Abstract
Fire Blight is a destructive disease of apple and pear caused by the enteric bacterial pathogen, Erwinia amylovora. E. amylovora initiates infection by colonizing the stigmata of apple and pear trees, and entering the plants through natural openings. Epiphytic populations of the related enteric bacterium, Pantoea, reduce the incidence of disease through competition and antibiotic production. In this study, we identify an antibiotic from Pantoea ananatis BRT175, which is effective against E. amylovora and select species of Pantoea. We used transposon mutagenesis to create a mutant library, screened approximately 5,000 mutants for loss of antibiotic production, and recovered 29 mutants. Sequencing of the transposon insertion sites of these mutants revealed multiple independent disruptions of an 8.2 kb cluster consisting of seven genes, which appear to be coregulated. An analysis of the distribution of this cluster revealed that it was not present in any other of our 115 Pantoea isolates, or in any of the fully sequenced Pantoea genomes, and is most closely related to antibiotic biosynthetic clusters found in three different species of Pseudomonas. This identification of this biosynthetic cluster highlights the diversity of natural products produced by Pantoea.
Introduction
Fire blight is a destructive disease known to plague many rosaceous plants, in particular apple and pear plants [1], [2], [3]. It has been a threat to the cultivation of these crop plants in various parts of the world, including North America and Europe [4], with first reports appearing as early as 1790 in North America [5]. The causative agent, Erwinia amylovora, is an epiphytic bacterium that relies on nutrients from the plants for survival [6]. The site of colonization is generally the stigmata of immature apple and pear blossoms, which provide a moist environment, rich in nutrients [4]. E. amylovora enters the plants through natural openings and spreads to various parts of the plants through the vascular system, allowing for widespread colonization [7]. Once E. amylovora is established, disease symptoms such as flower necrosis and fruit rot develop, ultimately resulting in major losses [7].
Methods of control for E. amylovora have been the subject of many studies. Initially, antibiotics like streptomycin were shown to be an effective means of controlling bacterial colonization when applied to plants during early bloom [8]. Long term use led to streptomycin resistant populations of E. amylovora [8], [9], which was a cause for concern in medical communities using the antibiotic for therapeutic purposes [8], [9]. Recent studies have shown that the application of the bacterial antagonist, Pantoea agglomerans (formerly Erwinia herbicola), to apple and pear trees in early bloom may help to reduce the disease caused by E. amylovora [1], [10], [11], [12]. Pantoea vagans C9-1 and P. agglomerans E325 have been commercialized as the active agents in BlightBan C9-1 and Bloomtime Biological, respectively [13], and are currently being used in several parts of the world as biocontrol agents for fire blight [14]. The antagonists not only compete for sites of colonization and plant nutrients, but also produce antibiotics that prevent the colonization of E. amylovora [13].
Pantoea isolates have been shown to produce a variety of antibiotics, such as pantocins [15], [16], [17], [18], herbicolins [19], [20], microcins [21], [22], [23], and phenazines [24], several of which target amino acid biosynthesis genes in E. amylovora. These antibiotics have been classified into five distinct groups based on whether they are effective in the presence of specific amino acids [25]. Group I antibiotics, which include pantocin A, herbicolin O, and microcin MccEh252, lose their toxicity in the presence of L-histidine, while groups II (pantocin B), III, and IV are neutralized in the presence of L-arginine, L-lysine, and L-asparagine, respectively. Interestingly, Group V (herbicolin I) is unaffected by the presence of any amino acid [25]. The specific targets of several of these have been determined. Pantocin B directly targets N-acetylornithine transaminase, the last step in arginine biosynthesis [26], while pantocin A acts against E. amylovora by inhibiting L-histidinol phosphate aminotransferase, a critical step in histidine biosynthesis, resulting in a histidine deficiency [2], [27].
In this study, we identify an antibiotic from Pantoea ananatis strain BRT175 (PanBRT175) that is effective against E. amylovora, which we have called Pantoea Natural Product 1 (PNP-1). We evaluate the impact of nutritional changes on production, including whether PNP-1 activity is affected by amino acids. We demonstrate that the activity appears to be relatively narrow spectrum affecting Erwinia and Pantoea, and that resistance in Pantoea appears to have evolved in the lineage leading to the Pantoea agglomerans and Pantoea eucalyptii groups. We also identify the biosynthetic cluster responsible for PNP-1 production, and evaluate its distribution in Pantoea and other species.
Materials and Methods
Bacterial Strains
All strains used were cultured and maintained on lysogeny broth (LB) media (Table 1). Pantoea and Erwinia strains were incubated at 30°C. E. coli strains VPE42 (pBSL118) and HB101 (RK600), Staphylococcus aureus K1-7, Pseudomonas aeruginosa ATCC 27853, Streptococcus mutans UAIS9:wt, and Lactococcus lactis were incubated at 37°C. Where appropriate, antibiotics were supplemented in the following final concentrations: kanamycin, 50 µg/ml, chloramphenicol, 50 µg/ml, and rifampicin, 50 µg/ml.
Table 1. List of isolates used in this study.
