1%) | 0(0.0%) | 3(1.6%) | 3(1.4%) | 2(1.2%) | 0(0.0%) | 0(0.0%) |
| Smoking | ||||||||
| Never smoker | 77(100%) | 36(12.5%) | 77(100%) | 0(0.0%) | 10(4.5%) | 0(0.0%) | 0(0.0%) | 0(0.0%) |
| Current/Ever smoker | 0(0.0%) | 240(83.3%) | 0(0.0%) | 188(100%) | 204(92.7%) | 169(100%) | 0(0.0%) | 0(0.0%) |
| Unknown | 0(0.0%) | 12(4.2%) | 0(0.0%) | 0(0.0%) | 6(2.7%) | 0(0.0%) | 75(100%) | 75(100%) |
Note: DASL -(cDNA-mediated Annealing Selection extension and Ligation) microarray
RNA-seq – RNA sequencing
IHC –immunohistochemistry
TCGA Dataset
The mRNA-seq data for lung adenocarcinoma and squamous carcinoma were downloaded from TCGA websites (http://tcga-data.nci.nih.gov/tcga/findArchives.htm). Both clinical and BCAR1 expression were retrieved from the datasets. The mRNA expression data was processed level 3 data (RNA-seq V2), which were normalized gene count to a fixed upper quantity value of 1000. We used log2 (normalized count) for the analysis. The clinical-demographical characteristics of the adenocarcinoma and squamous carcinoma are presented as Table 1.
2.1.2 Protein
We used tissue microarray and IHC to measure BCAR1-protein expression in 150 cases from Daping Hospital (Chongqing city, China). The tissue microarray was constructed in the Department of Pathology at Daping Hospital according to our published protocol 8. The clinical-demographical characteristics of adenocarcinoma and squamous carcinoma are presented in Table 1. All the study protocols were approved by Daping Hospital’s Institutional Review Board.
The sections were incubated with serum blocking solution and anti-BCAR1 antibody (BD Transduction Laboratories, USA, 1:100), biotinylated secondary antibody, and streptavidin-horseradish peroxidase. Diaminobenzidine solution was used as a chromogen. The slides were then counterstained in a hematoxylin solution.
The intensity and percentage of IHC staining in tumor cells were recorded. The intensity was scored from 0 to 3+ and defined as follows: 0, no staining; 1+, weak staining; 2+, moderate staining; 3+, strong staining. Additionally, Q score was adopted for IHC scoring by multiplying the percentage of positive cells by the intensity (Q = Percentage × Intensity; maximum = 300). Three observers scored the slides and the mean score was used for final analysis.
2.1.3 miRNA
The miRNA-seq dataset, including 39 adenocarcinoma and 35 squamous carcinoma and their matched normal tissues, was downloaded from the TCGA website. This was the level 3 normalized data using RPM (reads of mapped miRNA per million of sequence reads). Therefore, we used log2 (RPM) for the analysis.
miRNAs that target 3'UTR region of BCAR1 were identified using miRNA target prediction tools “miRWalk”- (http://www.umm.uni-heidelberg.de) 11. In the meanwhile, miRWalk website also provides the other prediction tools for crosschecking. These tools include “miRanda” 12, “Target scan”13, DIANAmT14, miRDB15, RNAhybrid16, PICTAR417, PITA18, and RNA2219.
2.1.4 Hazard Ratio
We used overall survival (OS) as a primary end point that was defined as the time from diagnosis to death. The patients whose follow-up time was less than 30 days or who died within 30 days after surgery were excluded. To avoid potential confounding from smoking, we separated the cases into never-smokers, current or former smokers. Mayo Clinic cases were all from never-smokers. Because there were only 26 adenocarcinoma and 8 squamous carcinoma cases from the never-smokers with valid follow-up information in TCGA, we only analyzed the prognostic value of BCAR1 in current or former smokers in TCGA-cases.
