Abstract
Candida albicans is known as a commensal microorganism but it is also the most common fungal pathogen in humans, causing both mucosal and systemic infections. Biofilm-associated C. albicans infections present clinically important features due to their high levels of resistance to traditional antifungal agents. Quorum sensing is closely associated with biofilm formation and increasing fungal pathogenicity. We investigated the ability of the novel bacterial quorum sensing quencher thiazolidinedione-8 (S-8) to inhibit the formation of, and eradication of mature C. albicans biofilms. In addition, the capability of S-8 to alter fungal adhesion to mammalian cells was checked. S-8 exhibited specific antibiofilm and antiadhesion activities against C. albicans, at four- to eightfold lower concentrations than the minimum inhibitory concentration (MIC). Using fluorescence microscopy, we observed that S-8 dose-dependently reduces C. albicans–GFP binding to RAW macrophages. S-8 at sub-MICs also interfered with fungal morphogenesis by inhibiting the yeast-to-hyphal form transition. In addition, the tested agent strongly affected fungal cell wall characteristics by modulating its hydrophobicity. We evaluated the molecular mode of S-8 antibiofilm and antiadhesion activities using real-time RT-PCR. The expression levels of genes associated with biofilm formation, adhesion and filamentation, HWP1, ALS3 and EAP1, respectively, were dose-dependently downregulated by S-8. Transcript levels of UME6, responsible for long-term hyphal maintenance, were also significantly decreased by the tested agent. Both signaling pathways of hyphal formation-cAMP-PKA and MAPK-were interrupted by S-8. Their upstream general regulator RAS1 was markedly suppressed by S-8. In addition, the expression levels of MAPK cascade components CST20, HST7 and CPH1 were downregulated by S-8. Finally, transcriptional repressors of filament formation, TUP1 and NRG1, were dramatically upregulated by our compound. Our results indicate that S-8 holds a novel antibiofilm therapeutic mean in the treatment and prevention of biofilm-associated C. albicans infections.
Introduction
The yeast Candida albicans is found as a commensal microorganism in the digestive tract of mammals [1], as well as in the oral cavity of humans [2]. It is also the most common fungal pathogen in humans, causing both mucosal and systemic infections, particularly in immunocompromised individuals [3]. Several factors can predispose individuals to candidiasis, such as prolonged treatment with antibiotics, corticosteroids, diabetes mellitus, nutritional deficiencies, immunosuppressive diseases or hormone therapy [4]. Several virulence factors that contribute to the development of candidal infection have been identified. They include (i) adhesins that allow adhesion to human cells with subsequent invasion [5], (ii) the ability to form biofilm on human mucosa [5] and on artificial surfaces such as catheters [6], [7] and dental devices [8], [9], and (iii) the ability to switch from yeast to hyphal form [10]. Biofilm formation is an important factor in C. albicans pathogenesis [11]; it involves attachment, colonization and the development of a mature biofilm structure composed of yeast, pseudo- and true hyphae, and extracellular matrix [11]–[14]. Like bacterial biofilm, and in contrast to the planktonic form, fungal biofilm is highly resistant to antifungal drugs. This resistance is multifactorial and complex, involving: (i) limited drug penetration into the biofilm due to the high density of extracellular matrix, (ii) drug absorption or binding by the biofilm extracellular matrix, (iii) decreased growth rate, (iv) overexpression of genes involved in drug resistance, particularly those encoding efflux pumps, (v) and multidrug tolerance due to persistant cells [11], [13], [15], [16]The outcome of immobilized fungi in biofilm in terms of pathogenicity and drug resistance emphasizes the need for new antibiofilm agents that can inhibit biofilm formation or destroy preformed biofilm without affecting fungal viability.
It has been found that microorganisms actually exchange information between themselves. This cross-talk is termed quorum sensing (QS). QSis associated with biofilm formation and fungi's increased pathogenicity [17]. Indeed, fungal biofilm integrity is highly dependent on QS. Recently, two main QS regulators-farnesol and tyrosol-have been described in C. albicans. Farnesol regulates C. albicans morphology and inhibits biofilm formation [18], [19], whereas tyrosol is associated with increased biomass of C. albicans biofilms, probably by stimulating hyphal growth [20]. Thiazolidinediones (TZDs) have been proposed as potential QS inhibitors in Vibrio harveyi [21]. Several TZD derivatives have also been tested for their ability to affect C. albicans pathogenicity [16]. The aim of this study was to investigate the antibiofilm effect of the TZD derivative S-8 on the fungal pathogen C. albicans. In addition, the molecular mechanism governing S-8's antibiofilm activity was evaluated.
Results
Effect of S-8 on C. albicans viability and biofilm formation
Planktonic C. albicans viability, as measured by 2,3-bis(2-methoxy-4-nitro-5-sulfo-phenyl)-2H-tetrazolium-5-carboxanilide (XTT) reduction assay, was not significantly affected by S-8 up to 32 µg/ml, while the MIC value was 64 µg/ml. This concentration of S-8 reduced fungal metabolic activity by more than 90% (P<0.05) (Fig. 1). In contrast, S-8 dose-dependently decreased biofilm metabolic activity, and even at 8 µg/ml it inhibited biofilm formation by 50% (P<0.05), which was recorded as the minimal biofilm inhibitory concentration 50 (MBIC50) (Fig. 1). The C. albicans cell can reduce its metabolic activity as a protective mechanism under adverse conditions [22], [23]. Therefore, we studied the effects of S-8 on morphology and viability of the biofilm cells. Based on these results, in subsequent experiments we used S-8 at concentrations of up to 16 µg/ml, which is four times lower than the MIC, to prevent fungicidal effects of S-8 on the planktonic cells.
Figure 1. Effect of S-8 on C. albicans viability and biofilm formation.
