Abstract
Pediatric rhabdomyosarcoma (RMS) is traditionally classified by histologic appearance into alveolar (ARMS) and embryonal (ERMS) subtypes. The majority of ARMS contain a PAX3-FOXO1 or PAX7-FOXO1 gene fusion, but about 20% do not. Intergroup Rhabdomyosarcoma Study (IRS) Stage and group-matched ARMS typically behaves more aggressively than the embryonal subtype, but recent studies have shown that it is, in fact, fusion status that drives outcome for RMS. Gene expression microarray data indicate that several genes discriminate between fusion positive and fusion negative RMS with high specificity. Using tissue microarrays containing a series of both ARMS and ERMS, we identified a panel of four immunohistochemical markers, myogenin, AP2β, NOS-1 and HMGA2, which can be used as surrogate markers of fusion status in RMS. These antibodies provide an alternative to molecular methods for identification of fusion positive RMS, particularly in cases where there is scant or poor quality material. Additionally, these antibodies, may be useful in fusion negative ARMS as an indicator that a variant gene fusion may be present.
Keywords: rhabdomyosarcoma, fusion status, immunohistochemistry
Introduction
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma of childhood and accounts for 5-10% of childhood malignancies.[1] RMS has been traditionally classified by histologic appearance into two major subtypes, alveolar (ARMS) and embryonal (ERMS).[2] Stage and Group-matched ARMS typically behaves more aggressively than ERMS.[3-5]
Most ARMS are associated with a novel fusion gene generated by a balanced chromosomal translocation between chromosomes 2 or 1 with FOXO1 on chromosome 13; these respectively fuse PAX3 or PAX7 with FOXO1.[6] Detection of the fusion gene by reverse transcriptase polymerase chain reaction (RT-PCR) or the t(2;13) or t(1;13) translocation by fluorescent in situ hybridization (FISH) is also used in diagnosis and classification of RMS.[7] Approximately 20% of ARMS cases lack evidence of a gene fusion by FISH or RT-PCR. Microarray-based gene expression profiling demonstrates a distinctive genetic signature in fusion gene-positive ARMS, while fusion gene-negative ARMS demonstrate a different expression pattern that resembles that of ERMS.[8, 9] Fusion gene expression confers distinct biological properties to fusion gene-positive ARMS cases, and it also has clinical and prognostic importance.[10] Skapek et al recently confirmed in a prospective study that PAX-FOXO1 fusion status drives unfavorable outcome for children with RMS.[11] Detection of fusion gene status is therefore critical to risk stratification of children with RMS, and molecular biologic strategies to identify fusion positive RMS (RMSp) versus fusion negative RMS (RMSn) are being incorporated into future Children's Oncology Group (COG) Soft Tissue Sarcoma protocols.
RT-PCR and FISH detect greater than 90% of RMSp; however, the clinical adaptability of these techniques is complicated by the need for high quality material. Commonly used RTPCR and FISH assays fail to detect rare variant translocations, such as PAX3/NCOA1 or PAX3/FOXO4.[12, 13] Immunohistochemistry (IHC) is widely accepted as a standard tool for diagnosis of RMS, and detection of myogenin expression both confirms the diagnosis of RMS and aids in histologic subclassification.[14] Published microarray data for RMS suggest several genes which discriminate between RMSp and RMSn subgroups with high specificity.[8, 9, 13, 15] Commercially available antibodies have been raised to many of the protein products of these genes, so that they are excellent candidates for IHC markers of fusion status in RMS. Development of an IHC panel as a surrogate marker of fusion status provides a useful, cost-effective method for RMS subclassification.
Materials and Methods
Tissue microarrays
Unstained slides from three RMS tissue microarrays (TMAs) were obtained from the Cooperative Human Tissue Network at the Biopathology Center at Nationwide Children's Hospital (Columbus, OH). Cases were selected for inclusion based on original Intergroup Rhabdomyosarcoma Study Group (IRSG)/Children's Oncology Group (COG) diagnosis, and all cases were sampled in duplicate or triplicate. Using the original study enrollment diagnosis, the ERMS TMA includes 95 tumor sections comprised of material from 36 individual ERMS, 6 ARMS, and 4 skeletal muscle control cases. The ARMS TMA includes 108 tumor sections from 39 ARMS, 5 ERMS controls and 5 skeletal muscle controls. The “Lookback” TMA includes 196 tumor sections comprised of material from 55 ARMS, 21 ERMS, 6 mixed RMS, 2 RMS not otherwise specified (NOS), 1 undifferentiated sarcoma and 6 skeletal muscle controls.