2.2 Statistical Analysis
The statistical analyses were performed using the Student’s t-test to analyze mRNA and protein differential expression between two groups, X2 or Fisher test for comparison of rates, Spearman’s rho for ordinal variables, e.g., correlation of mRNA/stage, and Pearson Correlation for continuous variables, e.g., correlation of mRNA/age. Prognostic factors were examined by univariate and multivariate analyses using a Cox proportional hazards model. All of the aforesaid calculations were performed using SPSS Version 11.0 software for Windows (SPSS, Inc., Chicago, USA) and Partek Genomics Suite 6.6 software (Partek Inc., Saint Louis, USA). A value of p<0.05 (two-sided) was considered statistically significant.
3. Results
3.1 Association between BCAR1-mRNA expression and clinical characteristics of NSCLC
Table 2 shows that in the 77 never-smokers from the Mayo Clinic dataset, a higher BCAR1-mRNA level was significantly correlated with advanced tumor-stage (Spearman's rho, P<0.001). Additionally, the BCAR1-mRNA levels in the cases with lymphatic metastasis were significantly higher than of those without metastasis (7.271±0.578 vs. 6.825±0.521, P<0.001, Table 2). Similar changes were observed in the TCGA RNA-seq dataset (Table 2). For squamous carcinoma, we did not see any significant association between BCAR1-mRNA and clinical-demographical characteristics (Table 2).
Table 2.
Associations between BCAR1 expression and clinical-demographical characteristics in NSCLC dataset
| Variables | Analysis | Adenocarcinoma-mRNA Mayo Clinic dataset DASL(N=77) |
Adenocarcinoma-mRNA TCGA dataset RNA-seq v2(N=288) |
Squamous Carcinoma-mRNA TCGA dataset RNA-seq V2(N=220) |
Adenocarcinoma-protein Daping Hospital IHC(N=75) |
Squamous carcinoma-protein IHC(N=75) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Valid Case (n) |
BCAR1 | Valid Case (n) |
BCAR1 | Valid Case (n) |
BCAR1 | Valid Case (n) |
BCAR1 (Q score) |
Valid Case (n) |
BCAR1 (Q score) |
||
| Age | Pearson Correlation | 77 | 0.047 | 268 | -0.028 | 211 | 0.041 | 74 | 0.061 | 75 | 0.021 |
| P value | 0.686 | 0.648 | 0.551 | 0.606 | 0.855 | ||||||
| Gender# | Male | 14 | 6.800±0.440 | 133 | 10.332±0.803 | 161 | 10.434±0.609 | 41 | 1.778±0.846 | 71 | 1.649±0.866 |
| Female | 63 | 7.002±0.612 | 154 | 10.448±0.717 | 58 | 10.282±0.883 | 34 | 1.927±0.875 | 4 | 1.100±0.702 | |
| P value | 0.197 | 0.151 | 0.459 | 0.161 | |||||||
| Smoking Status | Never-smoker | N.A. | 36 | 10.331±1.054 | 10 | 10.540±0.653 | N.A. | ||||
| Current or former smoker | 239 | 10.395±0.707 | 203 | 10.392±0.694 | |||||||
| P value | 0.989 | 0.509 | |||||||||
| Stage | Spearman's rho | 77 | 0.400** | 282 | 0.130* | 216 | 0.011 | 75 | 0.170 | 75 | 0.121 |
| P value | <0.001 | 0.029 | 0.877 | 0.145 | 0.300 | ||||||
| Lymphatic metastasis | No | 52 | 6.825±0.521 | 171 | 10.327±0.794 | 141 | 10.363±0.713 | 34 | 1.603±0.846 | 44 | 1.594±0.898 |
| Yes | 25 | 7.271±0.578 | 110 | 10.502±0.694 | 78 | 10.449±0.074 | 40 | 2.068±0.822 | 31 | 1.657±0.824 | |
| P value | 0.001** | 0.052* | 0.755 | 0.020* | 0.761 | ||||||
Shaded variables are statistically different (P<0.05).
P value is significant at the 0.01 level (2-tailed).
P value is significant at the 0.05 level (2-tailed).