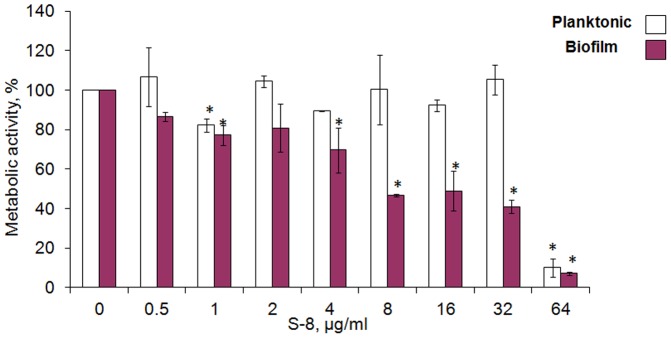
C. albicans cells were incubated with 0.5–64 µg/ml S-8 for 48 h. The metabolic activity of planktonic and biofilm cells was measured using the XTT reduction assay. The XTT values of the untreated control were set to 100%. The results are presented as means ± SD of three independent experiments. *Significantly lower than the untreated control (P<0.05).
S-8 affects fungal morphology in the biofilm
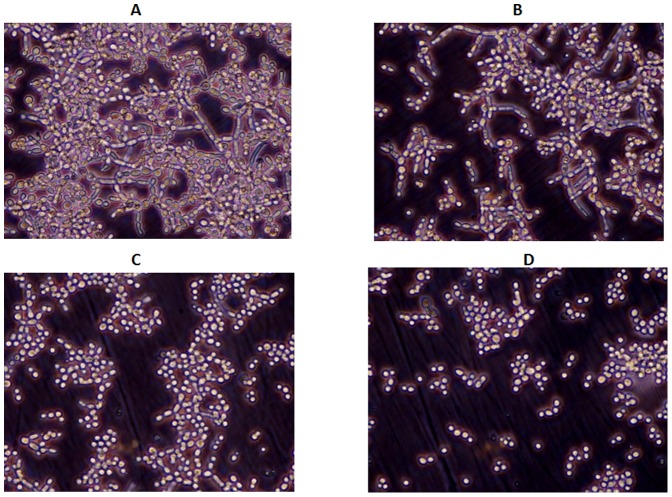
Microscopic observation showed that in addition to decreasing metabolic activity in the biofilms, S-8 also inhibits the yeast-to-hyphal form transition, and reduces the length of existing filaments of adhered fungi. As shown in Fig. 2, untreated control biofilm (Fig. 2A) consisted of mostly branched hyphae and pseudohyphae, and only a few yeast cells. S-8 at 1 µg/ml increased the average number of yeast cells by 40% as compared to the untreated control, while shortening the average filament length by 1.3-fold (130 µm vs. 170 µm in the control) (Fig. 2B). Increasing the concentration of S-8 to 4 µg/ml dramatically reduced the average length of filaments adhering to the surface by 8.5-fold (20 µm vs. 170 µm in the control), while increasing the average count of the yeast form by 5.3-fold as compared to the untreated control (Fig. 2C). Finally, the highest tested dose of S-8 (16 µg/ml) resulted in almost total disappearance of the hyphal form with mostly yeast cells observed (Fig. 2D). This increase in S-8 concentration resulted in a sparse, loosely attached monolayer of C. albicans yeast cells, indicating disruption of the yeast-to-hyphal form transition, without causing cell death. Moreover, mycelium density was dose-dependently decreased with increasing S-8 concentration (Fig. 2).
Figure 2. Effect of treatment with S-8 on C. albicans morphology.
The morphology of C. albicans biofilms after treatment with S-8 was visualized using phase-contrast microscopy. A. Untreated control. B, C and D. Biofilms developed with 1, 4 and 16 µg/ml S-8, respectively. Magnification: 400X. At least four random fields were observed. Three independent experiments were performed.
Treatment with S-8 disrupts preformed biofilm
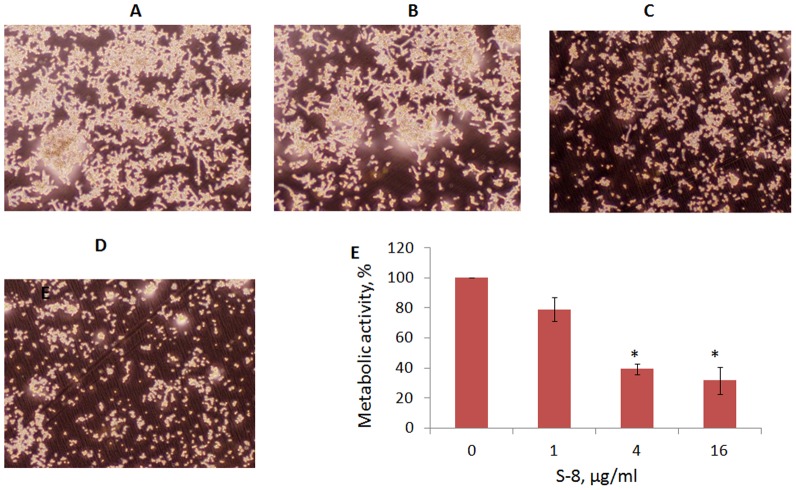
Preformed C. albicans biofilm was significantly attenuated by the presence of S-8 (Fig. 3). At concentrations of 1 µg/ml (Fig. 3B), 4 µg/ml (Fig. 3C) and 16 µg/ml (Fig. 3D), S-8 caused the detachment of preformed biofilms in a dose-dependent manner as compared to the control (Fig. 3A). In addition, it dramatically decreased candidal filamentation and branching in a dose-dependent manner (Fig. 3A–D). Quantitative analysis using XTT assay, in accordance with microscopic observations, revealed that S-8 at the above doses reduces metabolic activity in developed biofilms by 21%, 61% (P<0.05) and 70% (P<0.05), respectively, as compared to the untreated control (Fig. 4E).
Figure 3. Effect of treatment with S-8 on maintenance of preformed biofilms.
