Abstract
Background
Hypermethylation of CpG islands in tumor suppressor gene plays an important role in carcinogenesis. Many studies have demonstrated that hypermethylation in promoter region of RARβ gene could be found with high prevalence in tumor tissue and autologous controls such as corresponding non-tumor lung tissue, sputum and plasma of the NSCLC patients. But with the small number subjects included in the individual studie, the statistical power is limited. Accordingly, we performed this meta-analysis to further asses the relationship of methylation prevalence between the cancer tissue and atuologous controls (corresponding non-tumor lung tissue, sputum and plasma).
Methods
The published articles about RARβ gene promoter hypermethyltion were identified using a systematic search strategy in PubMed, EMBASE and CNKI databases. The pooled odds ratio (OR) of RARβ promoter methylation in lung cancer tissue versus autologous controls were calculated.
Results
Finally, eleven articles, including 1347 tumor tissue samples and 1137 autologous controls were included in this meta-analysis. The pooled odds ratio of RARβ promoter methylation in cancer tissue was 3.60 (95%CI: 2.46–5.27) compared to autologous controls with random-effect model. Strong and significant correlation between tumor tissue and autologous controls of RARβ gene promoter hypermethylation prevalence across studies (Correlation coefficient 0.53) was found.
Conclusion
RARβ promoter methylation may play an important role in carcinogenesis of the NSCLC. With significant methylation prevalence correlation between tumor tissue and autologous of this gene, methylation detection may be a potential method for searching biomarker for NSCLC.
Introduction
Lung cancer, accounting for 100 million deaths and 120 million incidence per year world-wide, is the leading cause of cancer related death[1]. Generally, lung cancer is divided in to non-small cell lung cancer (NSCLC) and small-cell lung cancer according to the pathology. The prognosis of NSCLC, accounting for 80–85% of the lung cancer, is poor for lack of effective early diagnosis methods[2]. Recently, CpG island methylation of tumor suppressor genes promoters have been found to be important for carcinogenesis in many kinds of carcinomas including NSCLC[3]. The DNA methylation is a biochemical process involving the addition of a methyl group to thecytosine or adenine DNA nucleotides. DNA methylation may affect the transcription of the tumor suppressor genes in two ways. Firstly, the methylated DNA may physically impede the binding of transcriptional proteins to the gene itsel[4]. Secondly, may be with more important, methylated DNA may be bound by proteins known as methyl-CpG-binding domain proteins (MBDs)[5]. MBD proteins then recruit additional proteins to the locus, such as histone deacetylases and other chromatin remodeling proteins that can modify histones, thereby forming compact, inactive chromatin, termed heterochromatin[6].
RARβ gene is a typical tumor suppressor gene encoding retinoic acid receptor beta, a member of the thyroid-steroid hormone receptor subfamily, which belongs to nuclear receptor superfamily. The protein encoded by this gene could bind retinoic acid, the biologically active form of vitamin A which mediates cellular signallings in embryonic morphogenesis, cell growth and differentiation[7]. Recent studies demonstrated that hypermethylation in promoter region of RARβ gene could be found with high prevalence in tumor tissue and autologous controls such as corresponding non-tumor lung tissues (CNTLT), sputum and plasma of the NSCLC patients. But with the small number subjects included in the individual studies, the statistical power is limited. Thus, a meta-analysis about the relationship of RARβ promoter methylation between lung cancer tissue and autologous controls was done in order to better identify the correlation of methylation status between cancer tissue and autologous samples.