N.A. Not available
In addition, there were no appreciable differences of BCAR1-mRNA between adenocarcinoma and squamous carcinoma in the TCGA dataset (t test, P=0.707).
3.2 Association between BCAR1-protein and clinical characteristics of NSCLC
BCAR1-protein was detected in cytoplasm/nucleus or cytoplasm in adenocarcinoma and squamous carcinoma, as described previously.8 Association between BCAR1-protein and clinical characteristics are presented in Table 2. The expression levels in the adenocarcinoma cases with lymphatic metastasis were significantly higher than of those without metastasis (2.068±0.822 vs 1.603±0.846, P=0.020). However no significant difference was seen in the squamous carcinoma cases. Furthermore, no appreciable difference of BCAR1-protein between adenocarcinoma and squamous carcinoma was detected (t test, P=0.138).
3.3 BCAR1-mRNA expression and NSCLC OS
The COX model showed that “tumor-stage” was an independent factor for prognosis in adenocarcinoma from the TCGA dataset (HR: 2.545, 95.0% CI for HR: 1.504–4.308, Table 3). However, “tumor stage” was not a prognostic factor in squamous carcinoma from the TCGA dataset (95.0% CI for HR: 0.872–2.459) and adenocarcinoma from the Mayo Clinic dataset (95.0% CI for HR: 0.754–5.357).
Table 3.
HR-analysis of BCAR1-mRNA in NSCLC by using COX model
| Dataset (Histology) |
Cases (N) |
COX model analysis of OS and BCAR1-mRNA |
||||
|---|---|---|---|---|---|---|
| Variables | P value | HR | 95.0% CI for HR | |||
| Lower | Upper | |||||
| Mayo Clinic (Adenocarcinoma) |
77 | BCAR1 | 0.325 | 1.439 | 0.697 | 2.968 |
| TCGA (Adenocarcinoma) |
188 | BCAR1 | 0.008 | 1.776 | 1.159 | 2.722 |
| Stage # | 0.001 | 2.545 | 1.504 | 4.308 | ||
| `TCGA (Squamous carcinoma) |
169 | BCAR1 | 0.017 | 1.566 | 1.082 | 2.266 |
The COX model did not show a significant association between BCAR1 expression and OS in the 77 never-smokers of the Mayo Clinic dataset (95.0% CI for HR: 0.697–2.968, Table 3). However, a significant association was observed in both adenocarcinoma (HR=1.776, 95.0% CI for HR: 1.159–2.722) and squamous carcinoma cases (HR=1.566, 95.0% CI for HR: 1.082–2.266) from the TCGA dataset (Table 3), suggesting 56% to 77% increased risk of death when the gene’s expression was doubled.
3.4 The decoupling of BCAR1-mRNA and –protein in tumor and normal matched tissues
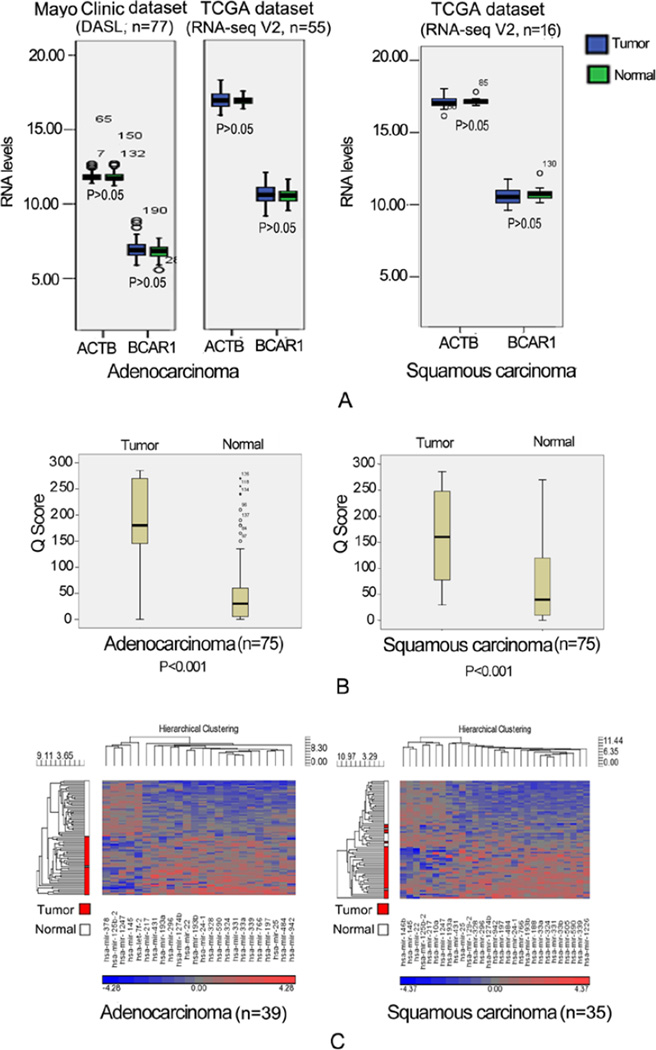
Figure 1A indicates there were no appreciable differences in BCAR1-mRNA expression between tumors and matched normal tissues in either the Mayo Clinic or TCGA dataset.