C. albicans biofilms were formed for 24 h followed by 24-h treatment with 1, 4 and 16 µg/ml S-8. Morphology of C. albicans in pre-formed biofilms exposed to S-8 was visualized using phase-contrast microscopy. A. Untreated control. B, C and D. Preformed biofilms exposed to 1, 4 and 16 µg/ml S-8, respectively. Magnification: 100X. At least four random fields were observed. Three independent experiments were performed. E. Biofilm metabolic activity was estimated using the XTT reduction assay. The XTT values of the untreated control were set to 100%. The results are presented as means ± SD of three independent experiments. *Significantly lower than the value for the untreated control (P<0.05).
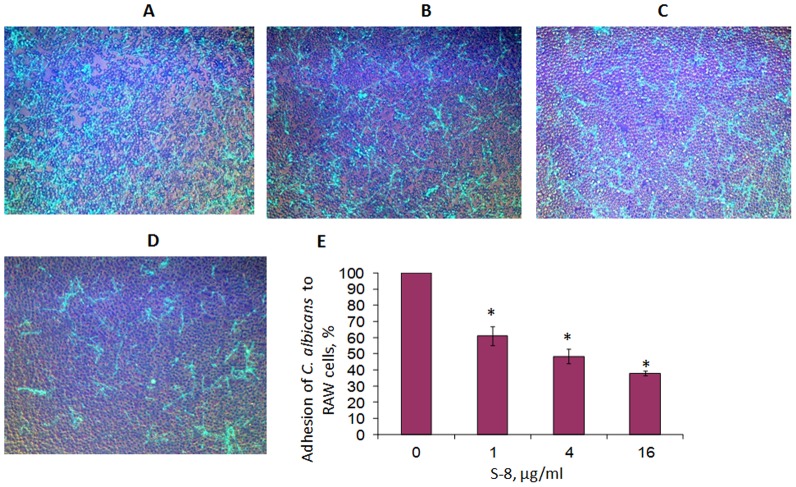
Figure 4. Effect of treatment with S-8 on adherence of C. albicans cells to RAW macrophages.
A–D. C. albicans–GFP adhered to RAW cells in the presence or absence of S-8. A. Untreated control. B, C and D. Samples exposed to 1, 4 and 16 µg/ml S-8, respectively. Magnification: 100X. Image processing was performed using fluorescence microscopy at excitation and emission wavelengths of 488 and 522 nm, respectively. E. Quantitative analysis of fluorescence images. A value of 100% was assigned to C. albicans adhered to RAW cells not treated with S-8. *Significantly lower than the value for the untreated control (P<0.05). At least four random fields were observed and analyzed. Assays were performed in triplicate and the means ± SD from three independent experiments were calculated.
Inhibition of fungal attachment to mammalian macrophages by S-8
Since interactions between C. albicans and macrophages involve initial binding of the pathogenic organism to the surface of the host cell, we investigated whether S-8 interferes with this process. Fluorescence microscopy observations demonstrated a marked dose-dependent reduction in the number of C. albicans attached to RAW 264.7 mouse macrophages in the presence of S-8 at a concentration of 1 µg/ml (Fig. 4B), 4 µg/ml (Fig. 4C) and 16 µg/ml (Fig. 4D) as compared to the untreated control (Fig. 4A). Quantitative analysis of fluorescence images demonstrated that at the lowest tested dose of 1 µg/ml, S-8 had significantly decreased fungal binding to RAW cells, by 39% (P<0.05) compared to control. Further elevation of S-8 at concentrations of 4 µg/ml and 16 µg/ml enhanced its antiadhesion effect up to 63% (P<0.05), as compared to untreated controls (Fig. 4E). Neither C. albicans nor S-8, alone or in combination, reduced the viability of epithelial cells as determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay (data not shown).
Effect of S-8 on cell-surface hydrophobicity of C. albicans
To gain further insight into the mechanism by which S-8 treatment reduces C. albicans biofilm formation, we investigated whether S-8 can modify the cell-surface hydrophobicity of C. albicans. The hydrophobicity index (HI) was significantly and dose-dependently decreased after a 30-min treatment of C. albicans with S-8 (Table 1).
Table 1. Effect of S-8 on cell-surface hydrophobicity of C. albicans.
| S-8, µg/ml | 0 | 1 | 4 | 16 |
| HI | 77% (+/−2%) | 68% (+/−4%) | 37% (+/−1.7%) | 21% (+/−0.15%) |
The hydrophobicity index (HI) of C. albicans cells incubated for 30 min with S-8 at 0, 1, 4, and 16 µg/ml. Assays were performed in triplicate and repeated three times.
Effect of S-8 on the expression of biofilm-associated genes in C. albicans
To elucidate the molecular mechanism underlying S-8's inhibitory effect on adhesion properties, biofilm formation and integrity, we analyzed changes in C. albicans genes' expression levels in biofilms exposed to S-8 (Fig. 5). Genes involved in biofilm development were analyzed, including those associated with adhesion: HWP1 (hyphal cell wall protein), ALS3 (agglutinin-like sequence), EAP1 (extracellular adhesion protein), and UME6 (hyphal formation and maintenance. Expression of hyphal transcriptional regulators in the mitogen-activated protein kinase (MAPK) pathway (RAS1, CST20, HST7 and CPH1) and the cAMP-dependent protein kinase A (PKA) pathway (RAS1 and EFG1) were also assayed, as were levels of transcriptional repressors of filamentation (TUP1 and NRG1). The transcriptional levels of genes involved in the adhesion process, such as the hyphal-specific genes HWP1 and ALS3, as well as EAP1, were dose-dependently downregulated by S-8 (Fig. 5A). These genes' products are known to be associated with attachment to abiotic and mammalian cell surfaces. EAP1 encodes a protein involved in adhesion to various surfaces, an initial and critical step in biofilm formation. We therefore consider it a biofilm-associated gene. In addition, UME6, a key filament-specific transcriptional regulator which is involved in long-term hyphal maintenance, was strongly suppressed by S-8 (Fig. 5B). Interestingly, expression of EFG1—part of the cAMP-PKA signaling cascade—was not altered by treatment with S-8, whereas three regulatory genes involved in the MAPK pathway—CST20, HST7 and CPH1—were downregulated by this treatment (Fig. 5B). In addition, the level of RAS1, an upstream regulator of both pathways, was significantly decreased (Fig. 5B). Finally, transcriptional repressors of hyphal-specific gene expression, TUP1 and NRG1, were dramatically upregulated by S-8 in a dose-dependent manner (Fig. 5C) which strongly correlates with alteration of candidal morphogenesis.