| Species | Isolation Source/Genotype | Source |
| Enterobacter sp. | ||
| TX1 | human, CF sputum | [59] |
| Erwinia amylovora | ||
| EA321 | hawthorn | [59] |
| Escherichia coli | ||
| DH5a | fhuA2 lac(del)U169 phoA glnV44 Φ80′ lacZ(del)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | Dr. David Guttman, University of Toronto |
| HB101 (RK600) | conjugative plasmid | Dr. David Guttman, University of Toronto |
| VPE42 (pBSL118) | mini-Tn5 transposon | [60] |
| Lactococcus lactis | ||
| HD1 | Heather Dietz, University of Regina | |
| Pantoea agglomerans | ||
| 83 | wheat | ICMP |
| 788 | green bean | ICMP |
| 1512 | green bean | ICMP |
| 1574 | unidentified | ICMP |
| 3581 | oat seed | ICMP |
| 5565 | soybean | ICMP |
| 7373 | onion | ICMP |
| 7612 | grass grub | ICMP |
| 12531 | Gypsophila (Baby's Breath) | ICMP |
| 12534 | human, knee laceration | ICMP |
| 13301 | Golden Delicious apple | ICMP |
| 17124 | olive | ICMP |
| 770398 | human, female, blood | [59] |
| 240R | pear flower | [59] |
| 308R | pear flower | [59] |
| B015092 | human, female, urine midstream | [59] |
| B016395 | human, female, superficial wound | [59] |
| B025670 | human, female, superficial wound | [59] |
| B026440 | human, male, superficial wound | [59] |
| BB834250 | human, female, sputum, aortic aneurysm | [59] |
| DB522094 | human, elbow sore | [59] |
| DC432 | maize | [61] |
| DC434 | maize | [61] |
| DC556 | Gypsophila (Baby's Breath) | [61] |
| Eh318 | apple leaf | CUCPB 2140; Dr. Brion Duffy [18] |
| G4032547 | human, ear | [59] |
| H42501 | human, male, blood | [59] |
| SN01080 | slug | [59] |
| SN01121 | bee | [59] |
| SN01122 | bee | [59] |
| SN01170 | caterpillar | [59] |
| SP00101 | raspberry | [59] |
| SP00202 | apple | [59] |
| SP00303 | raspberry | [59] |
| SP01201 | strawberry leaf | [59] |
| SP01202 | strawberry leaf and stem | [59] |
| SP01220 | healthy rose bush | [59] |
| SP01230 | Virginia creeper leaves and stem | [59] |
| SP02022 | thistle | [59] |
| SP02230 | diseased tree leaf | [59] |
| SP02243 | unidentified tree | [59] |
| SP03310 | diseased tree leaf | [59] |
| SP03383 | diseased maize leaf | [59] |
| SP03412 | diseased bean leaf | [59] |
| SP04010 | tomato leaf | [59] |
| SP04011 | tomato leaf | [59] |
| SP04021 | tomato leaf | [59] |
| SP04022 | tomato leaf | [59] |
| SP05051 | tomato leaf | [59] |
| SP05052 | tomato leaf | [59] |
| SP05061 | tomato leaf | [59] |
| SP05091 | tomato leaf | [59] |
| SP05092 | tomato leaf | [59] |
| SP05120 | diseased maize leaf | [59] |
| SP05130 | diseased maize stamen | [59] |
| SS02010 | soil - ground squirrel burrow | [59] |
| SS03231 | soil - ground squirrel burrow | [59] |
| TX10 | human, CF sputum | [59] [62] |
| Pantoea ananatis | ||
| 15320 | rice | [59] |
| 17671 | rice | [59] |
| 26SR6 | maize leaf | Dr. Steven Lindow, UC Berkeley [59] |
| B7 | maize, rifR derivative of M232A | Dr. Steven Lindow, UC Berkeley [59] |
| BRT175 | strawberry | Dr. Gwyn Beattie, Iowa State [59] [28] |
| BRT98 | strawberry | Dr. Steven Lindow, UC Berkeley [59] |
| Cit30-11 | naval orange leaf | Dr. Steven Lindow, UC Berkeley [59] |
| M232A | maize | Dr. Steven Lindow, UC Berkeley [59] |
| Pantoea anthophila | ||
| 1373 | balsam | [59] |
| Pantoea brenneri | ||
| 91151 | human | [59] |
| B011483 | human, female, superficial wound | [59] |
| B014130 | human, male, superficial wound | [59] |
| B016381 | human, female, groin | [59] |
| B024858 | human, female, breast abscess | [59] |
| Pantoea calida | ||
| B021323 | human, female, urine midstream | [59] |
| BB957621A1 | human, male, CAPD dialysate, peritonitis | [59] |
| BB957621A2 | human, male, CAPD dialysate, peritonitis | [59] |
| BB957621B1 | human, male, CAPD dialysate, peritonitis | [59] |
| BB957621B2 | human, male, CAPD dialysate, peritonitis | [59] |
| BB957621C1 | human, male, CAPD dialysate, peritonitis | [59] |
| BB957621C2 | human, male, CAPD dialysate, peritonitis | [59] |
| Pantoea conspicua | ||
| B011017 | human, female, superficial wound | [59] |
| Pantoea dispersa | ||
| 625 | sorghum | ICMP |
| M1657A | human, male, blood | [59] |
| M1657B | human, male, blood | [59] |
| Pantoea eucalyptii | ||
| 299R | pear flower | Dr. Steven Lindow, UC Berkeley |
| B011489 | human, female, superficial wound | [59] |
| F9026 | human, male, blood | [59] |
| SM03214 | goose feces | [59] |
| SP02021 | thistle leaf | [59] |
| SP03372 | diseased maize leaf | [59] |
| SP03391 | diseased bean leaf | [59] |
| SP04013 | tomato leaf | [59] |
| Pantoea eucrina | ||
| 6686 | human, headache | [59] |
| TX5 | human, blood | [59] |
| TX6 | human, blood | [59] |
| Pantoea septica | ||
| 81828 | human, post hemicholectomy | [59] |
| 101150 | human | [59] |
| 062465A | human, cerebellar CVA (stroke) | [59] |
| 062465B | human, cerebellar CVA (stroke) | [59] |
| 091957A | human, renal failure | [59] |
| 091957B | human, renal failure | [59] |
| B016375 | human, female, finger | [59] |
| BB350028A | human, female, blood culture, fever | [59] |
| BB350028B | human, female, blood culture, fever | [59] |
| BE528629 | human, peritoneal dialysis | [59] |
| G2291404 | human | [59] |
| G3271436 | human, urine | [59] |
| G4071105 | human, urine | [59] |
| M1517 | human, female, blood | [59] |
| M41864 | human, female, blood | [59] |
| TX3 | human, blood | [59] |
| TX4 | human, blood | [59] |
| VB38951A | human, female, blood culture, sore throat | [59] |
| VB38951B | human, female, blood culture, sore throat | [59] |
| X44686 | human, female, blood | [59] |
| Pantoea stewartii | ||
| 626 | maize | ICMP |
| DC283 | maize | [63] |
| Pseudomonas aeruginosa | ||
| ATCC 27853 | clinical | Heather Dietz, University of Regina |
| Pseudomonas syringae pv. s yringae | ||
| ES4326 | radish pathogen | Dr. David Guttman, University of Toronto |
| Staphylococcus aureus | ||
| K1-7 | clinical | Dr. Chris Yost, University of Regina |
| Streptococcus mutans | ||
| UAIS9:wt | clinical | Heather Dietz, University of Regina |
Effects of Nutrition on Antibiotic Activity
The antibiotic activity of PanBRT175 was evaluated using the agar overlay method on E. coli minimal media with peptone concentrations of 2.5 mg/ml, 5.0 mg/ml, 10.0 mg/ml, and 15.0 mg/ml, and tryptone concentrations of 5.0 mg/ml, 10.0 mg/ml, 15.0 mg/ml, and 20.0 mg/ml, both with and without glucose. Antibiotic activity was also evaluated in the presence of the following amino acids: alanine, β-alanine, asparagine, aspartic acid, arginine, cystine, glutamic acid, glutamine, glycine, histidine, isoleucine, leucine, lysine, methionine, orinithine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, and valine. All amino acids were obtained from Sigma-Aldrich®, except for β-alanine, asparagine, and arginine, which were obtained from Alfa Aesar®. The amino acids were added into separate 5X Glycerol Salt Solutions at a concentration of 1.5 mg/ml, which was then added into the top layer.
Transposon Mutagenesis and Screening
Transposon mutagenesis of PanBRT175 was carried out with a triparental mating involving the helper E. coli HB101 (RK600) and the mini-Tn5 donor E. coli VPE42 (pBSL118). Overnight LB broth cultures of each strain were aliquoted in 1 mL volumes, centrifuged at 13,500 g for 1 minute, and resuspended in 100 µL of 10 mM Mg2SO4. All three were combined, vortexed briefly, and three 100 µL volumes were spotted on to a single LB agar plate, which was incubated overnight at 30°C. Following 16-24 hours of incubation, small samples of each triparental spot were spread on to individual LB agar plates containing kanamycin and incubated at 30°C for 48 hours.