Figure 1. BCAR1-mRNA,-protein and potential miRNA in tumor vs. matched normal tissue.
A: There were no appreciable differences in BCAR1-mRNA expression between tumors and matched normal tissues in either the Mayo Clinic or TCGA dataset; B: IHC stain of BCAR1-protein demonstrated remarkably higher levels of Q scores as compared with matched normal tissues (adenocarcinoma vs. normal tissues: 185±86 vs. 56±72, P<0.001; squamous carcinoma vs. normal tissues: 162±86 vs 75±83, P<0.001); C: The potential BCAR1-miRNAs predicted by tools were significantly different between tumor and normal tissues. 5 in adenocarcinoma and 7 in squamous carcinoma were down-regulated in the tumors.
However, at protein expression by IHC, Figure 1B indicates both adenocarcinoma and squamous carcinoma demonstrated remarkably higher levels of Q scores as compared with matched normal tissues (Adenocarcinoma vs normal tissues: 185±86 vs 56±72, P<0.001; Squamous carcinoma vs normal tissues: 162±86 vs 75±83, P<0.001).
3.5 Potential microRNAs (miRNAs) of BCAR1
We obtained 112 miRNAs that potentially target BCAR1, which are supported by at least 2 prediction tools. We then retrieved the data of these 112 miRNAs from the TCGA miRNA-seq dataset and analyzed the differentially expressed miRNAs in adenocarcinoma and squamous carcinoma separately.
The miRNAs which were significantly different between tumor and normal tissues in either adenocarcinoma or squamous carcinoma subset are presented in Figure 1C.
Because of the decoupling of mRNA and protein shown above, we were interested in miRNAs that were down-regulated in the tumors. Five miRNAs in adenocarcinoma and seven miRNAs in squamous carcinoma met such criteria (Table 4). Let-7f-2 and miR-22 differed the most between tumors and normal tissues (Table 4 and Figure 1C). mir-125b-2, mir-1247, and miR-145 overlapped between adenocarcinoma and squamous carcinoma (Table 4 and Figure 1C).
Table 4.