Methods
Studies identification
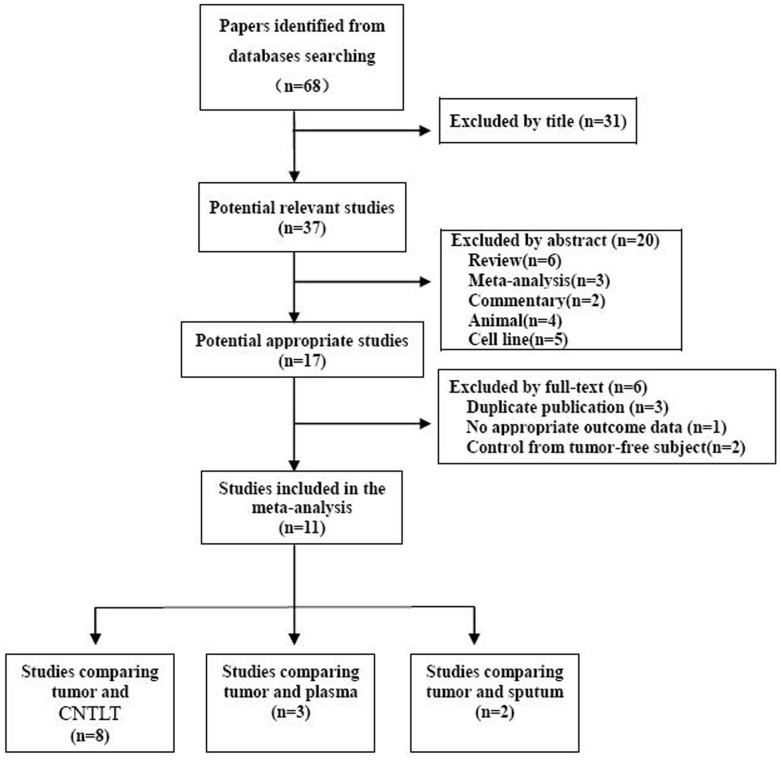
The selection procedure of the relevant articles was demonstrated in figure 1. Papers about RARβ gene promoter CpG island hypermethylation in NSCLC, published before February 2013, were identified by searching Pubmed, EMBASE and CNKI databases. The “Non-small cell lung carcinoma/NSCLC” “methylaiton/hypermethlation” and “RARβ/RAR-beta” were used as the MeSH and free text word when searching the databases. The inclusion criteria: non-small cell lung cancer patients with histology or cytology conformation; The methylation was detected by methylation-specifec PCR (MSP) or quantitative MSP (q-MSP); The methylation rate in the cancer tissue and autologous controls would be drawn in the original studies included in this meta-analysis. All potentially relevant articles were assessed in full-text paper and all references of included articles were scanned for additional analysis according to the Cochrane handbook for systematic review[8].
Figure 1. Flowchart of the literature search strategy (Data from some studies was used more than once).
Data extraction
Information about the general characteristics and methylation prevalence of the original included studies were retrieved. For general characteristics: The first author, year of publication, journals, race of the included subjects, mean or median age of the included patients, methylation detection methods were all recorded. For methylation prevalence in cancer tissue and autologous controls: the RARβ promoter methylation rate or number of methylated (M) and unmethylated (U) samples in cancer tissue and autologous controls were all retrieved from the included studies. The methylation prevalence (MP) was calculated according to the formula PM = M/(M+U)×100%. All of the information details were extracted by two reviewers (FH and NZF) and then checked by the third reviewer (JDG).
Statistical analysis
The odds ratio (OR) for each individual study was calculated by the formula OR = (a/b)/(c/d). (a = number of methylated samples in tumor tissue b = number of unmethylated samples in tumor tissue; c = number of methylated in control tissue and d = number of unmethylated in control tissue.) The OR and its 95% confidence intervals (CI) of RARβ promoter hypermethylation in cancer tissue compared to autologous controls (CNTLT, sputum and plasma) for the included studies was pooled by STATA/SE 11.0(StataCorp LP, http://www.stata.com)statistic software. Statistical heterogeneity among articles included in this meta-analysis was evaluated by I2 [9].Without significant heterogeneity (I2>50%), the fixed-effect method was used to pool the data. Inversely, with significant heterogeneity among studies, random-effect method (Dersimonian-Laird method) was taken to pool the OR. The methylation prevalence correlation between cancer tissue and autologous clinical control (CNTLT, sputum and plasma) was evaluated by Spearman's rank correlation test. And the publication bias was assessed by Begg's funnel plot and Egger's test [10].