Fusion status
For 133 total cases (including all three TMAs), evaluation for a rearrangement of the FOXO1 (13q14), PAX3 (2q35) and PAX7 (1p36) loci by FISH on representative formalin-fixed, paraffin-embedded (FFPE) TMA slides was performed using a FOXO1 Dual Color Break Apart Probe (Abbott Molecular, Inc., Des Plaines, IL) and custom-designed probes as previously described[7].
Briefly, 4-μm thick sections were deparaffinized with CitriSolv and rinsed with 95% ethanol. The slides were then pretreated in 0.2N hydrochloric acid for 20 min at room temperature, followed by pretreatment reagent (Abbott Molecular, Inc., Des Plaines, IL) at 80°C for 30-60 min and protease digestion for 30-60 min at 37°C. Following pretreatment, the slides were post-fixed with 1% formaldehyde for 5 min at room temperature, dehydrated in an ethanol series, and air dried. The diluted probe was added to the slide and co-denatured with DNA at 80°C for 5-10 min and incubated at 37°C overnight using the HYBrite™ denaturation/hybridization system. Note, a subset of specimens required more than a single pretreatment. Unbound probe was removed by washing with 2x SSC/0.1% NP-40 at 72°C for 2 min, then at room temperature for 2 min. The slides were air-dried in the dark and counterstained with DAPI II (Abbott Molecular, Inc., Des Plaines, IL). Hybridization signals were assessed in interphase nuclei with strong, well-delineated signals and distinct nuclear borders. An interphase cell specimen was interpreted as abnormal if a split of flanking probe signals was detected in more than 10% of the cells evaluated (more than two standard deviations above the average false-positive rate).
Fusion status for 100 total cases (including all three TMAs) had been previously analyzed by quantitative RT-PCR assay to assess expression of a PAX3-FOXO1 (P3F) or PAX7-FOXO1 (P7F) fusion transcript using frozen or formalin-fixed, paraffin-embedded material as described previously.7 Following reverse transcription from a FOXO1-specific primer, the assay incorporates consensus 5’ PAX3/PAX7 and 3’ FOXO1 primers and gene-specific PAX3 and PAX7 probes, thus determining both the presence and subtype of the fusion. In a second reaction, expression of the GAPDH gene was quantified to assess the quality of the RNA. GAPDH expression level equivalent to that found in 0.5 ng of a control rhabdomyosarcoma cell line was the minimum cutoff for a satisfactory result in a sample with a negative fusion result. Eighty cases were examined by both RT-PCR and FISH.
Immunohistochemistry
Five micron-thick sections of the multi-tumor TMAs were stained with primary antibodies to myogenin (1:100, BD Biosciences), activating enhancer binding protein 2 beta (AP2β) (1:25, Santa Cruz Biotechnology), neuronal nitric oxide synthase, or nitric oxide synthase 1 (NOS-1) (1:250, Santa Cruz Biotechnology), and high mobility group AT-hook 2 (HMGA2) (1:500, BioCheck). (Table 1). For myogenin and HMGA2, deparaffinized slides were rehydrated followed by heat-induced epitope retrieval in a steamer (30 minute incubation in 96° EDTA buffer, followed by 25 minute incubation in citrate buffer). After incubation with normal horse serum, sections were then incubated with the primary antibody for 30 minutes, detected using Vectastain Elite ABC kit, and visualized via color development with diaminobenzidine (Vector Laboratories, Burlingame, CA). Slides were counterstained with hematoxylin. For AP2β and NOS-1, staining was performed using the Ventana Benchmark XT (Ventana Medical Systems, Tuscon, AZ, USA). Slides were heated at 42° C with cell conditioner 1 (standard manufacturer's protocol) for antigen retrieval, and they were incubated with the primary antibody for 32 minutes.
Table 1.