Screening for PanBRT175 mutants was performed using an agar overlay method. The bottom agar was E. coli Minimal Medium (per liter: 0.25 g yeast extract, 1.72 g KH2PO4, 4.0 g K2HPO4, 0.5 g NaCl, 0.2 g sodium citrate, 2.0 g (NH4)2SO4, 0.002 g MgSO4*7H2O, 20 mL glycerol) with a top layer of 0.9% agar containing 5X Glycerol-Arginine Salt Solution (per 100 mL: 5.57 g K2HPO4, 2.25 g KH2PO4, 0.06 g MgSO4*7H2O, 10.0 g glucose, 0.025 g nicotinic acid, 0.15 g L-asparagine) seeded with E. amylovora EA321, resuspended in 300 µL of 5 mM K2HPO4. PanBRT175 mutants were picked from the triparental spread plates based on pigmentation and spotted on to the E. coli minimal media plate. The plates were incubated at 30°C overnight. Mutants that did not inhibit the EA321 indicator in the agar overlay were selected, retested, and cultured on LB agar containing kanamycin.
Inverse PCR, Sequencing, and Sequence Analysis
All selected PanBRT175 mutants were inoculated into 3 mL of LB broth and incubated overnight at 30°C. Genomic DNA was then extracted using E.Z.N.A. Bacterial DNA Kit (Omega Bio-Tek®). The genomic DNA was digested in a reaction containing 5 units HincII (New England Biolabs), 0.2 µL 10x Buffer 3, 2.0 µL BSA (10 mg/ml), 13.8 µL dH2O, 2.0 µL gDNA (280-440 ng/µL) and a unimolecular ligation performed in a reaction containing 3 units T4 DNA Ligase (NEB), 162.0 µL dH2O, 20.0 µL T4 DNA Ligase Buffer, and 15.0 µL digested gDNA. Samples at both steps were cleaned using E.Z.N.A. Purification Kit (Omega Bio-Tek®). An inverse PCR was then performed (8.6 µL dH2O, 0.2 µL EconoTaq DNA Polymerase (Lucigen) (5 U/µL), 0.5 µL forward primer (50 µM), 0.5 µL reverse primer (50 µM), 2.0 µL 10X EconoTaq Buffer, 1.2 µL 25 mM MgCl2, 2.0 µL dNTPs (200 µM each), 5.0 µL template DNA) and resulting samples were purified directly, or extracted from the gel using Omega E.Z.N.A. Gel Extraction Kit. Sequencing was performed by Eurofins MGW Operon (Huntsville, AL, USA).
Genetic and Genomic Analyses
Mutant sequences were queried against GenBank at NCBI using the Basic Local Alignment Search Tool (BLAST). Mutant sequences were also queried against a draft sequence of the PanBRT175 genome (Accession: ASJH00000000) [28] using standalone BLAST to identify the location of the gene cluster. Once identified, each gene was queried against the draft genomes of Pantoea septica X44686, Pantoea agglomerans Eh318, Pantoea ananatis BRT98, Pantoea agglomerans TX10, Pantoea eucalyptii F9026, Pantoea ananatis 15320, Pantoea agglomerans DC432, Pantoea dispersa 625, Pantoea eucalyptii 299R, Pantoea eucalyptii SP03391, Pantoea agglomerans SP04022, Pantoea eucalyptii SP03372, Pantoea eucalyptii SP04013, Pantoea eucalyptii SP02021, Pantoea agglomerans SP00101, Pantoea agglomerans VB39851-A, Pantoea calida B021323, Pantoea eucalyptii B011489, Pantoea agglomerans B025670, Pantoea brenneri B024858, Pantoea brenneri B016381, Pantoea dispersa M1657A, and Pantoea calida BB957621-B2 using standalone BLAST with an e-value cutoff of 0.0001 to identify even the most divergent potential homologs. The NCBI Conserved Domain Database (CDD) was used to assign each protein to a family.
Pantoea agglomerans Eh318 Genome Sequencing
Total DNA was sequenced using Illumina HiSeq 2000, 100-bp paired-end sequencing, resulting in 16,055,112 reads, with an average Phred quality score of 32. ABySS version 1.3.5 was used for de novo paired-end assembly using the default parameters and an optimized k-mer value of 87. This resulted in 51 contigs with an N50 of 376,883 bp and an estimated genome size of 5,036,004 bp at 319X coverage. Contigs that were 200 bp or larger (37 total) were submitted to the NCBI Prokaryotic Genome Automatic Annotation Pipeline version 2.0. The genome has been deposited under accession number AXOF00000000.
Distribution of the PNP-1 Cluster in Pantoea
PCR primers (Table 2) specific to pnp1A, pnp1C, pnp1D, and pnp1F were used to examine the distribution of the biosynthetic cluster in Pantoea. Colony PCR was performed for each Pantoea isolate by toothpick inoculation of a single colony into a PCR reaction containing 13.6 µL dH2O, 0.2 µL EconoTaq DNA Polymerase (5 U/µL), 0.5 µL forward primer (50 µM), 0.5 µL reverse primer (50 µM), 2.0 µL 10X EconoTaq Buffer, 1.2 µL 25 mM MgCl2, and 2.0 µL dNTPs (200 µM each), on each Pantoea strain (Table 1).
Table 2. List of primers used in this study.
| Gene | Primer | Sequence | Length | Annealing Temp |
| pnp1A | GntR+11 | GTGCTGTCGATACAGACGGCGCAT | 24 | 68.0 |
| GntR-700 | GACGTGATCCTGCGGGCTTACTGTC | 25 | 69.5 | |
| pnp1C | 07851+59 | GCAAGCTCAACCGTAGGATATTCTC | 25 | 64.6 |
| 07851-680 | AATTAGACTGTCAAGAGAGAATGGT | 29 | 66.0 | |
| pnp1D | Carb+64 | ACAAGCACTGAGCAGCCCGTCAGC | 25 | 69.5 |
| Carb-836 | TGCAGCATCAGTGACTGGTGAGAGA | 25 | 66.2 | |
| pnp1F | 07881+95 | GATCCGGGTGATGCGTGGCCAGAG | 25 | 72.8 |
| 07881-555 | TGAAACAGCGGTGATCCGGTTCGT | 25 | 66.2 | |
| mini-Tn5 transposon | npt-41 | AGCCGAATAGCCTCTCCACCCAAG | 24 | 68.0 |
| npt+772 | TTCGCAGCGCATCGCCTTCTATC | 23 | 66.3 |
Results and Discussion
Effects of Nutrition on Antibiotic Activity
Antibiotic production assays of PanBRT175 were carried out on E. coli minimal medium supplemented with glucose, which resulted in a zone of inhibition (ZOI) in a lawn of E. amylovora. To determine how antibiotic production and/or activity was affected by nutritional changes, the assay was attempted on the rich medium, LB. The ZOI was reduced to approximately half the size of that formed on minimal medium with glucose (Figure S1). The assay was repeated with varying concentrations of either peptone or tryptone in minimal salt medium, with and without glucose. Antibiotic production was greatest at the lowest concentrations of 2.5 mg/ml peptone and 5.0 mg/ml tryptone (approximately double the ZOI formed on minimal medium) whether or not glucose was present. At the higher concentrations, 10.0 mg/ml peptone, 15.0 mg/ml peptone and 15.0 mg/ml tryptone, and in the presence of glucose, antibiotic production was still approximately double, but the edge of the ZOI was less defined (Figure S1). The remaining peptone and tryptone concentrations with and without glucose had similar PNP-1 production as minimal medium with glucose. In general, these results suggest that PNP-1 biosynthesis is not repressed significantly under nutrient-rich conditions, as has been reported for several antibiotic biosynthetic pathways [29]. Antibiotics like streptothricin from Streptomyces lavendulae and streptomycin from Streptomyces griseus, are also produced in abundance on tryptone-starch and nutrient medium, respectively [30]. Micronutrients and salts can also alter antibiotic production with phenazine production and accumulation in Pseudomonas being affected by the presence of boric acid, iron and magnesium sulfate [31].