Potential miRNA target BCAR1 in adenocarcinoma and squamous carcinoma
| MiRNA | Predictor Tools |
Mean (Tumor) |
Mean (Normal) |
Mean Ratio (tumor/normal) |
P value |
|
|---|---|---|---|---|---|---|
| Adenocarcinoma | miR-125b-2 | Target scan, PICTAR 5 |
3.288 | 4.156 | 0.548 | <0.001 |
| miR-1247 | miRanda, miRWalk |
3.217 | 4.869 | 0.318 | <0.001 | |
| miR-378 | miRanda, miRWalk |
6.575 | 7.296 | 0.606 | <0.001 | |
| miR-145 | miRanda, miRWalk |
9.103 | 9.535 | 0.742 | <0.001 | |
| Let-7f-2 | miRanda, miRWalk |
13.793 | 14.694 | 0.535 | <0.001 | |
| Squamous carcinoma | mir-217 | miRanda, PICTAR 5 |
3.561 | 4.392 | 0.562 | 0.002 |
| mir-125b-2 | miRanda, miRWalk PICTAR 5, Target scan |
4.027 | 4.708 | 0.623 | <0.001 | |
| mir-1247 | miRanda, miRWalk |
4.080 | 5.239 | 0.447 | <0.001 | |
| mir-146b | miRanda, miRWalk |
9.527 | 10.387 | 0.551 | 0.007 | |
| mir-145 | miRanda, miRWalk |
10.152 | 11.289 | 0.455 | 0.004 | |
| mir-10a | miRanda, PICTAR 5 |
14.421 | 14.885 | 0.725 | 0.012 | |
| mir-22 |
miRanda, miRWalk |
16.446 | 16.786 | 0.789 | 0.007 | |
4. Discussion
BCAR1 was initially found to allow breast cancer cells to escape antiestrogen therapy. 20 Subsequently, intensive studies concentrated on the carcinogenic mechanisms of BCAR1 in breast cancer.21–24 Breast cancer with elevated BCAR1 levels is prone to relapse and has poor prognosis. 25 As a scaffolding and adapter protein, BCAR1 has attracted growing attention due to its tumorigenic roles, e.g., invasion and migration, and has been implicated in the carcinogenesis and prognosis in a variety of malignancies, i.e., ovarian, liver, esophagus, and oral cavity.7, 26–28 Recent studies show BCAR1’s involvement in non-small-cell lung cancer (NSCLC).8, 9 BCAR1 mediates a cell survival signal from cell-matrix interaction and alteration, and prevents A549 lung adenocarcinoma cells from anoikis, hence contributing to anchorage independence and metastasis of cancer cells.29 BCAR1 knockdown caused cell migration inhibition of A549 cells.8
Additionally, our studies demonstrated BCAR1 potentially has important clinical implications in NSCLC.8, 9 For examples, serum BCAR1 levels are significantly higher in NSCLC than in the control group, increasing gradually with the progression of tumor staging and decreasing after the removal of the malignant lesions.9 In a small cohort of 151 Chinese patients with NSCLC, elevated BCAR1 protein expressions in tumor tissues predict poor prognosis (hazard ratio 1.777, P =0.028).8 Recently, a study in another small cohort of Chinese patients (n=105) with NSCLC also demonstrated BCAR1 over-expression predicts poor prognosis (HR=2.816, P=0.001). 30
However, many questions remain, for instance, the correlations between BCAR1-expression and clinical-demographical characteristics of NSCLC, e.g., smoking, cell type, and tumor staging. In addition, predictive power of BCAR1 over-expression for poor prognosis should be verified in a large sample of NSCLC. Additionally, little is known about the regulation of BCAR1 protein by-miRNA in cancer.
In our study, high level of BCAR1-mRNA or -protein in adenocarcinoma is significantly correlated with lymphatic metastasis and advanced tumor stage in adenocarcinoma, but not in squamous carcinoma. However, cases with metastasis (M1) in the study are too few to be analyzed because they all were from the surgical candidates.
There were no appreciable differences of the levels of BCAR1-mRNA between tumor and matched normal tissues. However, there is remarkable protein expression difference between tumor and normal tissues. The inconsistency between mRNA and protein level expression in tumor and normal tissues leads us to hypothesize the increased protein expression of BCAR1 in tumor but not in normal may be the result of reduced inhibition of miRNAs that target deactivation of mRNA and subsequent translation of protein level. We used multiple prediction tools and predicted several possible miRNAs of BCAR1 in adenocarcinoma and squamous carcinoma, respectively. However, the bioinformatics prediction and data observation can not establish causal relationship and further mechanistic studies should be performed to investigate the association between expression of these miRNAs and levels of BCAR1-protein. For example, miRNAs’ overexpression/knockdown experiments are critical to establish the regulatory relationship between the miRNAs and BCAR1 mRNA and protein. Following these assays, it is also necessary to elucidate the biological functions of these miRNAs via BCAR1 signaling in apoptosis, proliferation, cell migration and invasion. Furthermore, expression of these miRNAs and clinical outcomes of lung cancer patients should also be investigated.