Antibody information
| Antibody | Clone | Dilution | Company |
|---|---|---|---|
| Myogenin | 556358 | 1:100 | BD Biosciences, San Jose, CA |
| AP2β | H-87 | 1:25 | Santa Cruz Biotechnology, Santa Cruz, CA |
| NOS-1 | H299 | 1:250 | Santa Cruz Biotechnology, Santa Cruz, CA |
| HMGA2 | 59170AP | 1:500 | BioCheck, Foster City, CA |
IHC stains were scored on a 0-4+ scale as follows: 0, absent expression; 1+, less than 10% expression; 2+, 10-50% expression; 3+, 50-90% expression, 4+, greater than 90% expression. Reactivity was interpreted as specific only when found in the expected subcellular location, ie. in the nucleus for AP2β, HMGA2 and myogenin and in the cell membrane and cytoplasm for NOS-1. Each individual tissue core was scored. In cases where duplicate tumor samples had different scores, the highest score was used.
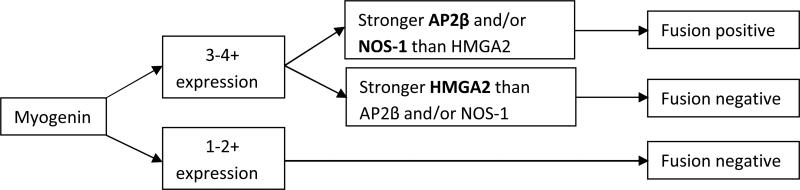
We developed an algorithm for prediction of fusion status based on IHC expression patterns (Figure 1). Previous data and the experience of COG central review pathologists (DP, LT, ER), have indicated that essentially no RMS cases with less than 3+ myogenin expression have evidence of a gene fusion.[16] Therefore, this observation provided the first cut point for prediction of fusion status, ie. cases with less than 50% myogenin expression were predicted to be fusion negative. For cases with strong myogenin expression, we then considered NOS-1, AP2β and HMGA2 expression. Stronger NOS-1 and/or AP2β expression (typically 3-4+) with weaker HMGA2 expression (typically 0-2+) supported fusion positivity. The inverse result, weaker NOS-1 and/or AP2β than HMGA2 expression supported fusion negativity. For cases where NOS-1 and AP2β scores were discrepant, the higher score was used. Using these guidelines, IHC stains were interpreted by two pathologists (ER and DP), blinded to histologic diagnosis or fusion status.
Figure 1.
Algorithm for prediction of fusion status in RMS using myogenin, AP2β, NOS-1 and HMGA2.
Statistics
Sensitivity and specificity of predicting fusion positivity was calculated for each antibody individually, as well as for the algorithmic prediction. For single antibody calculations, scores that predicted the presence of a fusion gene were considered “positive.” Therefore, this was considered to be 3+ or greater for myogenin, NOS-1 and AP2b, and 0-2+ for HMGA2. Sensitivity and specificity calculations were performed on the ARMS and ERMS TMAs combined, and the results were then confirmed in an independent data set using the Lookback TMA. Positive and negative predictive values were calculated assuming the expected prevalence of RMSp is 25% [70% ERMS, 25% ARMSp and 5% ARMSn]. The distributions of staining patterns between patients with RMSp and RMSn were compared using the Fisher's Exact test for each antibody.
Results
Tissue microarrays
A total of 28 cases from the ERMS TMA provided complete data on the four immunostains and, in addition, had known fusion status. (Table 2) Forty (40) total cases from the ARMS TMA provided complete data on the four immunostains and, in addition, had known fusion status. Seven (7) of these complete cases were present on both TMAs and one duplicate was excluded, leaving a total of 61 cases from the ERMS and ARMS TMAs for analysis. For the Lookback TMA, a total of 13 ERMS cases and 37 ARMS cases provided complete data on the four immunostains and, in addition, had known fusion status.
Table 2.