Pantoea antibiotics target specific amino acid biosynthesis pathways, and have been traditionally grouped by their loss of effectiveness in the presence of certain amino acids. To determine whether the activity of this antibiotic could be neutralized in the presence of amino acids, each amino acid was supplemented into the agar overlay on minimal salt medium. Antibiotic activity against E. amylovora was not affected by any of the tested amino acids, which is consistent with the Group V antibiotics, such as herbicolin I [19], [20]. Interestingly, some E. amylovora colonies were able to spontaneously acquire resistance to PNP-1, suggesting that a mutation in a single gene can result in antibiotic resistance. Retests using an agar overlay assay confirmed that the isolated spontaneous mutants were indeed resistant, as no ZOI formed.
Spectrum of Antibiotic Activity
The antibiotic produced by PanBRT175 showed a narrow spectrum of activity inhibiting E. amylovora, but not S. aureus, P. aeruginosa, S. mutans, E. coli, or L. lactis (Table 3). We extended this testing to 30 isolates of Pantoea representing 11 different species. Of these, 20 isolates were susceptible, while all resistant Pantoea strains fell into the closely related P. agglomerans and P. eucalyptii groups (Table 3). This suggests that resistance to PNP-1 was acquired by a common ancestor of these sister species [32], and that resistance is not necessarily directly tied to the biosynthetic cluster. The antibiotic showed activity against both clinical and environmental Pantoea isolates in the other species groups. Both P. ananatis and P. stewartii are known plant pathogens [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], and so PNP-1 has the potential to be used in its purified form for control of these strains.
Table 3. Evaluation of resistance to PNP-1 across non-Pantoea and Pantoea species.
| Strain | Source | Resistant (R)/Susceptible (S) |
| Erwinia amylovora | ||
| EA321 | Environmental | S |
| Escherichia coli | ||
| DH5a | Lab strain | R |
| Lactococcus lactis | ||
| HD1 | Environmental | R |
| Pantoea agglomerans | ||
| 1512 | Environmental | R |
| 83 | Environmental | R |
| 5565 | Environmental | R |
| Eh318 | Environmental | R |
| DC432 | Environmental | R |
| B016395 | Clinical | R |
| B025670 | Clinical | R |
| B026440 | Clinical | R |
| TX10 | Clinical | R |
| SP05130 | Environmental | R |
| SS02010 | Environmental | R |
| SP05120 | Environmental | R |
| SP04022 | Environmental | R |
| Pantoea eucalyptii | ||
| 299R | Environmental | R |
| F9026 | Clinical | R |
| B011489 | Clinical | R |
| SP03372 | Environmental | R |
| SP02021 | Environmental | R |
| SP04013 | Environmental | R |
| SP03391 | Environmental | R |
| Pantoea conspicua | ||
| B011017 | Clinical | S |
| Pantoea brenneri | ||
| B016381 | Clinical | S |
| B024858 | Clinical | S |
| Pantoea anthophila | ||
| 1373 | Environmental | S |
| Pantoea stewartii | ||
| DC283 | Environmental | S |
| 262 | Environmental | S |
| Pantoea ananatis | ||
| BRT98 | Environmental | S |
| 15320 | Environmental | S |
| 17671 | Environmental | S |
| Cit30-11 | Environmental | S |
| M232A | Environmental | S |
| B7 | Environmental | S |
| 26SR6 | Environmental | S |
| Pantoea calida | ||
| B021323 | Clinical | S |
| BB957621-B2 | Clinical | S |
| Pantoea septica | ||
| B016375 | Clinical | S |
| VB38951-A | Clinical | S |
| X44686 | Clinical | S |
| Pantoea dispersa | ||
| M1657A | Clinical | S |
| 625 | Environmental | S |
| Pantoea eucrina | ||
| 6868 | Clinical | S |
| Pseudomonas aeruginosa | ||
| ATCC 27853 | Clinical | R |
| Staphylococcus aureus | ||
| K1-7 | Clinical | R |
| Streptococcus mutans | ||
| UAIS9:wt | Clinical | R |
Identification of the Biosynthetic Cluster
Transposon mutagenesis was used to identify the antibiotic biosynthetic cluster in PanBRT175. Approximately 5000 mutants were screened using the agar overlay method, resulting in 41 independent mutants confirmed to be impaired in antibiotic production and/or secretion. We analyzed 29 of these mutants, and in four of them, the transposon had inserted into the disrupted carbamoyl-phosphate synthase large chain (carB) and phosphoribosylformylglycinamidine synthase (purL) genes, which are necessary components in purine and pyrimidine metabolism [45], [46] (Table 4). In the other 25 mutants, the transposon had inserted independently in six genes that were part of a 8.2 kb seven-gene cluster in the PanBRT175 genome [28] (Table 4).
Table 4. Recovered P. ananatis BRT175 mutants defective in PNP-1 production/export.