The BCAR1-associated signaling pathways contributing to metastasis are still unclear. Thus far, overexpression of BCAR1 has been found to be correlated with elevated EGFR expression levels in prostate cancer, 31 and elevated VEGF and p53 expression levels in esophageal cancer.7 Additionally, it has been proven to be associated with decreased KAI1 expression (a metastasis suppressor) in prostate cancer. 31 Recently, Miao et al 32 also found that overexpression of BCAR1 was significantly correlated with abnormal expression of E-cadherin and beta-catenin in 105 NSCLC tissues, suggesting BCAR1 holds an important role in epithelial–mesenchymal transition (EMT) and metastasis. The underlying mechanism of BCAR1-overexpression that predicts poor prognosis in NSCLC warrants further studies.
In this study, the COX model showed that “tumor-stage” was an independent factor for OS in adenocarcinoma from the TCGA dataset, but it was not a significant predictor for squamous carcinoma from the TCGA dataset and adenocarcinoma from Mayo Clinic dataset. When we used disease free survival (DFS) instead of OS to analyze HR in the Mayo Clinic dataset, tumor stage was a significant predictor for DFS (HR: 4.458, 95.0% CI for HR: 2.216–9.347, data not shown). Because most of the Mayo Clinic cases were in early stages and some deaths were not related to lung cancer after long-term follow-up, use of DFS as an outcome endpoint may provide more reliable results. Unfortunately, we did not have the exact cause of death information for the TCGA dataset for such analysis.
BCAR1-mRNA levels predicted poorer prognosis in former or current smokers with adenocarcinoma or squamous carcinoma cases but were not significant in the 77 never-smokers of the Mayo Clinic dataset even though it was associated with stage and lymphatic metastasis status. As discussed above, this may be the result of underpowered sample size using OS instead of DFS, or NSCLC being less affected by BCAR1 expression in never smokers, which warrants further investigation.
Acknowledgments
The work was supported by the National Natural Science Foundation of China (NSFC) (No. 81101782), NSF project CQ CSTC (No. CSTC2011BB5020), NSF Third Military Medical University (No. 2010XQN36 and MiaoPu project), USA National institute of health grants (R01 CA80127 and R01 CA84354), and Mayo Clinic Foundation.
The authors appreciate Professor. Li Zeng Peng and his staff for conducting IHC tests and reviewing slides from the Department of Pathology at Daping Hospital, Chongqing City, China. The authors also appreciate Susan Ernst, M.A., for her technical assistance with the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
References
- 1.Reynolds AB, Roesel DJ, Kanner SB, Parsons JT. Transformation-specific tyrosine phosphorylation of a novel cellular protein in chicken cells expressing oncogenic variants of the avian cellular src gene. Mol Cell Biol. 1989;9:629–638. doi: 10.1128/mcb.9.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanner SB, Reynolds AB, Parsons JT. Tyrosine phosphorylation of a 120-kilodalton pp60src substrate upon epidermal growth factor and platelet-derived growth factor receptor stimulation and in polyomavirus middle-T-antigen-transformed cells. Mol Cell Biol. 1991;11:713–720. doi: 10.1128/mcb.11.