Results
General characteristics of included studies
Finally, eleven articles[11]–[21], including 1347 tumor tissue samples and 1137 autologous controls, were included in this meta-analysis.(Figure 1). Of the 11 articles included in this meta-analysis, 6 were conducted in Chinese mainland, 2 were in Taiwan, 2 in USA and 1 in Italy. The median methylation prevalence of RARβ gene promoter was 47.56% and 17.50% in cancer tissue and autologous controls, respectively (table S1). The general characteristics of the included articles were showed in table1.
Table 1. General characteristics of included studies.
| Author | Year | Location | Age(y) | Gender M/F | Sample size (n) | Method | Control type | |
| T | C | |||||||
| Zhao[21] | 2011 | China | 59.4±9.6 | 68/12 | 80 | 80 | MSP | CNTLT |
| Hu[20] | 2011 | China | 59.6±9.3 | 102/18 | 120 | 120 | MSP | CNTLT |
| Hu[20] | 2011 | China | 59.6±9.3 | 102/18 | 120 | 120 | MSP | Plasma |
| Zhang[12] | 2011 | China | 61.0 | 162/38 | 200 | 200 | MSP | CNTLT |
| Song[11] | 2011 | China | 59(35–80) | NA | 78 | 78 | MSP | CNTLT |
| Zhang[19] | 2011 | China | 59(median) | 58/20 | 78 | 110 | MSP | Plasma |
| Feng[13] | 2008 | USA | NA | 24/25 | 49 | 49 | q-MSP | CNTLT |
| Hsu[14] | 2007 | Taiwan | 69(median) | 45/18 | 63 | 63 | MSP | CNTLT |
| Hsu[14] | 2007 | Taiwan | 69(median) | 45/18 | 63 | 63 | MSP | Plasma |
| Hsu[15] | 2007 | Taiwan | NA | NA | 82 | 82 | MSP | Sputum |
| Cirincione[16] | 2006 | Italy | NA | NA | 29 | 18 | Msp | Sputum |
| Yang[18] | 2005 | China | 56±11 | 34/15 | 49 | 49 | MSP | CNTLT |
| Toyooka[17] | 2003 | USA | NA | 355/159 | 514 | 84 | MSP | CNTLT |
M = male; F = female; T = tumor; C = control; CNTLT = Corresponding non-tumor lung tissues; NA = not aviable.
Pooled results of this meta-analysis
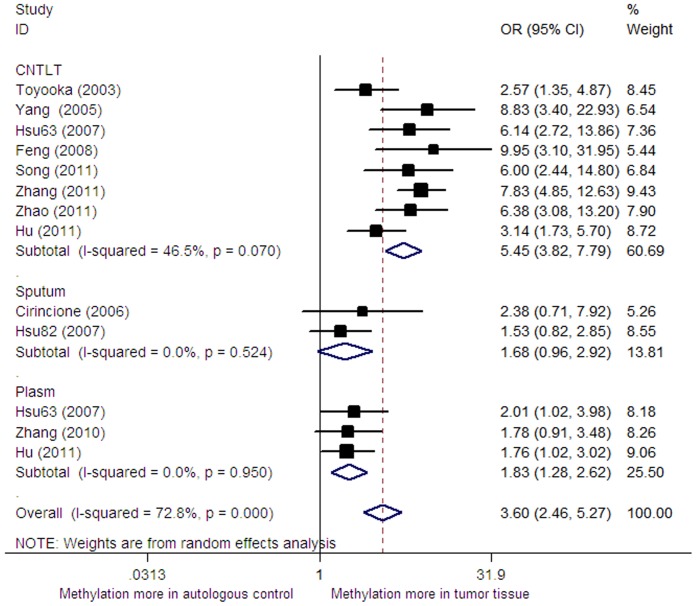
Significant heterogeneity was found across the included studies (I2 = 72.8%). Accordingly, the random-effect model was taken in pooling the OR. In this meta-analysis, the pooled odds ratio of RARβ promoter methylation in cancer tissue was 3.60 (95%CI: 2.46–5.27, p = 0.000) versus autologous controls with random-effect model (Figure 2). The sensitivity analysis was also done by omitting a single study under the random-effect model. But no significant changes of OR and it 95%CI was found in the sensitivity analysis which demonstrated that the pooled odds ratio was not sensitive to a single study.