| Mutant | Gene | Organism | Putative protein | Conserved domains | Length (nt) | % Similarity | Accession |
| 5, 6, 18 | pnp1A | P. ananatis BRT175 | GntR Transcriptional Regulator | Aspartate Aminotransferase Family, cd00609; Winged helix-turn-helix, cd07377. | 1404 | ERM15323 | |
| P. syringae pv. maculicola ES4326 | 1317 | 48 | EGH58727 | ||||
| 9, 16, 26 | pnp1B | P. ananatis BRT175 | Hypothetical protein | SGNH/GDSL hydrolase Family, pfam13472 | 972 | ERM14577 | |
| P. syringae pv. maculicola ES4326 | 993 | 75 | EGH58728 | ||||
| 4 | pnp1C | P. ananatis BRT175 | Hypothetical protein | N/A | 2111 | ERM15479 | |
| P. syringae pv. maculicola ES4326 | 2202 | 61 | EGH58729 | ||||
| 2, 3, 8, 10, 14, 22, 24, 27, 28 | pnp1D | P. ananatis BRT175 | Carbamoyltransferase | NodU/CmcH Family, cl12209 | 1830 | ERM15480 | |
| P. syringae pv. maculicola ES4326 | 927 | 70 | EGH58732 | ||||
| 1, 7, 11, 12, 15, 17, 25 | pnp1E | P. ananatis BRT175 | Hypothetical Protein | Aspartate Aminotransferase (AAT) Family, cl00321 | 1474 | ERM15482 | |
| P. syringae pv. maculicola ES4326 | 1317 | 72 | EGH58734 | ||||
| 13,23 | pnp1F | P. ananatis BRT175 | Hypothetical Protein | Formyltransferase Core Superfamily, cd08369 | 683 | ERM15483 | |
| P. syringae pv. maculicola ES4326 | 600 | 70 | EGH58735 | ||||
| N/A | pnp1G | P. ananatis BRT175 | MFS Transporter | MFS Superfamily, cd06174 | 560 | ERM15324 | |
| P. syringae pv. maculicola ES4326 | 627 | 61 | EGH58736 | ||||
| 19 | carB | P. ananatis BRT175 | Carb3 | ATP-grasp Superfamily, cl17255 | 3230 | ERM15221 | |
| P. ananatis LMG 5342 | 3228 | 83 | YP_005197068 | ||||
| 20, 21, 29 | purL | P. ananatis BRT175 | PurL | PurM-like Superfamily, cl10019 | 3890 | ERM12406 | |
| P. ananatis LMG 5342 | 3888 | 97 | AER31669 |
The first gene identified is the divergently transcribed pnp1A, which belongs to COG1167 (ARO8), a family whose members contain both a winged helix-turn-helix domain of the GntR family of transcriptional regulators (Conserved Domain Database Accession: cl17414), as well as an aspartate aminotransferase domain (CDD Accession:cl00321). The GntR transcriptional regulators have been implicated in bacterial growth and development, as well as antibiotic production [47]. Interestingly, antibiotic biosynthesis in one of the recovered pnp1A mutants could be restored with the exogenous supplementation of histidine. Transposon insertion in this mutant occurred in the aminotransferase region, possibly indicating that histidine is a PNP-1 precursor, and that the product of this gene plays a role in regulating histidine availability. The supplementation of histidine would have bypassed the requirement for this locus, thereby restoring antibiotic biosynthesis. The pdxR regulator of Streptomyces venezuelae, which also contains both an aminotransferase and helix-turn-helix domain was shown to complement pyridoxal (vitamin B6) auxotrophy by regulating the availability of pyridoxal phosphate and related co-enzymes [48]. None of the other 29 PNP-1 mutants could be rescued by histidine, or any other amino acids.
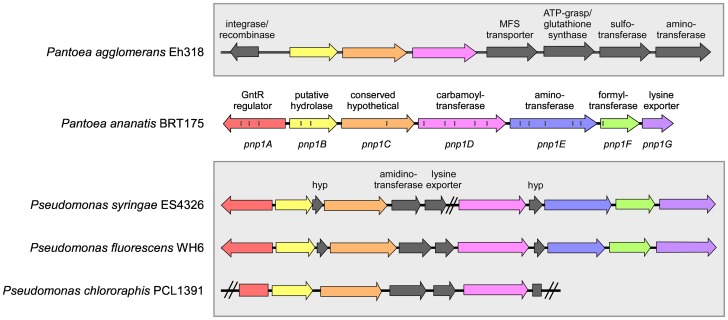
pnp1B, pnp1C, and pnp1D are the next three genes of the predicted biosynthetic cluster (Figure 1). pnp1B belongs to the SGNH/GDSL family of hydrolases (CDD Accession: cl01053), which include esterases and lipases that have known activity towards esters, acyl-CoAs, and amino acid derivatives. Applications for these hydrolytic enzymes include synthesis of pharmaceuticals, including phospholipase A and B [49]. pnp1C is a 2111 bp-long predicted coding region; however, it has no identifiable protein domain that could indicate biochemical function. Finally, pnp1D codes for a carbamoyltransferase of the NodU/CmcH family (CDD Accession: cl12209, pfam02543), which is responsible for post-transcriptional modifications of late gene products [50]. Carbamoyltransferases are key enzymes in β-lactam antibiotic production in certain species of Rhizobium, Bradyrhizobium, and Streptomyces [50]. Homologs of these three genes were identified in the gene clusters of Pseudomonas syringae pv. maculicola ES4326, Pseudomonas fluorescens WH6, and Pseudomonas chlororaphis PCL1391, as well as P. agglomerans Eh318 (discussed later) (Figure 1, Table 4).
Figure 1. The composition and organization of the 8.2-1 antibiotic biosynthetic cluster in P. ananatis BRT175 (Accession: ASJH00000000).
. The insertion sites of the mini-Tn5 transposon for mutants generated by transposon mutagenesis are indicated with lines within the cluster. Homologous biosynthetic gene clusters of P. syringae pv. maculicola ES4326 (11.6 kb) (Accessions: AEAK01000141, AEAK01000142), P. fluorescens WH6 (11.6 kb) (Accession: AEAZ01000041), and P. chlororaphis PCL1391 (7.5 kb) (Accession: DQ367408) are shown. A gene cluster in P. agglomerans Eh318 (10kb) (Accession: AXOF00000000) having several homologous genes is shown. Gene colours indicate homology between the biosynthetic clusters. Cluster in P. syringae pv. maculicola ES4326 overlaps two contigs.
The product of pnp1E falls under the aspartate aminotransferase (AAT) superfamily (fold type I) of pyridoxal phosphate (PLP)-dependent enzymes (Accession: cl00321), members of which are known to play a role in transamination, racemization, decarboxylation, and various other side chain reactions [51]. The product of pnp1F belongs to the formyltransferase, catalytic core domain superfamily (CDD Accession: cl00395), which is known to be involved in purine biosynthesis, formate biosynthesis, translation initiation, and modification of lipid A, leading to resistance to cationic antimicrobial peptides and clinical antimicrobials [51].
The final annotated gene in the PanBRT175 cluster, pnp1G, is a member of the MFS transporter family that includes the LysE type translocator family (pfam01810) and the RhtB homoserine/threonine transport family (TIGR00949, COG1280). LysE exporters have been noted to transport L-lysine, cadmium, and various quaternary amines in bacterial species such as E. coli, Mycobacterium tuberculosis, and Bacillus subtilis [52], while the RhtB protein family has been proposed to be involved in the excretion of metabolites [53]. This is consistent with the fact that transposon insertion mutants were not recovered for this gene, since disruption could result in metabolite accumulation and lethality. Alternatively, this could have been a consequence of our non-saturating genetic screen.