Rhabdomyosarcoma cases analyzed on tissue microarrays (TMA)
| ERMS TMA | ARMS TMA | Lookback TMA | Total | |
|---|---|---|---|---|
| Total number of tumors | 28 | 40 | 50 | 118* |
| Histologic subtype | ||||
| ERMS | 22 | 4 | 13 | 39 |
| ARMS | 6 | 36 | 37 | 79 |
| Fusion positive | 5 | 26 | 24 | 55 |
| Fusion negative | 1 | 10 | 13 | 24 |
Seven cases were sampled on both the ERMS and ARMS TMAs
Genetic analysis
Two cases had discordant results by RT-PCR and FISH. Both of these cases were fusion positive by FISH, but negative for both the PAX3-FOXO1 and PAX7-FOXO1 fusion transcript by RT-PCR performed on FFPE tissue. In each case, however, there was a low GAPDH expression level, so that false-negativity secondary to inadequate RNA cannot be excluded.
FISH was equivocal for fusion status in three cases represented on the ARMS TMA. One case had low but detectable signal for PAX7-FOXO1 by RT-PCR performed on FFPE tissue. Although there was low internal GAPDH expression, the results were interpreted as positive for PAX7-FOXO1 translocation. RT-PCR performed on frozen tissue in the second case showed no evidence of a fusion transcript. RT-PCR on frozen tissue from the third case generated a light positive signal that was shown to be a PAX3-FOXO4 (AFX) by sequencing followed by reanalysis with a RT-PCR assay specific for this fusion.[17]
Immunohistochemistry
The combined ARMS and ERMS TMAs and the Lookback TMA showed differential expression patterns for all four antibodies studied. (Table 3). Sensitivities and specificities for the combined ARMS/ERMS TMAs and the Lookback TMA are reported for each single antibody in Table 4 (p<0.0001). In our series, myogenin had 96-100% sensitivity for detecting fusion status, but poor (51-65%) specificity as a single marker. HMGA2, the only marker of fusion negativity, had 83-89% sensitivity and 73-80% specificity for predicting fusion status. NOS-1 had 71-83% sensitivity, but 94-96% specificity for fusion positivity. AP2β was the most predictive single IHC marker of fusion status, with 91-93% sensitivity and 93-94% specificity. The positive predictive value of strong (3+ or greater) AP2β staining was 81%, with a 97% negative predictive value. Use of the four-marker algorithm increased the sensitivity of predicting fusion positivity to 96% with 91-92% specificity (Table 5). This corresponds to a positive predictive value of 78-80% and a negative predictive value of 99%.
Table 3.
Range of immunohistochemistry (IHC) scores for single antibodies
| IHC score | |||||
|---|---|---|---|---|---|
| AP2beta | 0 | 1 + | 2+ | 3+ | 4+ |
| ERMS/ARMS TMA | |||||
| Translocation negative (N=33) | 85% | 6% | 3% | 0% | 6% |
| Translocation positive (N=28) | 0% | 4% | 4% | 11% | 82% |
| Lookback TMA | |||||
| Translocation negative (N=26) | 77% | 4% | 12% | 8% | 0% |
| Translocation positive (N=24) | 0% | 8% | 0% | 8% | 83% |
| NOS-1 | |||||
| ERMS/ARMS TMA | |||||
| Translocation negative (N=33) | 27% | 48% | 18% | 3% | 3% |
| Translocation positive (N=28) | 7% | 3% | 7% | 29% | 54% |
| Lookback TMA | |||||
| Translocation negative (N=26) | 77% | 19% | 0% | 0% | 4% |
| Translocation positive (N=24) | 17% | 8% | 4% | 29% | 42% |
| Myogenin | |||||
| ERMS/ARMS TMA | |||||
| Translocation negative (N=33) | 0% | 21% | 30% | 39% | 9% |
| Translocation positive (N=28) | 0% | 0% | 0% | 21% | 79% |
| Lookback TMA | |||||
| Translocation negative (N=26) | 0% | 27% | 38% | 23% | 12% |
| Translocation positive (N=24) | 0% | 4% | 0% | 13% | 83% |
| HMGA2 | |||||
| ERMS/ARMS TMA | |||||
| Translocation negative (N=33) | 9% | 18% | 0% | 21% | 52% |
| Translocation positive (N=28) | 61% | 21% | 7% | 7% | 4% |
| Lookback TMA | |||||
| Translocation negative (N=26) | 8% | 12% | 0% | 15% | 65% |
| Translocation positive (N=24) | 37% | 42% | 4% | 13% | 4% |
Table 4.