Figure 5. Quantitative real time RT-PCR analysis of C. albicans specific genes.
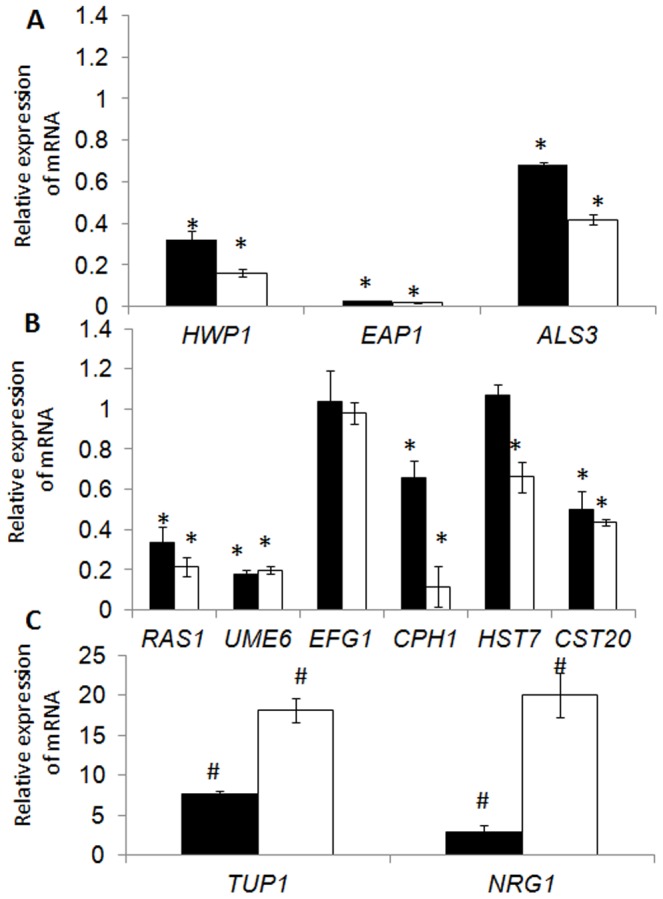
C. albicans biofilms were formed in the presence or absence of 4 and 16 µg/ml S-8 and expression of the target genes was determined by quantitative real-time RT-PCR. Housekeeping gene 18S rRNA was used for normalization. A. Hyphal-specific and biofilm-associated genes involved in the adhesion process. B. Hyphal transcriptional regulator genes; C. Transcriptional repressors of hyphal development. Black columns: 4 µg/ml S-8; white columns: 16 µg/ml S-8. The expression level of the untreated sample was set to 1 for each gene. *Significantly lower than the value for the untreated control (P<0.05). #Significantly higher than the value for the untreated control (P<0.05). The assays were performed in triplicate and the means ± SD from three independent experiments were calculated.
Discussion
The decreasing efficacy of today's antifungal treatments and the emergence of multidrug-resistant Candida strains call for a new strategy in treatment perception. One venue is developing novel agents interfering with biofilm formation [24]–[26] not necessarily affecting viability of the Candida.
The TZD derivative S-8, synthesized in our laboratory, demonstrated a specific antibiofilm effect. We showed that this agent inhibited C. albicans biofilm formation by 50%, at a concentration eight fold lower than the MIC for planktonic cells. In addition, the tested agent disrupted developed biofilm in a dose-dependent manner. Mature biofilms are generally difficult to eradicate and are more resistant to antifungal treatments. C. albicans cells growing in a biofilm environment exhibit much greater resistance to antifungal agents [27], [28]. Moreover, since mature biofilms are strongly related to device-associated Candida infections, their elimination is critical, presenting a therapeutic challenge for management of the disease [29]. At a fourfold lower concentration than the MIC, S-8 decreased preformed biofilm by 70%, while other antifungals such as fluconazole and amphotericin B can only affect mature biofilms at doses that are 100-fold and 30-fold higher, respectively, than the MIC for planktonic cells [27], [28]. The above findings clearly indicate a specific antibiofilm effect of S-8.
Inhibition of biofilm formation by S-8 was strongly associated with alteration of fungal morphology. C. albicans is a dimorphic fungal pathogen which is capable of reversible transitions between yeast and hyphal forms. Morphogenesis is an important and critical determinant of C. albicans virulence. Strains that are defective in morphogenesis also exhibit attenuated virulence in systemic candidiasis models [30], [31]. Indeed, various studies have found that hyphal development is necessary for C. albicans to evade macrophages [32], escape from blood vessels [33], and colonize medical devices by forming biofilms [9], [34]. It could be that S-8 caused the detachment of hyphal form cells, and, therefore, we observed mostly firmly attached yeast cells. Numerous small molecules have been reported to regulate the yeast-to-hypha transition in C. albicans, and they are part of the current trend in antifungal drug development [35].
C. albicans can adhere to a variety of different tissues in the human body, thus facilitating its strong presence of many host niches [36]. Our study showed that S-8 significantly and dose-dependently inhibits C. albicans adherence to RAW macrophages. S-8's ability to inhibit C. albicans adherence to immune cells suggests that this compound may prevent a key and early event in C. albicans pathogenicity and consequently, could minimize the fungus's virulence.
Further experiments were performed to evaluate the mechanism underlying S-8's antibiofilm and antiadhesion activity. We observed a marked modification of the C. albicans cell surface properties from highly hydrophobic to hydrophilic following exposure to S-8. Hydrophobic interactions are generally considered to play an important role in C. albicans adherence to mammalian cells and also to various inert surfaces [37]. Therefore, agents that can modify the surface characteristics of C. albicans may alter its adherence capacity, thereby preventing biofilm formation and subsequently, invasion of the host cells [24]. Whether this effect on hydrophobicity is a surface effect, or an effect on the expression of hydrophobic proteins in the cell surface remains to be elucidated.