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim W, Seok KY, Soo KJ, Shin NY, Hanks SK, Song WK. The integrin-coupled signaling adaptor p130Cas suppresses Smad3 function in transforming growth factor-beta signaling. Mol Biol Cell. 2008;19:2135–2146. doi: 10.1091/mbc.E07-10-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chodniewicz D, Klemke RL. Regulation of integrin-mediated cellular responses through assembly of a CAS/Crk scaffold. Biochim Biophys Acta. 2004;1692:63–76. doi: 10.1016/j.bbamcr.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Cabodi S, Tinnirello A, Bisaro B, Tornillo G, del PCM, Forni G, et al. p130Cas is an essential transducer element in ErbB2 transformation. FASEB J. 2010;24:3796–3808. doi: 10.1096/fj.10-157347. [DOI] [PubMed] [Google Scholar]
- 6.Cabodi S, Tinnirello A, Di SP, Bisaro B, Ambrosino E, Castellano I, et al. p130Cas as a new regulator of mammary epithelial cell proliferation, survival, and HER2-neu oncogene-dependent breast tumorigenesis. Cancer Res. 2006;66:4672–4680. doi: 10.1158/0008-5472.CAN-05-2909. [DOI] [PubMed] [Google Scholar]
- 7.Huang W, Deng B, Wang RW, Tan QY, Jiang YG. Expression of breast cancer anti-estrogen resistance 1 in relation to vascular endothelial growth factor, p53, and prognosis in esophageal squamous cell cancer. LID - 10.1111/j.1442-2050.2012.01376.x [doi] Dis Esophagus. 2012 doi: 10.1111/j.1442-2050.2012.01376.x. [DOI] [PubMed] [Google Scholar]
- 8.Huang W, Deng B, Wang RW, Tan QY, He Y, Jiang YG, et al. BCAR1 protein plays important roles in carcinogenesis and predicts poor prognosis in non-small-cell lung cancer. PLOS ONE. 2012;7:e36124. doi: 10.1371/journal.pone.0036124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng B, Huang W, Tan QY, Fan XQ, Jiang YG, Liu L, et al. Breast cancer anti-estrogen resistance protein 1 (BCAR1/p130cas) in pulmonary disease tissue and serum. Mol Diagn Ther. 2011;15:31–40. doi: 10.1007/BF03257191. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Sheu CC, Ye Y, de Andrade M, Wang L, Chang SC, et al. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol. 2010;11:321–330. doi: 10.1016/S1470-2045(10)70042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke C, Henry M, Doolan P, Kelly S, Aherne S, Sanchez N, et al. Integrated miRNA, mRNA and protein expression analysis reveals the role of post-transcriptional regulation in controlling CHO cell growth rate. BMC Genomics. 2012;13:656. doi: 10.1186/1471-2164-13-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maragkakis M, Vergoulis T, Alexiou P, Reczko M, Plomaritou K, Gousis M, et al. DIANA-microT Web server upgrade supports Fly and Worm miRNA target prediction and bibliographic miRNA to disease association. Nucleic Acids Res. 2011;39:W145–W148. doi: 10.1093/nar/gkr294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng Y, Kuang W, Hao Y, Zhang D, Lei M, Du L, et al. Downregulation of miR-27a* and miR-532-5p and upregulation of miR-146a and miR-155 in LPS-induced RAW264.7 macrophage cells. Inflammation. 2012;35:1308–1313. doi: 10.1007/s10753-012-9443-8. [DOI] [PubMed] [Google Scholar]
- 16.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D, Lu Z, Jia J, Zheng Z, Lin S. Changes in microRNAs associated with podocytic adhesion damage under mechanical stress. J Renin Angiotensin Aldosterone Syst. 2012 doi: 10.1177/1470320312460071. [DOI] [PubMed] [Google Scholar]
- 18.Ullah AZ, Sahoo S, Steinhofel K, Albrecht AA. Derivative scores from site accessibility and ranking of miRNA target predictions. Int J Bioinform Res Appl. 2012;8:171–191. doi: 10.1504/IJBRA.2012.048966. [DOI] [PubMed] [Google Scholar]
- 19.Loher P, Rigoutsos I. Interactive exploration of RNA22 microRNA target predictions. Bioinformatics. 2012;28:3322–3323. doi: 10.1093/bioinformatics/bts615. [DOI] [PubMed] [Google Scholar]