Figure 2. Forest plot of pooled OR for RARβ gene promoter methylation in cancer tissue versus autologous controls.
Subgroup analysis
With significant heterogeneity across the included studies (I2 = 72.8%), the subgroup analysis of this meta-analysis was performed according to race (East-asia, Caucasian) and control types (corresponding non-tumor lung tissues, sputum and plasma). In the subgroup analysis, the significant odds of the RARβ promoter methylation in tumor tissue was only changed when comparing to sputum (OR = 1.33, 95%CI: 0.98–1.80, P = 0.120) and in subjects with Caucasian ethnicity (OR = 2.38, 95%CI: 0.71–7.92, P = 0.159, Table 2). However, the conclusion of the sputum control and Caucasian ethnicity subgroup should be interpreted with caution as only a two and one articles with small subjects were included in these subgroups analysis.
Table 2. Subgroup analysis.
| Subgroup | NSCLC | Control | OR | 95%CI | p | ||
| M+ | Total | M+ | Total | ||||
| Race | |||||||
| ast-asian | 459 | 946 | 209 | 986 | 3.59 | 2.30–5.61 | 0.000 |
| aucasus | 19 | 29 | 8 | 18 | 2.38 | 0.71–7.92 | |
| Control type | |||||||
| NTLT | 491 | 1160 | 116 | 735 | 5.45 | 3.82–7.79 | 0.000 |
| lasma | 106 | 268 | 78 | 302 | 1.83 | 1.28–2.62 | 0.001 |
| putum | 58 | 110 | 39 | 100 | 1.68 | 0.96–2.92 | 0.067 |
Methylation prevalence correlation
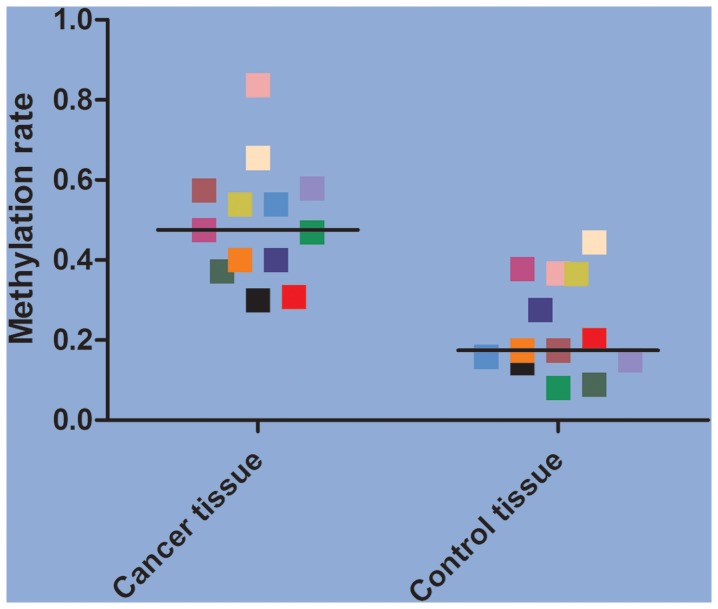
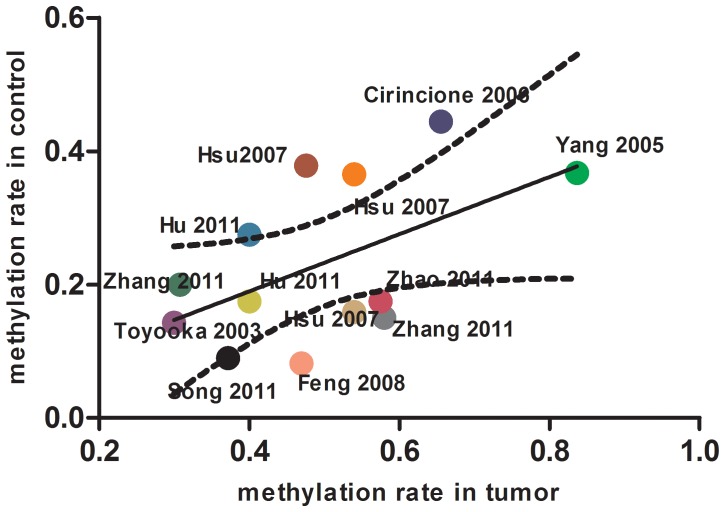
The methylation prevalence was much higher in tumor tissue (meidan methylation prevalence 47.56%) compared to autologous controls (meidan methylation prevalence 17.50%) of RARβ gene (Figure 3). And we also found significant RARβ gene promoter hypermethylation prevalence correlation between tumor tissue and autologous controls among the included studies (Correlation coefficient 0.53) (Figure 4). It means that the more methylation percentage in tumor tissue and the more methylation percentage in autologous controls.