Sensitivity and Specificity of single antibodies for predicting fusion status in rhabdomyosarcoma
| Antibody | Sensitivity | Specificity |
|---|---|---|
| ARMS/ERMS TMA | ||
| AP2beta | 93% | 94% |
| HMGA2 | 89% | 73% |
| Myogenin | 100% | 51% |
| NOS1 | 83% | 94% |
| Lookback TMA | ||
| AP2beta | 91% | 93% |
| HMGA2 | 83% | 80% |
| Myogenin | 96% | 65% |
| NOS1 | 71% | 96% |
Table 5.
Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of 4-antibody algorithm for predicting fusion status in rhabdomyosarcoma
| Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|
| ARMS/ERMS TMA | 96% | 91% | 78% | 99% |
| Lookback TMA | 96% | 92% | 80% | 99% |
There was excellent interobserver agreement for predicting fusion status using the proposed algorithm. The two pathologists (ER and DP) had 100% concordance on independent review of the 111 cases analyzed on the TMAs.
Discussion
Presence of a PAX-FOXO1 gene fusion drives poor outcome in patients with RMS, regardless of histologic subtype.[11] While RT-PCR and FISH are the gold standard for identifying the gene fusion, sufficient quality or quantities of tissue may not be available for analysis, particularly if needle biopsy samples are collected. IHC is the most common technique used to aid in the differential diagnosis of small round cell tumors, and myogenin IHC is routinely performed on FFPE tissue to confirm a diagnosis of RMS.
Gene expression microarray studies have detected numerous marker genes with fusion specific expression patterns, and many are detectable at the protein level by IHC.[8, 9, 13, 15] Based on these data, we selected four markers (including myogenin) with available IHC antibodies. Myogenin, AP2β and NOS-1 are markers of RMSp, while HMGA2 is a marker of RMSn. Strong, diffuse myogenin expression by immunoperoxidase stains has been shown to correlate with alveolar histology and poor outcome.[18-20] AP2β is highly expressed in fusion positive tumors with mean expression levels greater than 200-fold relative to RMSn.[8] Similarly, unpublished COG data shows NOS-1 is also expressed at much higher levels in RMSp (communication from JA). HMGA2 shows over eightfold greater expression levels in RMSn (both ERMS and ARMSn), although there is partial overlap with RMSp.[8]
Several studies have examined the sensitivity and specificity of single IHC antibodies for prediction of outcome and/or fusion status in RMS. As a single marker, Hostein et al showed that myogenin has 82% specificity for ARMS and 76% specificity for ERMS.[19] Diffuse myogenin expression has also been correlated with poor outcome [20], although myogenin expression is imperfect as a single test.[16] Davicioni et al validated their oligonucleotide microarray findings using IHC antibodies for AP2β and HMGA2, which demonstrated 90% sensitivity/100% specificity and 79% sensitivity/86% specificity, respectively for these two markers. Additionally, Wachtel et al and Grass et al described a panel of fusion-predictive IHC antibodies that included AP2β and P-cadherin for RMSp and epidermal growth factor receptor (EGFR) and fibrillin 2 for ERMS.[21, 22] As single markers, sensitivities ranged from 64% to 84% and specificities ranged from 81% to 92%. The specificity improved to 98% for RMSp and 90% for ERMS when using a combination of two markers, although the sensitivity decreased.
Our algorithm based on the 4-antibody IHC panel showed greater sensitivity and specificity than either individual antibodies or previously described panels. Our algorithmic approach showed 96% sensitivity for fusion positivity and predicted RMSp with 99% accuracy. We had superior interobserver agreement, and we selected antibodies with predominantly nuclear expression and widely differential staining intensities that allowed greater ease of interpretation.
TMAs offer the advantage of cost-effective screening of large numbers of cases, but they may not adequately reflect variation in staining seen on single sections. To address this variability, all cases were sampled in duplicate or triplicate. Additionally, Grass et al. showed an increase in sensitivity when looking at single tumor sections vs. TMA sections, but there was no decrease in specificity.[22] Prospective examination of these staining patterns in whole tissue sections will allow validation of the test as a surrogate marker for fusion status in RMS.