- 20.Brinkman A, der Flier Sv, Kok EM, Dorssers LC. BCAR1, a human homologue of the adapter protein p130Cas, and antiestrogen resistance in breast cancer cells. J Natl Cancer Inst. 2000;92:112–120. doi: 10.1093/jnci/92.2.112. [DOI] [PubMed] [Google Scholar]
- 21.Brinkman A, de Jong D, Tuinman S, Azaouagh N, van AT, Dorssers LC. The substrate domain of BCAR1 is essential for anti-estrogen-resistant proliferation of human breast cancer cells. Breast Cancer Res Treat. 2010;120:401–408. doi: 10.1007/s10549-009-0403-4. [DOI] [PubMed] [Google Scholar]
- 22.Cunningham-Edmondson AC, Hanks SK. p130Cas substrate domain signaling promotes migration, invasion, and survival of estrogen receptor-negative breast cancer cells. Breast Cancer (London) 2009;2009:39–52. doi: 10.2147/BCTT.S6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konstantinovsky S, Davidson B, Reich R. Ezrin and BCAR1/p130Cas mediate breast cancer growth as 3-D spheroids. Clin Exp Metastasis. 2012;29:527–540. doi: 10.1007/s10585-012-9468-2. [DOI] [PubMed] [Google Scholar]
- 24.Kumbrink J, Kirsch KH. Regulation of p130(Cas)/BCAR1 expression in tamoxifen-sensitive and tamoxifen-resistant breast cancer cells by EGR1 and NAB2. Neoplasia. 2012;14:108–120. doi: 10.1593/neo.111760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorssers LC, Grebenchtchikov N, Brinkman A, Look MP, van BSP, de Jong D, et al. The prognostic value of BCAR1 in patients with primary breast cancer. Clin Cancer Res. 2004;10:6194–6202. doi: 10.1158/1078-0432.CCR-04-0444. [DOI] [PubMed] [Google Scholar]
- 26.Guo C, Liu QG, Yang W, Zhang ZL, Yao YM. Relation among p130Cas, E-cadherin and beta-catenin expression, clinicopathologic significance and prognosis in human hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2008;7:490–496. [PubMed] [Google Scholar]
- 27.Nick AM, Stone RL, Armaiz-Pena G, Ozpolat B, Tekedereli I, Graybill WS, et al. Silencing of p130cas in ovarian carcinoma: a novel mechanism for tumor cell death. J Natl Cancer Inst. 2011;103:1596–1612. doi: 10.1093/jnci/djr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Z, Liu Y, Johnson JJ, Stack MS. Urinary-type plasminogen activator receptor (uPAR) modulates oral cancer cell behavior with alteration in p130cas. Mol Cell Biochem. 2011;357:151–161. doi: 10.1007/s11010-011-0885-3. [DOI] [PubMed] [Google Scholar]
- 29.Wei L, Yang Y, Zhang X, Yu Q. Anchorage-independent phosphorylation of p130(Cas) protects lung adenocarcinoma cells from anoikis. J Cell Biochem. 2002;87:439–49. doi: 10.1002/jcb.10322. [DOI] [PubMed] [Google Scholar]
- 30.Miao Y, Li AL, Wang L, Fan CF, Zhang XP, Xu HT, et al. Expression of p130cas, E-cadherin and beta-catenin and their correlation with clinicopathological parameters in non-small cell lung cancer: p130cas over-expression predicts poor prognosis. Folia Histochem Cytobiol. 2012;50:392–397. doi: 10.5603/11957. [DOI] [PubMed] [Google Scholar]
- 31.Fromont G, Vallancien G, Validire P, Levillain P, Cussenot O. BCAR1 expression in prostate cancer: association with 16q23 LOH status, tumor progression and EGFR/KAI1 staining. Prostate. 2007;67:268–273. doi: 10.1002/pros.20516. [DOI] [PubMed] [Google Scholar]
- 32.Miao Y, Li AL, Wang L, Fan CF, Zhang XP, Xu HT, et al. Expression of p130cas, E-cadherin and beta-catenin and their correlation with clinicopathological parameters in non-small cell lung cancer: p130cas over-expression predicts poor prognosis. Folia Histochem Cytobiol. 2012;50:392–397. doi: 10.5603/11957. [DOI] [PubMed] [Google Scholar]