Figure 3. The distribution of methylation rate for each included study (the paired tissue were in the same colour).
Figure 4. Correlation of RARβ gene promoter methylation between tumor tissue and autolougs controls.
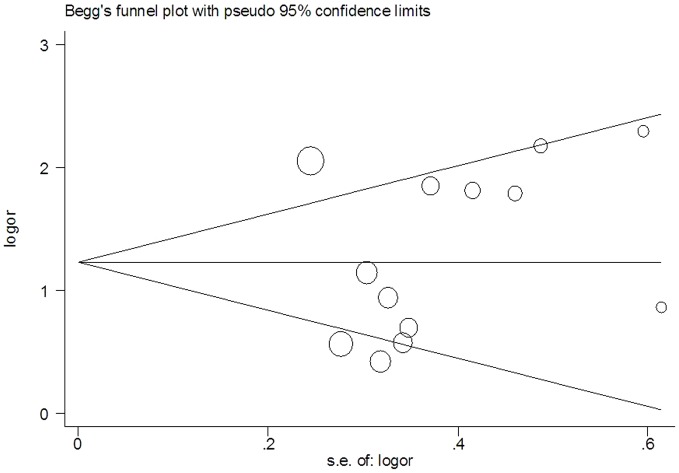
Publication bias
No statistical significant publication bias was detected by Begger's funnel plot and Egger's tests (t = 0.70, p = 0.500) (Figure 5).
Figure 5. Begg's funnel plot for assessment of publication bias.
Discussion
Lung cancer is leading cause of cancer related mortality world-wide with 100 million deaths and 120 million diagnoses in year 2009[22]. Without effective early diagnosis method, the prognosis of NSCLC is relative poor with a 5-year survival rate of less than 15%[2]. CpG island methylation of tumour suppressor genes promoters have been found to be important for carcinogenesis and early events in many kinds of carcinomas including NSCLC. RARβ gene is a typical anti-tumor gene encoding retinoic acid receptor beta, a member of the thyroid-steroid hormone receptor subfamily of nuclear receptor superfamily. The protein encoded by this gene could bind retinoic acid, the biologically active form of vitamin A which mediates cellular signallings in embryonic morphogenesis, cell growth and differentiation. This gene was always been silence by its promoter DNA hypermethylation on its promoter region which can physically impede the binding of transcriptional proteins to the gene itself.