To test whether the antibiofilm effect of S-8 is also manifested at the transcriptional level, we tested a few different biological function genes associated with biofilm formation. The expressions of genes encoding cell-wall proteins, which play an important role in C. albicans' adhesion process, such as HWP1, ALS3 and EAP1, were downregulated by S-8 (Fig. 5). ALS and HWP1 have been associated with attachment to human cells and biofilm formation [9], [38], [39], while EAP1 has been shown to play a role in adhesion to both mammalian cells and polystyrene [40]. We propose that S-8 reduced, at least in part, fungal binding to macrophages as well as C. albicans adhesion with subsequent biofilm formation on polystyrene plates due to a dramatic decrease in EAP1 transcript. In addition, a marked reduction in the hyphal form after treatment with S-8 could be explained by alteration of HWP1 expression. This observation is in accordance with a study showing that HWP1 is not expressed during the yeast growth phase but is strongly expressed on hyphal surfaces [41]. Indeed, an hwp1/hwp1 mutant contained exclusively yeast cells in vitro, while in vivo this mutation resulted in an absolute inability to form biofilm, indicating that the adherence of hyphae, but not of yeast, is dependent on HWP1 expression [9]. In addition, UME6, which is responsible for hyphal formation and maintenance, was strongly suppressed by S-8. It has been shown that although a ume6/ume6 mutant is able to initiate germ tubes, it is restricted in the maintenance of hyphal growth [42]. Similar results were obtained in our study: when fungal biofilm was exposed to even the lowest tested dose of S-8 (1 µg/ml), it consisted mostly of shortened filaments and the yeast form. Moreover, UME6 is known to induce a variety of hyphal-specific transcripts, including ALS3 and HWP1 [43]. Therefore, S-8's interruption of ALS3- and HWP1-associated filament formation and maintenance could be attributed to downregulation of UME6 expression. There are two major pathways regulating hyphal transformation in C. albicans: the MAPK (via CPH1) and cAMP-PKA (via EFG1) pathways. Three genes involved in the MAPK pathway, CST20, HST7 and CPH1, were differently suppressed by S-8. On the other hand, although the expression level of EFG1 was not altered by S-8, its upstream regulator and a major component of the signaling pathway of hyphal formation, RAS1, was markedly inhibited by the tested agent. It seems that S-8 affects both pathways. In addition, the hyphal-suppressor gene TUP1 and the DNA-binding protein NRG1 which functions with TUP1 were dramatically upregulated in the presence of S-8. Studies have shown that the QS inhibitor farnesol, which inhibits filamentation [44] and biofilm formation [19] in C. albicans, exerts its activity by repressing both the MAPK and cAMP-PKA signaling pathways and by stimulating the expression of hyphal suppressor genes such as TUP1 [45], [46]. TUP1 mutants are unable to grow as yeast and instead remain locked in the filamentous form [47]. Moreover, induction of TUP1 transcription-repressor complexes results in the downregulation of hyphal-specific gene expression [47]. These observations strongly support our hypothesis that in addition to interference with MAPK and cAMP-PKA signaling pathways, S-8 inhibits the yeast-to-hypha transition via induction of the TUP1–NRG1 transcriptional repressor complex. In conclusion, S-8 exhibits nonfungicidal antibiofilm activity against C. albicans at several levels: inhibition of hyphal formation, and modification of the expression of biofilm-related functional genes and cell-surface characteristics. This alteration of fungal morphogenesis leads to impaired biofilm formation and easier eradication of a developed biofilm, as well as a reduction in the fungus's binding capacity to mammalian cells. As the TZDs are new potential antibiofilm agents, further experiments are required to evaluate the mechanism underlying S-8's antibiofilm and antiadhesion properties. Our results suggest that S-8 has potential as a promising therapeutic agent in the treatment and prevention of biofilm-associated C. albicans infections.
Materials and Methods
Synthesis of S-8
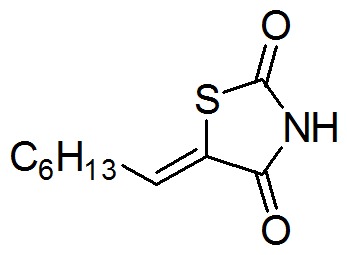
S-8 (Fig. 6) was prepared according to a previously described method [16], [21] with some modifications. Briefly, 0.085 g (1 mmol) piperidine was added to 0.170 g (1 mmol) thiazolidine-2,4-dione in 8 ml ethanol, followed by addition of 0.098 g (1 mmol) decanal. The mixture was stirred for 30 min at room temperature then it was maintained in refluxed ethanol for 24 h. After cooling to 0°C, 1 M HCl solution was added to the mixtures and it was maintained in the refrigerator for few days. Then the precipitate was filtered, washed with petroleum ether, dried and was analyzed by NMR (File S1) and melting point to give 60% yield. The purity of the compound was above 95%.
Figure 6. The structure of S-8.
Fungal strains and growth conditions
C. albicans ATCC 90028, C. albicans SC5314 and C. albicans SC5314 carrying the GFP reporter gene (C. albicans–GFP) [48], kindly provided by J. Berman (Tel Aviv University, Israel), were grown for 24–48 h at 37°C on Sabouraud dextrose agar (SDA; Novamed, Jerusalem, Israel) plates. To prepare a standard cell suspension, a single colony was inoculated into YNB medium (0.67% yeast nitrogen base without amino acids, 2% dextrose) (Difco, Sparks, MD) and incubated for 18 h at 30°C with agitation. The fungal cells were harvested by centrifugation, washed twice in PBS (pH 7.4), and resuspended at 1×107 cell/ml. RPMI medium (Biological Industries, Beit Haemek, Israel) supplemented with 2% glucose was used for biofilm and adhesion assays at 37°C, 5% CO2.