Distribution of the PNP-1 Biosynthetic Cluster
To determine whether this antibiotic cluster was present in other Pantoea strains, a PCR-based survey was conducted on four of the seven genes (pnp1A, pnp1C, pnp1D, and pnp1F). A total of 117 strains were evaluated, all of which were negative for all four genes in the antibiotic cluster. Standalone BLASTs were also performed for each gene in the PNP-1 cluster against a draft genome sequence of P. agglomerans Eh318, an isolate known to produce at least two antibiotics, pantocins A and B, which are effective against E. amylovora [18]. Interestingly, homologs of pnp1B, pnp1C, and pnp1D were identified as the first genes in a seven-gene 10 kb cluster in Eh318, while the remaining genes of the cluster include an MFS transporter, ATP-grasp/glutathione synthase, sulfotransferase, and aminotransferase (Figure 1). Although this suggests that this Eh318 gene cluster may direct the synthesis of another natural product, there is no evidence that this product is antimicrobial.
Because the PNP-1 cluster was unique and not found complete in any other isolate in our collection, we examined its distribution in other species. We queried GenBank and identified similar gene clusters in Pseudomonas syringae pv. maculicola ES4236, Pseudomonas fluorescens WH6 and Pseudomonas chlororaphis PCL1391 (Table 4). The clusters in ES4326 and WH6 were 11.6 kb in size and consisted of a total of eleven genes, seven of which corresponded to those found in the PNP-1 biosynthetic cluster (Figure 1). Four genes present in the Pseudomonas clusters that are annotated as hypothetical protein, inosamine-phosphate amidinotransferase, lysine exporter, and hypothetical protein, are absent from the PNP-1 cluster (Figure 1). Since there was substantial similarity between PNP-1 and clusters of PC1391, ES4326 and WH6, we performed some preliminary experiments on ES4326 to evaluate whether it can also antagonize Erwinia. An overlay assay with ES4326 against E. amylovora showed that P. syringae had no inhibitory activity against E. amylovora under the same media conditions tested (data not shown). These results suggest that if the cluster in ES4326 also produces an antimicrobial compound, it may be differentially regulated, such that it is not induced under the same conditions as PNP-1. Alternatively, the chemical structure and/or spectrum may be different than PNP-1, due to the differing composition of their gene clusters.
The chemical nature of the PNP-1 antibiotic is still not known. The homologous cluster in P. chlororaphis PCL1391 directs the production of a phenazine antibiotic, phenazine-1-carboxamide (Figure 1). Phenazines are heterocyclic compounds with various residue substitutions around the rings, and as a result produce a range of pigments from dark red to bright yellow to bright blue [54]. Those that are antimicrobial tend to be broad-spectrum, and have many ecological roles [55]. Phenazine-1-carboxamide (PCN) from P. chlororaphis PCL1391 has been shown to inhibit the effects of tomato foot and root rot, caused by Fusarium oxysporum [56]. In addition, PCN enhances P. chlororaphis colonization of the rhizosphere by eliminating all competition for nutrients and space [57]. Although this cluster is homologous to that of PNP-1, PNP-1 does not appear to be broad-spectrum like PCN. P. agglomerans EH1087 has been reported to produce a phenazine antibiotic; D-alanylgriseoluteic acid (AGA) [24], [58]. AGA, however, is a broad-range antibiotic effective against a wide range of Gram-negative and Gram-positive bacteria, unlike PNP-1 [24]. In addition, AGA synthesis has been attributed to a sixteen gene cluster, approximately 14 kb in size [24], and is not homologous to the PNP-1 biosynthetic cluster. Characteristic of a phenazine antibiotic, the activity of both AGA and PNP-1 is not abolished in the presence of amino acids [24].
This work has identified an antibiotic in P. ananatis BRT175 whose biosynthesis is determined by an 8.2 kb biosynthetic gene cluster consisting of seven genes. The loss of antibiotic production and/or export in 24 independent transposon mutants is strong evidence that the seven-gene cluster identified is likely responsible for the production of the antibiotic. Although this cluster has yet to be reported within Pantoea, the identification of homologous antibiotic biosynthetic gene clusters in Pseudomonas that have been shown to produce an antimicrobial phenazine adds further support to our conclusion. Although there are several conserved genes between this cluster and those of the pseudomonads, differences in the genetic composition of the clusters, along with the spectrum of activity, suggest that PNP-1 is likely structurally and functionally different from phenazines. Still, further characterization of the gene cluster, including its role in antibiotic biosynthesis, and the determination of the biosynthetic steps and chemical structure of PNP-1 are presently being pursued. PNP-1 has the potential to be used to control E. amylovora, as well as clinically and agriculturally relevant Pantoea isolates.
Supporting Information
Effects of nutrition on antibiotic production. A) LB medium with and without glucose, B) E. coli minimal medium, with and without glucose; C) E. coli minimal medium with varying peptone concentrations, with and without glucose; D) E. coli minimal medium with varying tryptone concentrations, with and without glucose.
(DOC)
Funding Statement
AW was supported by scholarships from the University of Regina Faculty of Graduate Studies and Research, and awards from the Rx&D Health Research Foundation. DDNS was funded by a Master's Award from the Canadian Institutes of Health Research. This work was funded by a Discovery Grant from the Natural Sciences and Engineering Council of Canada (386654-2010), an LOF from the Canada Foundation for Innovation (28591), and funding from the University of Regina Faculty of Science. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Beer SV, Rundle JR (1984) Interaction between Erwinia amylovora and Erwinia herbicola in vitro, in immature pear fruits and in apple blossums. Acta Horti 151: 203–204. [Google Scholar]
- 2. Jin M, Liu L, Wright SAI, Beer SV, Clardy J (2003) Structural and functional analysis of pantocin A: An antibiotic from Pantoea agglomerans discovered by heterologous expression of cloned genes. Angew Chem Int Ed 42: 2898–2901. [DOI] [PubMed] [Google Scholar]
- 3.Johnson KB, Stockwell VO (2000) Fire blight: the disease and its causative agent, Erwinia amylovora Fire Blight: The Disease and Its Causative Agent, Erwinia amylovora. Wallingford: CAB International. pp. 319–338.
- 4. Giddens SR, Houliston GJ, Mahanty HK (2003) The influence of antibiotic production and pre-emptive colonization on the population dynamics of Pantoea agglomerans (Erwinia herbicola) Eh1087 and Erwinia amylovora in planta. Environ Microbiol 5: 1016–1021. [DOI] [PubMed] [Google Scholar]
- 5.Bonn WG, van der Zwet T (2000) Distribution and economic importance of fire blight. Fire Blight: The Disease and Its Causative Agent, Erwinia amylovora. Wallingford: CAB International. pp. 37–53.