The relationship between cancer tissue and autologous controls such as CNTLT, plasma and sputum for this gene was not clear with small subjects in the individual studies. So, we performed this meta-analysis, which can strength the statistical power to quantify the hypermethylation-disease association by pooling data from open published articles. In this meta-analysis, we finally included eleven articles with 1347 tumor tissue samples and 1137 autologous controls to pool the odds ratio of methylation prevelance. The pooled odds ratio of RARβ promoter methylation in cancer tissue was 3.60 (95%CI: 2.46–5.27) compared to autologous controls with random-effect model. The subgroup analysis of this meta-analysis was performed according to race (East-asian, Caucasian) and control types. In the subgroup analysis, the significant odds of the RARβ promoter methylation in tumor tissue was only changed when comparing to sputum (p = 0.120) and in subjects with Caucasian ethnicity (p = 0.159). However, the conclusion of the sputum control and Caucasian ethnicity subgroup should be interpreted with caution as only a two and one articles with small subjects were included in these subgroup analyses. We also found strong and significant correlation between tumor tissue and autologous controls of RARβ gene promoter hypermethylation prevalence across studies (r = 0.53). The correlation showed that, the higher methylation prevalence in plasma/sputum samples, the higher methylation prevalence could be found in cancer tissue in patients with NSCLC. And this indicated that detection methylation status in plasma or sputum could be a useful assay for diagnosis of NSCLC without or with mirror invasion. Nevertheless, in our meta-analysis, all of samples (tumor tissue, non-tumor lung tissue sputum and plasma) are all come from the patients with confirmed lung cancer. There was no relative healthy person controls. So, the lung cancer screening sensitivity and specificity was not possible to calculate according to the Bayes theorem.
Some limitations of this meta-analysis are need for further consideration. First, significant heterogeneity was found in this meta-analysis (I2 = 72.8%), which could decrease the statistical power of this study. Second, the co-variate methylaiton status of different tumor suppressor genes may also be linked and interact with each other, suggesting methylation analysis of a single gene may be not enough[23].
In conclusion, this meta-analysis showed RARβ promoter methylation was much higher in tumor tissue compared to autologous controls, which indicates promoter hypermethylation of tumor suppressor gene may play an important role in carcinogenesis of the NSCLC. With significant methylation prevalence correlation, methylation detection may be a potential stragegy for searching biomarker for NSCLC [24].
Supporting Information
The methylation percentage in each included study
(DOC)
Prisma checklist.
(DOC)
Funding Statement
This project was supported by the grants from National Natural Science Foundation of China (to XBL) (No. 81302002). Science and technology foundation of Tianjin Municipal Health Bureau (to JDG) (2010KZ040). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. eSantis C, Naishadham D, Jemal A (2013) Cancer statistics for African Americans, 2013. CA Cancer J Clin 63: 151–166. [DOI] [PubMed] [Google Scholar]
- 2. iegel R, Naishadham D, Jemal A (2012) Cancer statistics for Hispanics/Latinos, 2012. CA Cancer J Clin 62: 283–298. [DOI] [PubMed] [Google Scholar]
- 3. Hanabata T, Tsukuda K, Toyooka S, Yano M, Aoe M, et al. (2004) DNA methylation of multiple genes and clinicopathological relationship of non-small cell lung cancers. Oncol Rep 12: 177–180. [PubMed] [Google Scholar]
- 4. Choy MK, Movassagh M, Goh HG, Bennett MR, Down TA (2010) Genome-wide conserved consensus transcription factor binding motifs are hyper-methylated. BMC Genomics 11: 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miller CA, Sweatt JD (2007) Covalent modification of DNA regulates memory formation. Neuron 53: 857–869. [DOI] [PubMed] [Google Scholar]
- 6. El-Osta A (2003) DNMT cooperativity—the developing links between methylation, chromatin structure and cancer. Bioessays 25: 1071–1084. [DOI] [PubMed] [Google Scholar]
- 7. Mattei MG, de The H, Mattei JF, Marchio A, Tiollais P, et al. (1988) Assignment of the human hap retinoic acid receptor RAR beta gene to the p24 band of chromosome 3. Hum Genet 80: 189–190. [DOI] [PubMed] [Google Scholar]
- 8.Higgins JP, Green S (2008) Cochrane Handbook for Systematic Reviews of Interventions updated March 2011 John Wiley & Sons, Available: http://www.cochrane-handbook.org/.Accessed: 2011 DEC 12.