Microarray data shows consistent gene expression patterns in RMSp regardless of the PAX fusion partner, suggesting these genes may be downstream targets of the fusion proteins. Detection of unique protein expression patterns by IHC or other techniques therefore allows identification of not only the common PAX3(7)-FOXO1 fusion protein, but also may identify novel variants such as PAX3-FOXO4 or PAX3-NCOA1[12, 13, 21] that may not be detected by standard RT-PCR or FOXO1 breakapart FISH techniques. Supporting this finding, Ebauer et al showed that AP2β forms a direct target of the PAX/FOXO1 fusion protein and mediates the anti- apoptotic function of the chimeric transcription factor. Using gene expression pattern, Wachtel et al also identified several cases with an RMSp-like IHC pattern that were originally classified as RMSn; a fusion was subsequently found in some of these cases [21].
In our series, three cases with an RMSp protein expression pattern had equivocal FISH results. One case was shown to contain a PAX3-FOXO4 fusion transcript by RT-PCR and one harbored a low fusion signal for PAX7-FOXO1 by RT-PCR. The third case remains equivocal and is still being investigated for possible fusion variants. Although not included in the statistical analysis, IHC results were able to predict fusion status in 2 cases where both RT-PCR and FISH failed or had insufficient material for testing.
RT-PCR and FISH have a nearly 100% concordance rate in the cases where adequate material is available for testing by both methods.[16] However, two of our TMA cases showed discordant results with detection of a fusion gene by FISH and a negative PAX-FOXO1 RT-PCR result in the setting of low RNA isolated from FFPE tissue and thus interpreted as “possible false negative”. The inherent limitations of RT-PCR and FISH are well known.[17] RT-PCR is dependent on sufficient quantities of RNA, and even with high-sensitivity techniques, low levels of fusion transcript expression may not be detected. False positive results have also been reported to occur secondary to contamination.[23] FISH offers the advantage of histologic correlation, although histologic interpretation is limited on fluorescent stains. As with RT-PCR, however, FISH is dependent upon the specific probe set used. Since the majority of variant fusions reported to date involve PAX3 with a partner gene other than FOXO1, a FOXO1 breakapart probe set will not recognize the variant.[12, 13]
Emerging RNA expression array technologies, such as nCounter (Nanostring Technologies, Seattle, WA), show promise for identifying unique, prognostically significant expression patterns and distinguishing RMSn from RMSp.[24] As a research tool, these techniques offer the advantage of detecting many more analytes at a time than IHC. However, these techniques are limited by the inverse correlation between cost and turn-around time, as the test becomes cost-effective only when multiple cases are run together. With rare tumors, this approach would require centralized testing to accrue sufficient numbers of cases to make this a viable technique. IHC offers a rapid, inexpensive and reproducible technique that is available in most clinical centers. Use of specific IHC expression patterns to guide treatment is a well-accepted practice, and such tools are increasingly common in this burgeoning era of personalized medicine.
Cost analysis of IHC versus conventional molecular testing methods is difficult due to the number of permutations in molecular methods and billing systems that exist between institutions. For the minority of cases requiring testing [16], the clinical charges for 3 IHC antibodies range from $300-600, FISH using a single probe for FOXO1 breakapart is approximately $750, and RT-PCR is approximately $600. IHC may potentially save up to $300-$400/case, but it is roughly equivalent in overall costs. The value of our IHC panel comes not solely in cost savings, however. It also provides reliable interpretation on small amounts of tissue, wide availability and ease of use in regions without access to other molecular testing methods, and potential to detect variant fusion transcripts that may be missed by FISH or RT PCR. An alternative approach would be the use of AP2β in conjunction with conventional molecular methods to screen for cases with variant gene fusions. This would not add a significant cost, but would ensure patients with variant translocations are identified and receive appropriate therapy.Although histologic subtyping of RMS remains independent of fusion status and gene or protein expression patterns, IHC provides valuable information in the prognosis of rhabdomyosarcoma, regardless of histologic pattern. The use of a four antibody panel for predicting fusion status in RMS shows similar performance characteristics to molecular diagnostic approaches, with greater than 95% sensitivity for RMSp. The lower specificity of our panel may be skewed due to lack of detection of variant fusions in the “false” positive cases. Use of the IHC panel alone, or AP2β in conjunction with molecular techniques, should be considered for evaluation of fusion status in RMS and determination of risk stratification.