- 6. Pusey PL, Stockwell VO, Reardon CL, Smits THM, Duffy B (2011) Antibiosis activity of Pantoea agglomerans biocontrol strain E325 against Erwinia amylovora on apple flower stigmas. Phytopathology 101: 1234–1241. [DOI] [PubMed] [Google Scholar]
- 7. Kamber T, Smits THM, Rezzonico F, Duffy B (2011) Genomics and current genetic understanding of Erwinia amylovora and the fire blight antagonist Pantoea vagans . Trees 26: 227–238. [Google Scholar]
- 8. Johnson KB, Stockwell VO, McLaughlin RJ, Sugar D, Loper JE, et al. (1993) Effect of antagonistic bacteria on establishment of honey bee-dispersed Erwinia amylovora in pear blossums and on fire blight control. Phytopathology 83: 995–1002. [Google Scholar]
- 9. Ozaktan H, Bora T (2004) Biological control of fire blight in pear orchards with a formulation of Pantoea agglomerans strain Eh 24. Braz J Microbiol 35: 224–229. [Google Scholar]
- 10.Riggle JH, Klos EJ (1972) Relationship of Erwinia herbicola to Erwinia amylovora Can J Bot 50 : 1077-&. [Google Scholar]
- 11.Chatterjee AK, Gibbins LN, Carpente Ja (1969) Some observations on physiology of Erwinia herbicola and its possible implication as a factor antagonistic to Erwinia amylovora in fire blight syndrome. Can J Microbiol 15: : 640-&. [PubMed] [Google Scholar]
- 12.Thomson SV, Gouk SC, New Zealand Plant Protect SOC (1992) Interactions between an antagonist, Erwinia herbicola, and E. amylovora and potential for biological control of fire blight. Proceedings of the Forty-Fifth New Zealand Plant Protection Conference: 295–300.
- 13.Sholberg PL, Boule J (2008) Evaluation of antibiotics and plant extracts for control of streptomycin-resistant Erwinia amylovora. In: Johnson KB, Stockwell VO, editors. Proceedings of the Eleventh International Workshop on Fire Blight.pp. 423–428. [Google Scholar]
- 14. Pusey PL, Stockwell VO, Rudell DR (2008) Antibiosis and acidification by Pantoea agglomerans strain E325 may contribute to suppression of Erwinia amylovora . Phytopathology 98: 1136–1143. [DOI] [PubMed] [Google Scholar]
- 15. Wright SA, Beer SV (1996) The role of antibiotics in biological control of fire blight by Erwinia herbicola strain EH318. Acta Horti 411: 309–311. [Google Scholar]
- 16.Wright SAI, Beer SV (2002) Genes for biosynthesis of pantocin A and B by Pantoea agglomerans Eh318. In: Hale C, Mitchell R, editors. Proceedings of the IX International Workshop on Fire Blight.pp. 237–241.
- 17.Wright SAI, Jin M, Clardy J, Beer SV (2006) The Biosynthetic genes of pantocin A and pantocin B of Pantoea agglomerans Eh318. In: Bazzi C, Mazzucchi U, editors. Proceedings of the X International Workshop on Fire Blight.pp. 313–319.
- 18. Wright SAI, Zumoff CH, Schneider L, Beer SV (2001) Pantoea agglomerans strain EH318 produces two antibiotics that inhibit Erwinia amylovora in vitro. Appl Environ Microbiol 67: 284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smits THM, Rezzonico F, Kamber T, Blom J, Goesmann A, et al. (2011) Metabolic versatility and antibacterial metabolite biosynthesis are distinguishing genomic features of the fire blight antagonist Pantoea vagans C9-1. PloS One 6. [DOI] [PMC free article] [PubMed]
- 20. Smits THM, Rezzonico F, Kamber T, Goesmann A, Ishimaru CA, et al. (2010) Genome sequence of the biocontrol agent Pantoea vagans strain C9-1. J Bac 192: 6486–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanneste JL, Cornish DA, Yu J, Voyle MD (2002) The peptide antibiotic produced by Pantoea agglomerans Eh252 is a microcin. In: Hale C, Mitchell R, editors. Proceedings of the X International Workshop on Fire Blight. pp. 285–290.
- 22.Vanneste JL, Cornish DA, Yu J, Voyle MD (2000) A microcin produced by a strain of Erwinia herbicola is involved in biological control of fire blight and soft rot caused by Erwinia sp. In: Fokkema NJ, Beek MA, VanSteekelenburg NAM, Samyn G, Maas JL, et al., editors. Proceedings of the XXV International Horticultural Congress, Pt 3: Culture Techniques with Special Emphasis on Environmental Implications. pp. 39–46.
- 23.Vanneste JL, Yu J, Cornish DA (2008) Presence of genes homologous to those necessary for synthesis of microcin MccEh252 in strains of Pantoea agglomerans. In: Johnson KB, Stockwell VO, editors. Proceedings of the Eleventh International Workshop on Fire Blight. pp. 391–396.
- 24. Giddens SR, Feng YJ, Mahanty HK (2002) Characterization of a novel phenazine antibiotic gene cluster in Erwinia herbicola Eh1087. Mol Microbiol 45: 769–783. [DOI] [PubMed] [Google Scholar]
- 25. Elgoorani MA, Hassanein FM, Shoeib AA (1992) Antibacterial and antifungal spectra of antibiotics produced by different strains of Erwinia herbicola (Pantoea agglomerans). J Phytopathology 136: 335–339. [Google Scholar]
- 26. Brady SF, Wright SA, Lee JC, Sutton AE, Zumoff CH, et al. (1999) Pantocin B, an antibiotic from Erwinia herbicola discovered by heterologous expression of cloned genes. J Am Chem Soc 121: 11912–11913. [Google Scholar]
- 27. Jin M, Wright SAI, Beer SV, Clardy J (2003) The biosynthetic gene cluster of pantocin A provides insights into biosynthesis and a tool for screening. Angew Chem Int Ed 42: 2902–2905. [DOI] [PubMed] [Google Scholar]
- 28.Smith DDN, Kirzinger MWB, Stavrinides J (2013) Draft genome sequence of the antibiotic-producing epiphytic isolate Pantoea ananatis BRT175. Genome Announc 1. [DOI] [PMC free article] [PubMed]
- 29. Sanchez S, Chavez A, Forero A, Garcia-Huante Y, Romero A, et al. (2010) Carbon source regulation of antibiotic production. J Antibiot 63: 442–459. [DOI] [PubMed] [Google Scholar]
- 30. Schatz A, Bugie E, Waksman SA (2005) Streptomycin, a substance exhibiting antibiotic activity against gram positive and gram-negative bacteria. Clin Orthop Relat Res 437: 3–6. [DOI] [PubMed] [Google Scholar]
- 31. Slininger PJ, Jackson MA (1992) Nutritional factors regulating growth and accumulation of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2-79. Appl Microbiol Biotechnol 37: 388–392. [Google Scholar]
- 32. Brady CL, Venter SN, Cleenwerck I, Engelbeen K, Vancanneyt M, et al. (2009) Pantoea vagans sp nov., Pantoea eucalypti sp nov., Pantoea deleyi sp nov and Pantoea anthophila sp nov. Int J Syst Evol Microbiol 59: 2339–2345. [DOI] [PubMed] [Google Scholar]
- 33. Coutinho TA, Preisig O, Mergaert J, Cnockaert MC, Riedel KH, et al. (2002) Bacterial blight and dieback of Eucalyptus species, hybrids, and clones in South Africa. Plant Dis 86: 20–25. [DOI] [PubMed] [Google Scholar]
- 34. Kido K, Adachi R, Hasegawa M, Yano K, Hikichi Y, et al. (2008) Internal fruit rot of netted melon caused by Pantoea ananatis ( = Erwinia ananas) in Japan. J Gen Plant Path 74: 302–312. [Google Scholar]
- 35. Walcott RR, Gitaitis RD, Castro AC, Sanders FH, Diaz-Perez JC (2002) Natural infestation of onion seed by Pantoea ananatis, causal agent of center rot. Plant Dis 86: 106–111. [DOI] [PubMed] [Google Scholar]
- 36. Gitaitis RD, Gay JD (1997) First report of a leaf blight, seed stalk rot, and bulb decay of onion by Pantoea ananas in Georgia. Plant Dis 81: 1096–1096. [DOI] [PubMed] [Google Scholar]
- 37. Fucikovsky L, Aranda S (2006) Pantoea ananas a new pathogen of agave in Mexico. Phytopathology 96: S37–S37. [Google Scholar]
- 38.Bell AA, Medrano EG, Jones MA. Frequency and pathogenicity of microorganisms associated with cotton seed rot in South Carolina; 2004; San Antonio, TX .