Effect of S-8 on C. albicans biofilm formation and preformed biofilms
Fungal biofilms were prepared on commercially available, presterilized, flat-bottomed 96-well polystyrene microtiter plates (Thermo Scientific, Waltham, MA). A standard cell suspension of C. albicans ATCC 90028 (200 µl) was transferred to the wells and incubated in RPMI medium supplemented with 2% glucose in the presence of different concentrations of S-8 (0.5 µg/ml to 64 µg/ml) for 24 h at 37°C, 5% CO2. To investigate the effect of S-8 on preformed biofilms, C. albicans biofilms were allowed to mature for 24 h at 37°C, 5% CO2 as described above. The wells were washed twice with PBS, fresh RPMI medium containing sub-MICs of S-8 was added and the plate was further incubated for 24 h at 37°C, 5% CO2. The metabolic activity of the C. albicans biofilms as well as planktonic fungi was determined quantitatively using a standard XTT reduction assay (Biological Industries) [16]. The assays were performed in triplicate and repeated three times.
XTT reduction assay
Prior to each assay, XTT solution (Biological Industries) was thawed and mixed with a N-methyl dibenzopyrazine methyl sulfate (PMS) (Biological Industries) solution at 50∶1 (v/v). The biofilms were washed and then incubated with 60 µl of the XTT–PMS solution in 2 ml of PBS. To determine planktonic cell viability, 60 µl of the XTT–PMS solution was added to the supernatants aspirated from the biofilms and placed in a new 96-well plate. Plates containing biofilms as well as supernatants were then incubated in the dark for 3 h at 37°C. Following incubation, the color change in the solution was measured spectrophotometrically at 492 nm using a GENios plate reader (Tecan, Salzburg, Austria).
Effect of S-8 on C. albicans morphology
Fungal biofilms developed in the presence of S-8 or preformed biofilms after exposure to S-8 were washed with PBS and immediately processed for observation. Morphology of fungi in untreated and S-8-treated biofilm was visualized under an Olympus CKX41 inverted microscope (Tokyo, Japan) and photographed with a DP72 camera. Calculation of average filament length and yeast-form number in each sample was performed using Image J v3.91 software (http://rsb.info.nih.gov/ij). At least four random fields were observed. Three independent experiments were performed at different times. Assays were performed in triplicate and repeated three times.
Effect of S-8 on adhesion of C. albicans to RAW macrophages
This assay was performed using the modified method of Feldman et al. [24]. Briefly, RAW 264.7 mouse macrophages were cultured in Dulbecco's Modified Eagle's Medium (DMEM) (Sigma–Aldrich, St. Louis, MO) supplemented with 10%fetal bovine serum (FBS), penicillin (1 IU/ml), streptomycin (1 µg/ml) and L-glutamine (200 mM) in a humidified incubator at 37°C with 5% CO2 until 80% confluence. The cells were seeded at a concentration of 4×105 cell/ml under the above conditions in sterile 96-well clear-bottom black microplates (Greiner Bio One, Frickenhausen, Germany) and incubated until confluence. Then, the wells were washed with DMEM-1% heat-inactivated FBS, blocked with 1% bovine serum albumin (BSA) to prevent nonspecific fungal attachment, and treated with S-8 diluted in DMEM-1% heat-inactivated FBS medium at sub-MIC for 1 h in a 5% CO2 atmosphere at 37°C. Untreated control wells (no S-8) were also prepared. Subsequently, cells from an overnight culture of C. albicans–GFP at 107 cell/ml in RPMI were applied at a multiplicity of infection (MOI) of 10 (10 C. albicans per mammalian cell) to S-8-pretreated or control RAW cells and incubated for 30 min at 37°C. All incubation and following washing steps were carried out in the dark. After incubation, unbound C. albicans was aspirated and wells were washed three times with PBS. Image processing was performed using an Olympus CKX41 inverted microscope (Tokyo, Japan) supported with fluorescence filters and photographed with a DP72 camera. Images of adhered C. albicans–GFP were observed at excitation and emission wavelengths of 488 and 522 nm, respectively. Image analysis was performed using Image J v3.91 software (http://rsb.info.nih.gov/ij). Three high-power fields (100X) were selected for analysis of each tested condition. The assays were performed in triplicate and repeated three times.
Evaluation of S-8 and C. albicans cytotoxicity
RAW cells were cultured as described above. For cytotoxicity assay, mammalian cells (2×104 cells; 100 µl) were seeded in 96-well microtiter plates for 24 h, then treated with increasing concentrations of S-8 (0, 1, and 16 µg/ml), with C. albicans at a MOI of 10, or with both S-8 and C. albicans. Cell viability was determined by MTT assay (Roche Diagnostics, Mannheim, Germany) after 24 h. MTT solution (5 mg/ml) was prepared by dissolving MTT powder in PBS, and filter-sterilizing (0.22 µm pore-size filter) to remove insoluble residue. At the end of the incubation, the supernatant was aspirated and MTT solution (20 µl) was added to each well and incubated for 4 h at 37°C. The solution was replaced with 1 ml DMSO (Sigma–Aldrich) to dissolve the dark blue formazan crystals. After incubation for 15 min at room temperature, the absorbance was read in a GENios plate reader at 570 nm. The assays were performed in triplicate and repeated three times.
Effect of S-8 on C. albicans cell-surface hydrophobicity
This assay was performed according to the method described by Feldman et al. [24], using xylene as the organic solvent. Briefly, C. albicans ATCC 90028 at a concentration of 107 cell/ml was incubated for 30 min at 37°C with S-8 at 0, 1, 4, and 16 µg/ml. Fungal cells were then washed with PBS, suspended in the same buffer, and the optical density was determined spectrophotometrically at 660 nm. The cells were mixed with xylene (2.5∶1, v/v), shaken for 2 min, and the tube was left for 20 min at room temperature for phase separation. The turbidity of the aqueous phase was read at 660 nm. The hydrophobicity index was calculated as HI = (ODcontrol - ODtest) × 100/ODcontrol, where ODcontrol = optical density at 660 nm before xylene treatment and ODtest = optical density at 660 nm after xylene treatment. Assays were performed in triplicate and repeated three times.