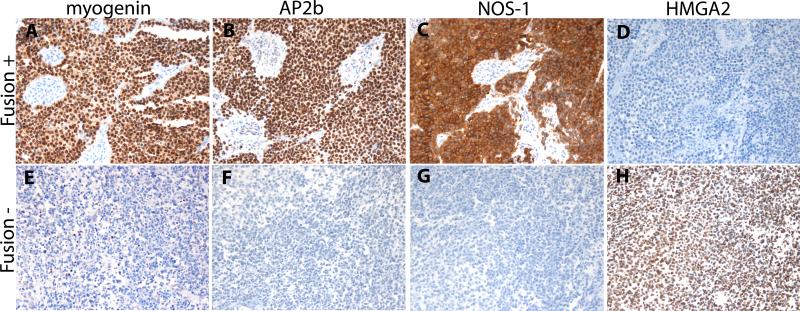
Image 1.
Fusion positive RMS demonstrate strong myogenin (A, 200x), AP2β (B, 200x), and NOS-1 (C, 200x) expression with weaker or absent HMGA2 expression (D, 200x). In contrast, fusion negative RMS demonstrate weak to moderate myogenin expression (E, 200x), weak or absent AP2β (F, 200x) and NOS-1 (G, 200x) expression and strong HMGA2 expression (H, 200x).
Acknowledgements
The authors would like to thank Dr. Xiao-qiong Liu (University of Nebraska Medical Center) for her valuable technical FISH contributions.
Research Support:
St. Baldrick's Foundation Childhood Cancer Research Grant, 179772 (ERR); NIH-RC2-CA148216 (SXS, JAB, FGB, JGF); FGB is supported by the Intramural Research Program of the NCI
Footnotes
Disclosures: The authors have no conflicts of interest or funding to disclose.
References
- 1.Pappo AS. Rhabdomyosarcoma and other soft tissue sarcomas of childhood. Curr Opin Oncol. 1995;7(4):361–6. doi: 10.1097/00001622-199507000-00012. [DOI] [PubMed] [Google Scholar]
- 2.Gurney JG,YJ, Roffers SD, et al. Soft Tissue Sarcomas. SEER Pediatric Monograph. 1999 [Google Scholar]
- 3.Meza JL, et al. Analysis of prognostic factors in patients with nonmetastatic rhabdomyosarcoma treated on intergroup rhabdomyosarcoma studies III and IV: the Children's Oncology Group. J Clin Oncol. 2006;24(16921036):3844–3851. doi: 10.1200/JCO.2005.05.3801. [DOI] [PubMed] [Google Scholar]
- 4.Tsokos M, et al. Rhabdomyosarcoma. A new classification scheme related to prognosis. Arch Pathol Lab Med. 1992;116(1497467):847–855. [PubMed] [Google Scholar]
- 5.Leuschner I, et al. Spindle cell variants of embryonal rhabdomyosarcoma in the paratesticular region. A report of the Intergroup Rhabdomyosarcoma Study. Am J Surg Pathol. 1993;17(8434703):221–230. doi: 10.1097/00000478-199303000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Barr FG. Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene. 2001;20(40):5736–46. doi: 10.1038/sj.onc.1204599. [DOI] [PubMed] [Google Scholar]
- 7.Nishio J, et al. Use of a novel FISH assay on paraffin-embedded tissues as an adjunct to diagnosis of alveolar rhabdomyosarcoma. Lab Invest. 2006;86(16607381):547–556. doi: 10.1038/labinvest.3700416. [DOI] [PubMed] [Google Scholar]
- 8.Davicioni E, et al. Molecular classification of rhabdomyosarcoma--genotypic and phenotypic determinants of diagnosis: a report from the Children's Oncology Group. Am J Pathol. 2009;174(2):550–64. doi: 10.2353/ajpath.2009.080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williamson D, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol. 2010;28(20351326):2151–2158. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]
- 10.Missiaglia E, et al. PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. J Clin Oncol. 2012;30(14):1670–7. doi: 10.1200/JCO.2011.38.5591. [DOI] [PubMed] [Google Scholar]
- 11.Skapek SX, et al. PAX-FOXO1 Fusion Status Drives Unfavorable Outcome for Children With Rhabdomyosarcoma: A Children's Oncology Group Report. Pediatr Blood Cancer. 2013 doi: 10.1002/pbc.24532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barr FG, et al. Genetic heterogeneity in the alveolar rhabdomyosarcoma subset without typical gene fusions. Cancer Res. 2002;62(12183429):4704–4710. [PubMed] [Google Scholar]