- 39. Cother EJ, Reinke R, McKenzie C, Lanoiselet VM, Noble DH (2004) An unusual stem necrosis of rice caused by Pantoea ananas and the first record of this pathogen on rice in Australia. Australas Plant Path 33: 495–503. [Google Scholar]
- 40. Roper MC (2011) Pantoea stewartii subsp. stewartii: lessons learned from a xylem-dwelling pathogen of sweet corn. Mol Plant Path 12: 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pérez-y-Terrón R, Villegas MC, Cuellar A, Muñoz-Rojas J, Castañeda-Lucio M, et al. (2009) Detection of Pantoea ananatis, causal agent of leaf spot disease of maize, in Mexico. Australas Plant Dis Notes 4: 96–99. [Google Scholar]
- 42. Azad HR, Holmes GJ, Cooksey DA (2000) A new leaf blotch disease of sudangrass caused by Pantoea ananas and Pantoea stewartii . Plant Dis 84: 973–979. [DOI] [PubMed] [Google Scholar]
- 43. Cha JS, Pujol C, Ducusin AR, Macion EA, Hubbard CH, et al. (1997) Studies on Pantoea citrea, the causal agent of pink disease of pineapple. J Phytopathology 145: 313–319. [Google Scholar]
- 44. Marin-Cevada V, Vargas VH, Juarez M, Lopez VG, Zagada G, et al. (2006) Presence of Pantoea citrea, causal agent of pink disease, in pineapple fields in Mexico. Plant Path 55: 294–294. [Google Scholar]
- 45. Makoff AJ, Radford A (1978) Genetics and biochemistry of carbamoyal phosphate biosynthesis and its utilization in the pyrimidine biosynthetic pathway. Microbiol Rev 42: 307–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gu ZM, Martindale DW, Lee BH (1992) Isolation and complete sequence of the purL gene encoding FGAM synthase II in Lactobacillus casei . Gene 119: 123–126. [DOI] [PubMed] [Google Scholar]
- 47. Resch M, Schiltz E, Titgemeyer F, Muller YA (2012) Insight into the induction mechanism of the GntR/HutC bacterial transcriptional regulator YvoA. Nuc Acids Res 38: 2485–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Magarvey N, He J, Aidoo KA, Vining LC (2001) The pdx genetic marker adjacent to the chloramphenicol biosynthesis gene cluster in Streptomyces venezuelae ISP5230: functional characterization. Microbiol 147: 2103–2112. [DOI] [PubMed] [Google Scholar]
- 49. Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF (2004) GDSL family of serine esterases/lipases. Prog Lip Res 43: 534–552. [DOI] [PubMed] [Google Scholar]
- 50. Martin JR (1998) New aspects of genes and enzymes for β-lactam antibiotic synthesis. Appl Microbiol Biotechnol 50: 1–15. [DOI] [PubMed] [Google Scholar]
- 51. Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, et al. (2013) CDD: conserved domains and protein three-dimensional structure. Nuc Acids Res 41: 348–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vrljic M, Garg J, Bellmann A, Wachi S, Freudl R, et al. (1999) The LysE superfamily: topology of the lysine exporter LysE of Corynebacterium glutamicum, a paradyme for a novel superfamily of transmembrane solute translocators. J Mol Microbiol Biotechnol 1: 327–336. [PubMed] [Google Scholar]
- 53. Aleshin VV, Zakataeva NP, Livshits VA (1999) A new family of amino-acid-efflux proteins. Trends Biochem Sci 24: 133–135. [DOI] [PubMed] [Google Scholar]
- 54. Price-Whelan A, Dietrich LEP, Newman DK (2006) Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol 2: 71–78. [DOI] [PubMed] [Google Scholar]
- 55. Mavrodi DV, Parejko JA, Mavrodi OV, Kwak YS, Weller DM, et al. (2013) Recent insights into the diversity, frequency and ecological roles of phenazines in fluorescent Pseudomonas spp . Environ Microbiol 15: 675–686. [DOI] [PubMed] [Google Scholar]
- 56. Chin-A-Woeng TFC, Bloemberg GV, Mulders IHM, Dekkers LC, Lugtenberg BJJ (2000) Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol Plant Microbe Interact 13: 1340–1345. [DOI] [PubMed] [Google Scholar]
- 57. Girard G, Rigaliz S (2011) Role of the phanzine-inducing protein Pip in stress resistance of Pseudomonas chlororaphis . Microbiol 157: 393–407. [DOI] [PubMed] [Google Scholar]
- 58. Kearns LP, Mahanty HK (1998) Antibiotic production by Erwinia herbicola Eh1087: Its role in inhibition of Erwinia amylovora and partial characterization of antibiotic biosynthesis genes. Appl Environ Microbiol 64: 1837–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nadarasah G, Stavrinides J (2014) Quantitative evaluation of the host-colonizing capabilites of the of the enteric bacterium Pantoea using plant and insect hosts. Microbiol 160: 602–615. [DOI] [PubMed] [Google Scholar]
- 60. Alexeyev M, Shokolenko I, Croughan T (1995) New mini-Tn5 derivatives for insertion mutagenesis and genetic engineering in Gram-negative bacteria. Can J Microbiol 41: 1053–1055. [DOI] [PubMed] [Google Scholar]
- 61. Coplin DL, Majerczak DR, Zhang YX, Kim WS, Jock S, et al. (2002) Identification of Pantoea stewartii subsp stewartii by PCR and strain differentiation by PFGE. Plant Dis 86: 304–311. [DOI] [PubMed] [Google Scholar]
- 62.Smith D, Kirzinger M, Stavrinides J (2013) Draft genome sequence of the antibiotic-producing cystic fibrosis isolate Pantoea agglomerans Tx10. Genome Announcements 1. [DOI] [PMC free article] [PubMed]
- 63. Coplin D, Frederick R, Majerczak D, ES H (1986) Molecular cloning of virulence genes from Erwinia stewartii J Bacteriol. 168: 619–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effects of nutrition on antibiotic production. A) LB medium with and without glucose, B) E. coli minimal medium, with and without glucose; C) E. coli minimal medium with varying peptone concentrations, with and without glucose; D) E. coli minimal medium with varying tryptone concentrations, with and without glucose.
(DOC)