Quantitative real time RT-PCR analysis of C. albicans specific genes
Biofilms of C. albicans SC5314 were grown in the presence of S-8 in 6-well plates under the conditions described above. After washing with PBS, biofilm cells were removed from the bottom of the plates with a sterile scraper following disruption in a Fast Prep Cell Disrupter (Bio 101, Savant Instruments, Inc., NY, USA). Total RNA was extracted from fungal biofilms using Tri-Reagent (Sigma–Aldrich) by a previously described method [19]. RNA concentration was determined spectrophotometrically using a Nanodrop ND-1000 Instrument (Wilmington, DE, USA). 2 µg of template was reverse-transcribed with Super Script First Strand (Invitrogen, Life Technologies, Carlsbad, CA, USA). The integrity and purity of the RNA was assessed using an Agilent 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA, USA). Expression of hyphal-specific and biofilm-associated genes (ALS3, EAP1, HWP1) and their transcriptional regulators (RAS1, UME6, CPH1, CST20, HST7, EFG1, TUP1, NRG1) was analyzed. The relative expression levels of the target genes were analyzed using an ABI-Prism 7300 Instrument (Applied Biosystems, Foster City, CA, USA). Platinum SYBR Green PCR Master Mix (Invitrogen) was used to monitor the amplified product in real time, following the manufacturer's protocol. Primers for the tested genes were taken from the literature (Table S1). For each set of primers, a standard amplification curve (critical threshold cycle vs. exponential of concentration) was plotted, and only those with slope ≈ -3 were considered reliable. The PCR consisted of denaturation at 95°C for 10 min, followed by 40 cycles of amplification (95°C for 10 s, 55°C for 10 s, and 72°C for 10 s) and quantification. The expression of 18S rRNA was used for normalization and to calculate the relative changes in target gene expression. Control reactions were also performed with RNA that had not been reverse-transcribed to ensure that no genomic DNA was amplified during the PCRs. Gene expression is given in relative values, setting the expression level of the untreated control to 1 for each gene. The assays were performed in triplicate and repeated three times.
Statistical analysis
Means ± standard deviation were calculated. The statistical analysis was performed using Student's t-test with a significance level of P<0.05.
Supporting Information
(DOC)
(DOCX)
Acknowledgments
We wish to thank Dr. Sarah Kagan for critical review of the manuscript. This article is in the memory of our distinguished colleague, the late Prof. Morris Srebnik.
Funding Statement
This work was partially supported by The International Association for Dental Research (IADR) and GlaxoSmithKline (GSK) Dental Care Innovation Award No. 0304914 and by internal Hebrew University sources. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nadăş GC, Taulescu MA, Ciobanu L, Fiţ NI, Flore C, et al. (2013) The interplay between NSAIDs and Candida albicans on the gastrointestinal tract of guinea pigs. Mycopathology 175: 221–230. [DOI] [PubMed] [Google Scholar]
- 2. Pereira CA, Toledo BC, Santos CT, Pereira Costa AC, Back-Brito GN, et al. (2013) Opportunistic microorganisms in individuals with lesions of denture stomatitis. Diagn Microbiol Infect Dis 76: 419–424. [DOI] [PubMed] [Google Scholar]
- 3. Pfaller MA, Diekema DJ (2007) Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20: 133–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mayer FL, Wilson D, Hube B (2013) Candida albicans pathogenicity mechanisms. Virulence 4: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wachtler B, Wilson D, Haedicke K, Dalle F, Hube B (2011) From attachment to damage: defined genes of Candida albicans mediate adhesion, invasion and damage during interaction with oral epithelial cells. PLoS One 6: e17046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ramage G, Martinez JP, Lopez-Ribot JL (2006) Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res 6: 979–986. [DOI] [PubMed] [Google Scholar]
- 7. Kojic EM, Darouiche RO (2004) Candida infections of medical devices. Clin Microbiol Rev 17: 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li L, Finnegan MB, Ozkan S, Kim Y, Lillehoj PB, et al. (2010) In vitro study of biofilm formation and effectiveness of antimicrobial treatment on various dental material surfaces. Mol Oral Microbiol 25: 384–390. [DOI] [PubMed] [Google Scholar]
- 9. Nobile CJ, Nett JE, Andes DR, Mitchell AP (2006) Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot Cell 5: 1604–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Umeyama T, Kaneko A, Nagai Y, Hanaoka N, Tanabe K, et al. (2005) Candida albicans protein kinase CaHsl1p regulates cell elongation and virulence. Mol Microbiol 55: 381–395. [DOI] [PubMed] [Google Scholar]
- 11. Finkel JS, Mitchell AP (2011) Genetic control of Candida albicans biofilm development. Nat Rev Microbiol 9: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blankenship JR, Mitchell AP (2006) How to build a biofilm: a fungal perspective. Curr Opin Microbiol 9: 588–594. [DOI] [PubMed] [Google Scholar]
- 13. Ramage G, Saville SP, Thomas DP, Lopez-Ribot JL (2005) Candida biofilms: an update. Eukaryot Cell 4: 633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Srinivasan A, Uppuluri P, Lopez-Ribot J, Ramasubramanian AK (2011) Development of a high-throughput Candida albicans biofilm chip. PLoS One 6: e19036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ramage G, Rajendran R, Sherry L, Williams C (2012) Fungal biofilm resistance. Int J Microbiol 2012: 528521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kagan S, Jabbour A, Sionov E, Al-Quntar AA, Steinberg D, et al. (2014) Anti Candida albicans biofilm effect of novel heterocyclic compounds. J Antimicrob Chemother 69: 416–427. [DOI] [PubMed] [Google Scholar]