- 9. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ (Clinical research ed.) 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Egger M, Davey SG, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical research ed.) 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song H, Yi J, Zhang Y, Wang R, Chen L (2011) DNA methylation of tumor suppressor genes located on chromosome 3p in non-small cell lung cancer. Chinese journal of lung cancer 14: 233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang CY, Jin YT, Xu HY, Zhang H, Zhang WM, et al. (2011) Relationship between promoter methylation of p16, DAPK and RAR beta genes and the clinical data of non-small cell lung cancer. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 28: 23–28. [DOI] [PubMed] [Google Scholar]
- 13. Feng Q, Hawes SE, Stern JE, Wiens L, et al. (2008) DNA methylation in tumor and matched normal tissues from non-small cell lung cancer patients. Cancer Epidemiol Biomarkers Prev 17: 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hsu HS, Chen TP, Hung CH, Wen CK, Lin RK, et al. (2007) Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer 110: 2019–2026. [DOI] [PubMed] [Google Scholar]
- 15. Hsu HS, Chen TP, Wen CK, Hung CH, Chen CY, et al. (2007) Multiple genetic and epigenetic biomarkers for lung cancer detection in cytologically negative sputum and a nested case-control study for risk assessment. J Pathol 213: 412–419. [DOI] [PubMed] [Google Scholar]
- 16. Cirincione R, Lintas C, Conte D, Mariani L, Roz L, et al. (2006) Methylation profile in tumor and sputum samples of lung cancer patients detected by spiral computed tomography: a nested case-control study. Int J Cancer 118: 1248–1253. [DOI] [PubMed] [Google Scholar]
- 17. Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, et al. (2003) Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer 103: 153–160. [DOI] [PubMed] [Google Scholar]
- 18.Yang ZH, Cai YY, Sun LH (2005).Promoter hypermethylation of 11 tumor-related genes in non-small cel lung cancer. J Clin Inter Med, 22: , 708–10. [Google Scholar]
- 19. Zhang Y, Wang R, Song H, Huang G, Yi J, et al. (2011) Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett 303: 21–28. [DOI] [PubMed] [Google Scholar]
- 20.Hu HL,Wang L,Chen SH (2011) Detection and clinical significance of RAR-beta gene methylation in plasma of no-small cell lung cancer patients. Chin J Clin Oncol, 38: ,890–893. [Google Scholar]
- 21. Zhao XD, Xue SL, Jin YT (2011) The detection and clinical implication of methylation of RAR-beta gene in patients with non-small cell lung cancer. Acta Universitatis Medicinalis Anhui 46: 409–411. [Google Scholar]
- 22. Jemal A, Siegel R, Ward E, Hao Y, Xu J, et al. (2009) Cancer statistics, 2009. CA Cancer J Clin 59: 225–249. [DOI] [PubMed] [Google Scholar]
- 23. Lou J, Xue J, Lin Q (2011) Transcriptome-wide network analysis of squamous lung cancer reveals potential methylation genes. Asian Pac J Cancer Prev 12: 2349–2352. [PubMed] [Google Scholar]
- 24. Gu J, Wen Y, Zhu S, Hua F, Zhao H, et al. (2013) Association between P(16INK4a) promoter methylation and non-small cell lung cancer: a meta-analysis. PLoS One 8: e60107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The methylation percentage in each included study
(DOC)
Prisma checklist.
(DOC)