- 13.Wachtel M, et al. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64(16):5539–45. doi: 10.1158/0008-5472.CAN-04-0844. [DOI] [PubMed] [Google Scholar]
- 14.Kumar S, et al. Myogenin is a specific marker for rhabdomyosarcoma: an immunohistochemical study in paraffin-embedded tissues. Mod Pathol. 2000;13(9):988–93. doi: 10.1038/modpathol.3880179. [DOI] [PubMed] [Google Scholar]
- 15.Lae M, et al. Global gene expression profiling of PAX-FKHR fusion-positive alveolar and PAX FKHR fusion-negative embryonal rhabdomyosarcomas. J Pathol. 2007;212(2):143–51. doi: 10.1002/path.2170. [DOI] [PubMed] [Google Scholar]
- 16.Rudzinski ER TL, Anderson JR, et al. Dense pattern of embryonal rhabdomyosarcoma: a lesion easily confused with alveolar rhabdomyosarcoma. american journal of clinical pathology. 2013 doi: 10.1309/AJCPA1WN7ARPCMKQ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barr FG, et al. Genetic heterogeneity in the alveolar rhabdomyosarcoma subset without typical gene fusions. Cancer Res. 2002;62(16):4704–10. [PubMed] [Google Scholar]
- 18.Dias P, et al. Strong immunostaining for myogenin in rhabdomyosarcoma is significantly associated with tumors of the alveolar subclass. Am J Pathol. 2000;156(10666368):399–408. doi: 10.1016/S0002-9440(10)64743-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hostein I, et al. Rhabdomyosarcoma: value of myogenin expression analysis and molecular testing in diagnosing the alveolar subtype: an analysis of 109 paraffin-embedded specimens. Cancer. 2004;101(15536621):2817–2824. doi: 10.1002/cncr.20711. [DOI] [PubMed] [Google Scholar]
- 20.Heerema-McKenney A, et al. Diffuse myogenin expression by immunohistochemistry is an independent marker of poor survival in pediatric rhabdomyosarcoma: a tissue microarray study of 71 primary tumors including correlation with molecular phenotype. Am J Surg Pathol. 2008;32(10):1513–22. doi: 10.1097/PAS.0b013e31817a909a. [DOI] [PubMed] [Google Scholar]
- 21.Wachtel M, et al. Subtype and prognostic classification of rhabdomyosarcoma by immunohistochemistry. J Clin Oncol. 2006;24(5):816–22. doi: 10.1200/JCO.2005.03.4934. [DOI] [PubMed] [Google Scholar]
- 22.Grass B, et al. Immunohistochemical detection of EGFR, fibrillin-2, P-cadherin and AP2beta as biomarkers for rhabdomyosarcoma diagnostics. Histopathology. 2009;54(7):873–9. doi: 10.1111/j.1365-2559.2009.03303.x. [DOI] [PubMed] [Google Scholar]
- 23.Ten Heuvel SE, Hoekstra HJ, Suurmeijer AJ. Diagnostic accuracy of FISH and RT-PCR in 50 routinely processed synovial sarcomas. Appl Immunohistochem Mol Morphol. 2008;16(3):246–50. doi: 10.1097/PAI.0b013e31815349f5. [DOI] [PubMed] [Google Scholar]
- 24.Siddappa CM, et al. Detection of disseminated tumor cells in the bone marrow of breast cancer patients using multiplex gene expression measurements identifies new therapeutic targets in patients at high risk for the development of metastatic disease. Breast Cancer Res Treat. 2013;137(1):45–56. doi: 10.1007/s10549-012-2279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]