- 17. Albuquerque P, Casadevall A (2012) Quorum sensing in fungi—a review. Med Mycol 50: 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nickerson KW, Atkin AL, Hornby JM (2006) Quorum sensing in dimorphic fungi: farnesol and beyond. Appl Environ Microbiol 72: 3805–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ramage G, Saville SP, Wickes BL, Lopez-Ribot JL (2002) Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl Environ Microbiol 68: 5459–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alem MA, Oteef MD, Flowers TH, Douglas LJ (2006) Production of tyrosol by Candida albicans biofilms and its role in quorum sensing and biofilm development. Eukaryot Cell 5: 1770–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brackman G, Al Quntar AA, Enk CD, Karalic I, Nelis HJ, et al. (2013) Synthesis and evaluation of thiazolidinedione and dioxazaborocane analogues as inhibitors of AI-2 quorum sensing in Vibrio harveyi . Bioorg Med Chem 21: 660–667. [DOI] [PubMed] [Google Scholar]
- 22. Gomes PN, da Silva WJ, Pousa CC, Narvaes EA, Del Bel Cury AA (2011) Bioactivity and cellular structure of Candida albicans and Candida glabrata biofilms grown in the presence of fluconazole. Arch Oral Biol 56: 1274–1281. [DOI] [PubMed] [Google Scholar]
- 23. Weber K, Schulz B, Ruhnke M (2010) The quorum-sensing molecule E, E-farnesol—its variable secretion and its impact on the growth and metabolism of Candida species. Yeast 27: 727–739. [DOI] [PubMed] [Google Scholar]
- 24. Feldman M, Tanabe S, Howell A, Grenier D (2012) Cranberry proanthocyanidins inhibit the adherence properties of Candida albicans and cytokine secretion by oral epithelial cells. BMC Complement Altern Med 12: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martinez LR, Mihu MR, Tar M, Cordero RJ, Han G, et al. (2010) Demonstration of antibiofilm and antifungal efficacy of chitosan against candidal biofilms, using an in vivo central venous catheter model. J Infect Dis 201: 1436–1440. [DOI] [PubMed] [Google Scholar]
- 26. Privett BJ, Nutz ST, Schoenfisch MH (2010) Efficacy of surface-generated nitric oxide against Candida albicans adhesion and biofilm formation. Biofouling 26: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ramage G, Vande Walle K, Wickes BL, Lopez-Ribot JL (2001) Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Chemother 45: 2475–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baillie GS, Douglas LJ (1998) Effect of growth rate on resistance of Candida albicans biofilms to antifungal agents. Antimicrob Agents Chemother 42: 1900–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kojic EM, Darouiche RO (2004) Candida infections of medical devices. Clin Microbiol Rev 17: 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, et al. (1997) Nonfilamentous C. albicans mutants are avirulent. Cell 90: 939–949. [DOI] [PubMed] [Google Scholar]
- 31. Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL (2003) Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell 2: 1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lorenz MC, Bender JA, Fink GR (2004) Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 3: 1076–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Phan QT, Belanger PH, Filler SG (2000) Role of hyphal formation in interactions of Candida albicans with endothelial cells. Infect Immun 68: 3485–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nobile CJ, Mitchell AP (2005) Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol 15: 1150–1155. [DOI] [PubMed] [Google Scholar]
- 36. Hobson RP, Munro CA, Bates S, MacCallum DM, Cutler JE, et al. (2004) Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J Biol Chem 279: 39628–39635. [DOI] [PubMed] [Google Scholar]
- 37. de Souza RD, Mores AU, Cavalca L, Rosa RT, Samaranayake LP, et al. (2009) Cell surface hydrophobicity of Candida albicans isolated from elder patients undergoing denture-related candidosis. Gerodontology 26: 157–161. [DOI] [PubMed] [Google Scholar]
- 38. Coleman DA, Oh SH, Zhao X, Zhao H, Hutchins JT, et al. (2009) Monoclonal antibodies specific for Candida albicans Als3 that immunolabel fungal cells in vitro and in vivo and block adhesion to host surfaces. J Microbiol Methods 78: 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hoyer LL, Green CB, Oh SH, Zhao X (2008) Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med Mycol 46: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grubb SE, Murdoch C, Sudbery PE, Saville SP, Lopez-Ribot JL, et al. (2008) Candida albicans-endothelial cell interactions: a key step in the pathogenesis of systemic candidiasis. Infect Immun 76: 4370–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sundstrom P (2002) Adhesion in Candida spp. Cell Microbiol 4: 461–469. [DOI] [PubMed] [Google Scholar]
- 42. Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, et al. (2008) UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol Biol Cell 19: 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zeidler U, Lettner T, Lassnig C, Muller M, Lajko R, et al. (2009) UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans . FEMS Yeast Res 9: 126–142. [DOI] [PubMed] [Google Scholar]
- 44. Oh KB, Miyazawa H, Naito T, Matsuoka H (2001) Purification and characterization of an autoregulatory substance capable of regulating the morphological transition in Candida albicans . Proc Natl Acad Sci U S A 98: 4664–4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sato T, Watanabe T, Mikami T, Matsumoto T (2004) Farnesol, a morphogenetic autoregulatory substance in the dimorphic fungus Candida albicans, inhibits hyphae growth through suppression of a mitogen-activated protein kinase cascade. Biol Pharm Bull 27: 751–752. [DOI] [PubMed] [Google Scholar]
- 46. Roman E, Alonso-Monge R, Gong Q, Li D, Calderone R, et al. (2009) The Cek1 MAPK is a short-lived protein regulated by quorum sensing in the fungal pathogen Candida albicans . FEMS Yeast Res 9: 942–955. [DOI] [PubMed] [Google Scholar]
- 47. Braun BR, Johnson AD (1997) Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277: 105–109. [DOI] [PubMed] [Google Scholar]
- 48. Gerami-Nejad M, Zacchi LF, McClellan M, Matter K, Berman J (2013) Shuttle vectors for facile gap repair cloning and integration into a neutral locus in Candida albicans . Microbiology 159: 565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOCX)