Abstract
An acute myeloid leukemia was suspected of having a t(8;16)(p11;p13) resulting in a KAT6A-CREBBP fusion because the bone marrow was packed with monoblasts showing marked erythrophagocytosis. The diagnostic karyotype was 46,XY,add(1)(p13),t(8;21)(p11;q22),der(16)t(1;16)(p13;p13)[9]/46,XY[1]; thus, no direct confirmation of the suspicion could be given although both 8p11 and 16p13 seemed to be rearranged. The leukemic cells were examined in two ways to find out whether a cryptic KAT6A-CREBBP was present. The first was the “conventional” approach: G-banding was followed by fluorescence in situ hybridization (FISH) and reverse transcription PCR (RT-PCR). The second was RNA-Seq followed by data analysis using FusionMap and FusionFinder programs with special emphasis on candidates located in the 1p13, 8p11, 16p13, and 21q22 breakpoints. FISH analysis indicated the presence of a KAT6A/CREBBP chimera. RT-PCR followed by Sanger sequencing of the amplified product showed that a chimeric KAT6A-CREBBP transcript was present in the patients bone marrow. Surprisingly, however, KATA6A-CREBBP was not among the 874 and 35 fusion transcripts identified by the FusionMap and FusionFinder programs, respectively, although 11 sequences of the raw RNA-sequencing data were KATA6A-CREBBP fragments. This illustrates that although many fusion transcripts can be found by RNA-Seq combined with FusionMap and FusionFinder, the pathogenetically essential fusion is not always picked up by the bioinformatic algorithms behind these programs. The present study not only illustrates potential pitfalls of current data analysis programs of whole transcriptome sequences which make them less useful as stand-alone techniques, but also that leukemia diagnosis still relies on integration of clinical, hematologic, and genetic disease features of which the former two by no means have become superfluous.
Introduction
The chromosome aberration t(8;16)(p11;p13) was first described in 1983 in an infant in whom the leukemic cells displayed prominent hemophagocytosis [1]. The recurrence of t(8;16)(p11;p13) in acute myeloid leukemia (AML) was independently established in 1987 by three groups. Bernstein et al [2] reported two infants with AML carrying the t(8;16)(p11;p13). Heim at al [3] described three cases of AML, two teenagers and one infant, with the t(8;16) as the sole chromosome abnormality. Lai et al [4] reported three more cases of t(8;16)-positive AML with additional structural chromosome aberrations present in two of them. Monocytic differentiation and phagocytosis were distinctive features of all the patients [2], [3], [4]. In the Mitelman Database of Chromosome Aberration and Gene Fusions in Cancer, there are now 116 cases of AML carrying the t(8;16)(p11;p13) chromosome abnormality (http://cgap.nci.nih.gov/Chromosomes/Mitelman, database last updated on August 14, 2013).
AML with t(8;16)(p11;p13) is now recognized as a distinct disease entity characterized by monocytic differentiation of the leukemic cells and marked erythrophagocytosis [5], [6], frequent skin involvement, and a tendency to develop diffuse intravascular coagulation. It often occurs at a young age and the response to treatment is poor resulting in short survival [5], [7].
The translocation t(8;16)(p11;p13) disrupts KAT6A (also known as MOZ and MYST3) on 8p11 and CREBBP (also named CBP) on 16p13 resulting in a fusion of the two genes [8], [9]. Although genomic rearrangements of KAT6A and CREBBP were repeatedly detected using fluorescence in situ hybridization (FISH) and Southern blot methodologies [8], [9], [10], attempts to amplify and further analyze chimeric KAT6A-CREBBP and CREBBP-KAT6A transcripts using reverse transcriptase-PCR (RT-PCR) analysis were long unsuccessful [8], [9], [10]. Various explanations such as low expression and/or instability of the chimeric transcripts were proposed [9]. Nevertheless, an RT-PCR strategy was developed to detect the KAT6A-CREBBP as well as CREBBP-KAT6A fusions [11] and two types of KAT6A-CREBBP fusion transcripts were identified: Type 1 was an in-frame transcript between codon 1117 of KAT6A and codon 29 of CREBBP, while type 2 was an out-of-frame transcript between exon codon 1117 of KAT6A and codon 267 of CREBBP. Subsequent studies confirmed the presence of KAT6A-CREBBP and CREBBP-KAT6A fusions, and it was shown that type 1 was the most frequent KAT6A-CREBBP fusion transcript [12], [13], [14]. Moreover, new KAT6A-CREBBP fusion transcript variants were described [15], [16] and real time PCR methodology (RT-PCR) was developed to monitor minimal residual disease status throughout the entire course of the treatment [17].
Recently, RNA-sequencing (RNA-seq, also known as whole transcriptome sequencing) was shown to be an efficient tool in the detection of fusion genes in cancer [18] and has created euphoria among those working with cancer fusion genes. The methodology is in principle simple: extracted RNA from cancer cells is massively sequenced, and then the raw data are analyzed with one or more programs specifically dedicated to the task of detecting fusion transcripts such as FusionMap and FusionFinder [19], [20]. However, it suffers from the shortcoming of identifying as “fusion genes” also many technical and perhaps also clinical “false positives,” thus making the assessment of which fusions are important and which are noise extremely difficult. We and others have used combinations of cytogenetics and RNA-seq to detect the “primary” fusion genes of neoplasms carrying only one or a few chromosomal rearrangements. This approach was used to identify the WWTR1-CAMTA1 and YWHAE-FAM22A/B chimeric genes in epithelioid hemangioendothelioma and high-grade endometrial stromal sarcomas, respectively [21], [22], ZC3H7-BCOR in endometrial stromal sarcomas [23], IRF2BP2-CDX1 in a mesenchymal chondrosarcoma [24], and EWSR1-YY1 in a subset of mesotheliomas [25]. In hematologic malignancies, an NFIA-CBFA2T3 (NFIA is located in 1p31) chimeric transcript was found in an acute erythroid leukemia with the translocation t(1;16)(p31;q24) [26], the ZMYND8-RELA fusion was detected in a congenital acute erythroid leukemia carrying a t(11;20)(p11;q13) translocation [27], and a cryptic FUS/ERG fusion gene was found in an acute myeloid leukemia with a rather complex karyotype [28].
We here describe a case of AML in which two translocations corresponding to a der(16)t(1;16)(p13;p13) and a t(8;21)(p11;q22) were found in the bone marrow cells. The patient had morphological features that invoked the suspicion of a t(8;16)(p11;p13) with KAT6A-CREBBP fusion in spite of the fact that the breakpoints suggested various other candidate genes. We therefore studied the patient's leukemic cells in two different ways looking for KAT6A-CREBBP: “conventionally” by karyotyping followed by FISH followed by RT-PCR, and in the modern manner using RNA-seq and programs specific for fusion genes.
Materials and Methods
Ethics Statement
The study was approved by the Regional Committee for Medical Research Ethics (Regional komité for medisinsk forskningsetikk Sør-Øst, Norge, http://helseforskning.etikkom.no). Written informed consent was obtained from the patient prior to his death. The ethics committee approval included a review of the consent procedure and all patient information has been anonymized and de-identified.
Case history
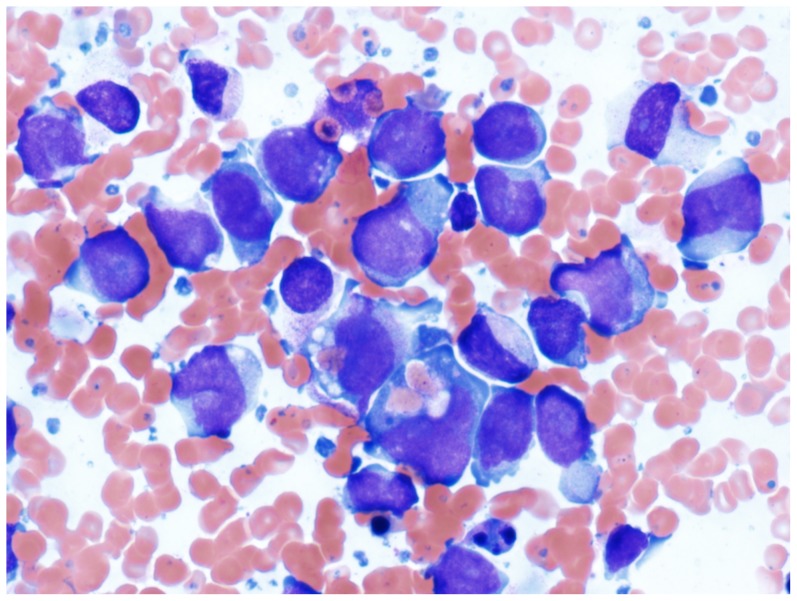
A 30 years old male was transferred to our institution with a preliminary diagnosis of acute myeloid leukemia. He presented with fever and lower back pain radiating to the left lower limb. The clinical examination was unremarkable except for the presence of gingival petecchiae. Gingival hyperplasia was not noted. Blood analysis revealed a severe thrombocytopenia and an elevated CRP and LDH, but all other parameters were normal. Magnetic resonance investigation (MRI) of the columna demonstrated a paramedian prolapse between the L5/S1 vertebrae. A chest radiography showed bilateral infiltrations in the lower pulmonary lobes. Examination of a bone marrow aspirate showed that normal hematopoiesis was replaced by intermediate to large monoblasts, often with prominent vacuolization and ingested red blood cells (Figure 1). Immunophenotypic analysis confirmed the monocytic origin of the blasts that were positive for HLA-DR antigens, CD15, CD13, CD33, and cyMPO. Molecular genetic analysis was negative for RUNX1-RUNX1T1 and CBFB-MYH11 fusion transcripts as well as FLT3 and NPM1 mutations. The patient received induction therapy for AML with daunorubicin 90 mg/m2 day 1–3 and cytarabin 200 mg/m2 day 1–7 after which he went into morphologic remission. He then received two cycles of consolidation with high dose cytarabin (3 g/m2×2 per day, day 1, 3 and 5). One month later, however, he relapsed. He now received reinduction treatment with M5A5E5 (Amsakrin 150 mg/m2 day 1–3, Cytarabin 200 mg/m2/24 hours and Etoposide 110 mg/m2 daily for 5 days). Although morphologic remission was obtained, complete hematological recovery was not achieved 2 months after re-induction treatment.The consolidation treatment M3A5E3 was therefore given with an intent to proceed to allogenic stem cell transplantation, but the patient died 3 weeks later because of liver failure and an acute abdomen caused by ischemic infarcts of the intestine and liver.
Figure 1. Bone marrow smear showing intermediate to large blasts with finely dispersed chromatin with variably abundant cytoplasm, vacuolization and phagocytosis of red blood cells (Wright-Giemsa 400 x).
G-banding and FISH
Bone marrow cells were cytogenetically investigated by standard methods. Chromosome preparations were made from metaphase cells of a 24-hours culture, G-banded using Leishman stain, and karyotyped according to ISCN 2009 guidelines [29]. FISH analysis was performed on metaphase plates.
BAC clones were retrieved from the Human genome high-resolution BAC re-arrayed clone set (the “32k set”; BACPAC Resources, http://bacpac.chori.org/pHumanMinSet.htm). The “32k set” is mapped on the UCSC Genome browser on Human May 2004 (NCBI/hg17) assembly. Mapping data for the 32k human rearray are available in an interactive web format (http://bacpac.chori.org/pHumanMinSet.htm, from the genomic rearrays page) and are obtained by activation of the ucsc browser track for the hg17 UCSC assembly from the “32k set” homepage (http://bacpac.chori.org/genomicRearrays.php). The BAC clones were selected according to physical and genetic mapping data on chromosomes 8 and 16 (see below) as reported on the Human Genome Browser at the University of California, Santa Cruz website (May 2004, http://genome.ucsc.edu/). In addition, FISH mapping of the clones on normal controls was performed to confirm their chromosomal location. The clones used were RP11-619A23 (chr16:3660077-3854572) and RP11-95J11 (chr16:3800375-3965511) mapping to 16p13.3 and which both contain the entire CREBBP gene (red), and RP11-642I24 (chr8:41795493-41975651) and RP11-589C21 (chr8:41992859-42155379) mapping to 8p11.21 for KAT6A (green). DNA was extracted and probes were labelled and hybridized according to Abbott Molecular recommendations (http://www.abbottmolecular.com/home.html). Chromosome preparations were counterstained with 0.2 µg/ml DAPI and overlaid with a 24×50 mm2 coverslip. Fluorescent signals were captured and analyzed using the CytoVision system (Applied Imaging, Newcastle, UK).
RT-PCR analyses
Total RNA was extracted from the patients bone marrow at the time of diagnosis using Trizol reagent according to the manufacturer's instructions (Invitrogen, Life Technologies, Oslo, Norway) and used for both RT-PCR and RNA-Seq analyses. For RT-PCR, one µg of total RNA was reverse-transcribed in a 20 µL reaction volume using iScript Advanced cDNA Synthesis Kit for RT-qPCR according to the manufacturer's instructions (Bio-Rad Laboratories, Oslo, Norway). The cDNA was diluted to 30 ng equivalent of RNA/µL and 2 µL were used as templates in subsequent PCR assays. The 25 µL PCR volume contained 12.5 µL Premix Ex Taq DNA Polymerase Hot Start Version (Takara Bio Europe/SAS, Saint-Germain-en-Laye, France), 2 µL of diluted cDNA, and 0.2 µM of each of the primers, the forward MOZ-3558F (5′-GAG GCC AAT GCC AAG ATT AGA AC-3′) and the reverse primer CBP-431R (5′-GTT GAT ACT AGA GCC GCT GCC TC-3′). The PCR was run on a C-1000 Thermal cycler (Bio-Rad Laboratories) with an initial denaturation at 94°C for 30 sec, followed by 35 cycles of 7 sec at 98°C, 30 sec at 55°C and 1 min at 72°C, and a final extension for 5 min at 72°C. Four µL of the PCR products were stained with GelRed (Biotium, VWR International, Oslo, Norway), analyzed by electrophoresis through 1.0% agarose gel, and photographed. The remaining PCR products were purified using the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel, VWR International, Oslo, Norway) and sequenced at GATC Biotech (Germany, http://www.gatc-biotech.com/en/home.html). The BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for computer analysis of sequence data.
RNA-sequencing (RNA-Seq)
Three µg of the total RNA extracted from the patients bone marrow at the time of diagnosis and used for RT-PCR analysis were sent for high-throughput paired-end RNA-sequencing at the Genomics Core Facility, The Norwegian Radium Hospital (http://genomics.no/oslo/). The Illumina software pipeline was used to process image data into raw sequencing data and only sequence reads marked as “passed filtering” were used in the downstream data analysis. A total of 53 million reads were obtained. The FASTQC software was used for quality control of the raw sequence data (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Two softwares were used for the discovery of fusion transcripts: FusionMap [30] (release date 2012-04-16) together with the pre-built Human B37 and RefGene from the FusionMap website (http://www.omicsoft.com/fusionmap/) and FusionFinder [31]. In addition, the “grep” command (http://en.wikipedia.org/wiki/Grep) was used to search the fastq files of the sequence data (http://en.wikipedia.org/wiki/FASTQ_format).
Results
G-banding and FISH
G-banding analysis yielded the diagnostic karyotype
46,XY,add(1)(p13),t(8;21)(p11;q22),der(16)t(1;16)(p13;p13)[9]/46,XY[1] (Figure 2A). Four months after diagnosis the bone marrow karyotype was: 47,Y,t(X;17)(p10;q10),der(1)(1qter->1q12::1p22->1q12::5q15->5qter),add(1)(p13),inv(2)(p21q13),der(5)t(1;5)(p22;q15), t(12;18)(q15;q21),+8,t(8;21)(p11;q22),del(13)(q21q31),der(16)t(1;16)(p13;p13)[5]/46,XY[11]. In addition to the primary t(1;16) and t(8;21) translocations, clearly many secondary aberrations had also accrued indicating clonal evolution.
Figure 2. Cytogenetic, FISH and RT-PCR analyses.
A) Karyotype at diagnosis showing the chromosome aberrations add(1)(p13), t(8;21)(p11;q22), and der(16)t(1;16)(p13;p13); breakpoint positions are indicated by arrows. B) Co-hybridization FISH analysis with the probes RP11-619A23/RP11-95J11 (red, for CREBBP) and RP11-642I24/RP11-589C21 (green, for KAT6A). A fusion signal of the KAT6A and CREBBP BACs is detected on the derivative chromosome 8, indicating the presence of a KAT6A/CREBBP chimera. C) Amplification of a 352 bp cDNA fragment using the primers MOZ-3558F and CBP-431R (lane 1); M, 1 Kb DNA ladder (GeneRuler, Fermentas); Lane 2, Blank, no RNA in cDNA synthesis. D) Partial sequence chromatogram of the 352 bp cDNA fragment showing that exon 16 of KAT6A is fused to exon 2 of CREBBP.
Co-hybridization FISH analysis with the probes RP11-619A23/RP11-95J11 (red, for CREBBP) and RP11-642I24/RP11-589C21 (green, for KAT6A) revealed a fusion signal of the KAT6A and CREBBP BACs on the derivative chromosome 8, indicating the presence of a KAT6A/CREBBP chimera (Figure 2B). No corresponding fusion signal was found on the der(16)t(1;16) suggesting that the translocation was accompanied by a deletion of the reciprocal CREBBP/KAT6A (Figure 2B).
RT-PCR
PCR with the MOZ3558F and CBP431R primer combination amplified a 352 bp fragment from the patients cDNA (Figure 2C). To verify the presence of a KAT6A-CREBBP chimeric transcript, the fragment was analyzed by direct sequencing which showed the presence in the patients bone marrow of a type 1 KAT6A-CREBBP chimeric transcript, i.e., the nt 3764 of mRNA of KAT6A (accession number NM_006766.3) was fused in-frame with the nt 290 of mRNA of CREBBP (accession number NM_004380.2) (Figure 2D).
RNA-Seq
Using FusionMap on the raw sequencing data obtained from the Genomics Core Facility, 874 fusion transcripts were found (Table S1). The KAT6A-CREBBP fusion transcript was not among them (Table S1). Instead, three other KAT6A (referred to as MYST3 in the FusionMap output) fusions were found: DTX3L-MYST3 ranking 91, MYST3-SLK ranking 193, and MYST-DNAJC14 ranking 606. Based on the map information on the genes, these fusions would have corresponded to the translocations t(3;8)(q21;p11), t(8;10)(p11;q24), and t(8;12)(p11;q13), respectively, none of which was seen by karyotyping. One CREBP fusion transcript was found, CREBBP-TTC28 which was ranked 401 in the list of fusion transcripts (Table S1). The transcript would have corresponded to a t(16;22)(p13;q12) which was not found by G-banding. The FusionFinder program detected 35 fusion transcripts none of which was KAT6A-CREBBP (Table S2).
Sequences which contained the first 20 nt of exon 2 of CREBBP (ATTTTGGATCATTGTTTGAC; nt 290–319 in sequence with accession number NM_004380.2) were retrieved from the raw sequencing data using the “grep” command. A total of 26 sequences were found (Table 1): 11 of them were KAT6A-CREBBP fusions, 15 sequences were exons 1–2 of CREBBP, and one was a genomic sequence from the chromosome band 16p13 containing the exon 2 of CREBBP. Similar to the results obtained by RT-PCR/Sanger sequencing, all 11 retrieved sequences showed fusion of 3764 nt of mRNA from KAT6A (accession number NM_006766.3) with nt 290 of mRNA from CREBBP (accession number NM_004380.2) (Table 1).
Table 1. The 26 retrieved sequences from the raw sequencing data which contained the first 20ATTTTGGATCATTGTTTGAC (in bold) of CREBBP.
| RETRIEVED SEQUENCES | KAT6A (NM_006766.3) | CREBBP (NM_004380.2) |
| CCCAAAAGAGCCAAACTCAGCTCGCCCGGTTTCTCGGCGAATGACAGCACAGATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGAT | 238–338 (exon 1–2) | |
| CGGCGAATGACAGCACAGATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAATGGAGGAGAATTAGGCCTTTTAAACAGT | 272–372 (exon 1–2) | |
| AAGAAGAAGATGAAGAGTCAGATGATGCTGATG ATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAATGGAGGAGAATTA | 3732–3764 (exon 16) | 290–357 (exon 2) |
| ACTTGCTGGACGGACCGCCCAACCCCAAAAGAGCCAAACTCAGCTCGCCCGGTTTCTCGGCGAATGACAGCACAGATTTTGGATCATTGTTTGACTTGGAA | 215–315 (exon 1–2) | |
| ATGCTGATG ATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAATGGAGGAGAATTAAGATCGGAAGAGCGTCGTGTAGGG | 3756–3764 (exon 16) | 290–357 (exon 2) |
| GTCAGATGATGCTGATG ATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAATGGAGGAGAATTAGGCCTTTTAAACAGTG | 3748–3764 (exon 16) | 290–373 (exon 2) |
| AAGAGTCAGATGATGCTGATG ATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAATGGAGGAGAATTAGGCCTTTTAAAC | 3744–3764 (exon 16) | 290–369 (exon 2) |
| AGAGTCAGATGATGCTGATG ATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAATGGAGGAGAATTAGGCCTTTTAAACA | 3745–3764 (exon 16) | 290–370 (exon 2) |
| AGAACTTGCTGGACGGACCGCCCAACCCCAAAAGAGCCAAACTCAGCTCGCCCGGTTTCTCGGCGAATGACAGCACAGATTTTGGATCATTGTTTGACTTG | 212–312 (exon 1–2) | |
| GACAGCACAGATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAATGGAGGAGAATTAGGCCTTTTAAACAGTGGGAACCT | 280–380 (exon 1–2) | |
| AACTCAGCTCGCCCGGTTTCTCGGCGAATGACAGCACAGATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAATGTAGGA | 251–351 (exon 1–2) | |
| CTGGACGGACCGCCCAACCCCAAAAGAGCCAAACTCAGCTCGCCCGGTTTCTCGGCGAATGACAGCACAGATTTTGGATCATTGTTTGACTTGGAAAATGA | 220–320 (exon 1–2) | |
| AGCCAAACTCAGCTCGCCCGGTTTCTCGGCGAATGACAGCACAGATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCAGATC | 246–341 (exon 1–2) | |
| GAAGATGAAGAGTCAGATGATGCTGATG ATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAATGGAGGGCAGATCGGAAG | 3737–3764 (exon 16) | 290–350 (exon 2) |
| GCCAAACTCAGCTCGCCCGGTTTCTCGGCGAATGACAGCACAGATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAATGG | 247–347 (exon 1–2) | |
| CTCAGGTGTCAGTCCTCTTCTAAGAGGAAGTCTAAAGATGAAGAAGAAGATGAAGAGTCAGATGATGCTGATG ATTTTGGATCATTGTTTGACTTGGAAAA | 3692–3764 (exon 16) | 290–317 (exon 2) |
| CTTGCTGGACGGACCGCCCAACCCCAAAAGAGCCAAACTCAGCTCGCCCGGTTTCTCGGCGAATGACAGCACAGATTTTGGATCATTGTTTGACTTGGAAA | 216–316 (exon 1–2) | |
| AAAGAGCCAAACTCAGCTCGCCCGGTTTCTCGGCGAATGACAGCACAGATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCC | 242–342 (exon 1–2) | |
| CAGTCCTCTTCTAAGAGGAAGTCTAAAGATGAAGAAGAAGATGAAGAGTCAGATGATGCTGATG ATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCC | 3701–3764 (exon 16) | 290–326 (exon 2) |
| GCCAAACTCAGCTCGCCCGGTTTCTCGGCGAATGACAGCACAGATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAATGG | 247–347 (exon 1–2) | |
| AAAAGAGCCAAACTCAGCTCGCCCGGTTTCTCGGCGAATGACAGCACAGATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACC | 241–341 (exon 1–2) | |
| GCCAAACTCAGCTCGCCCGGTTTCTCGGCGAATGACAGCACAGATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAGATC | 247–343 (exon 1–2) | |
| GATGTACTCAGGTGTCAGTCCTCTTCTAAGAGGAAGTCTAAAGATGAAGAAGAAGATGAAGAGTCAGATGATGCTGATG ATTTTGGATCATTGTTTGACTT | 3686–3764 (exon 16) | 290–311 (exon 2) |
| ATGATGCTGATG ATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAATGGAGGAGAATTAGGAGATCGGAAGAGCGTCGTG | 3753–3764 (exon 16) | 290–359 (exon 2) |
| GTAAAGGTTGCTTAGTTTCTCATTTCCATTTCTGTTTAATTTCTAGATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAA | 290–344 (exon 2) | |
| GATGATGCTGATG ATTTTGGATCATTGTTTGACTTGGAAAATGATCTTCCTGATGAGCTGATACCCAATGGAGGAGAATTAGGCCTTTTAAACAGTGGGAA | 3752–3764 (exon 16) | 290–377 (exon 2) |
KAT6A sequences are in italics.
Discussion
The present case of AML had hematologic features highly suggestive of AML with t(8;16)(p11;p13) and a KAT6A-CREBBP fusion gene. The leukemic karyotype at diagnosis had two chromosome translocations, a der(16)t(1;16)(p13;p13) and a t(8;21)(p11;q22), indicating the possible generation of KAT6A-CREBBP via a cryptic aberration since both chromosome bands 8p11, which contains the KAT6A gene, and 16p13, where CREBBP maps, were seen rearranged. However, all the chromosome breakpoints in this particular karyotype contain also other genes known to be involved in leukemogenesis. RBM15 in 1p13 is fused to MKL1 in AML with t(1;22)(p13;q13) [32], [33]. Likewise, chromosome band 16p13 contains, apart from CREBBP, also the CBFB gene which is fused to MYH11 in the subset of AML with inv(16)(p13q22) [34] as well as GLIS2 which is a partner in the fusion CBFA2T3-GLIS2 generated by inv(16)(p13q24) [35]. On 8p11, apart from KAT6A, FGFR1 is rearranged in fusions with several partner genes in the “8p11 myeloproliferative syndrome” [36], [37]. For example, the fusion genes ZNF198-FGFR1, CEP110-FGFR1, FOP-FGFR1, and BCR-FGFR1 result from the t(8;13)(p11;q12), t(8;9)(p11;q33), t(6;8)(q27;p11), and t(8;22)(p11q22) chromosome translocations, respectively [36], [37]. On 21q22, RUNX1 and ERG are fused to RUNX1T1 and FUS generating the RUNX1-RUNX1T1 and FUS-ERG fusion genes in AMLs carrying t(8;21)(p11;q22) and t(16;21)(p11;q22), respectively. In addition, both t(1;16)(p13;p13) and t(8;21)(p11;q22) could conceivably have generated novel leukemogenic fusion genes. Screening with FISH for all possibly rearranged genes associated with the present abnormal karyotype would have been a very laborious and time-consuming procedure. We therefore decided to perform two parallel investigations on the patients bone marrow. Based on the clinical and hematologic hunch that a KAT6A-CREBBP fusion was likely, we took the “conventional” approach - karyotyping and FISH followed by RT-PCR - to search for this known leukemogenic gene. In addition, we also performed RNA-Seq to search for this and other possible fusions concentrating exclusively on those fusion transcripts that had something to do with the chromosomal breakpoints. The FISH analysis showed a fusion signal for KAT6A- and CREBBP- specific probes on the derivative chromosome 8, indicating the presence here of a KAT6A/CREBBP chimera (Figure 2B). RT-PCR analysis followed by Sanger sequencing confirmed the presence of type 1 fusion KAT6A/CREBBP transcript [15] (Figures 2C and 2D). The transcript retains the part of the KAT6A gene encoding the C4HC3 and C2HC zinc fingers, two nuclear localization signals, the HAT domain, the MYST domain, and a portion of the acidic domain, whereas the retained part of CREBBP encodes a domain which binds to nuclear receptor RARA, the CREB-binding domain, the three cystein/histidine rich regions, the bromodomain, and the glutamine-rich domains [11].
Surprisingly, of the 874 fusion transcripts identified by the FusionMap program [30], none was the biologically important KAT6A-CREBBP, nor was KAT6A-CREBBP featured in the list of 35 fusion genes obtained using FusionFinder [31]. Moreover, none of the two programs offered other putative fusion genes generated by the translocations der(16)t(1;16)(p13;p13) and t(8;21)(p11;q22). To find out whether the raw sequencing data contained sequences which encompassed the junction between KAT6A and CREBBP, we retrieved sequences containing the first 20 nt of exon 2 of CREBBP from the raw sequencing data. The rationale behind this was that exon 2 is fused to KAT6A (Figures 2C and 2D). Thus, the retrieved sequences should contain both KAT6A-CREBBP fusion transcript and wild type KAT6A-CREBBP transcript. Indeed, among the altogether 26 retrieved sequences, 11 were KAT6A-CREBBP fusions whereas 15 sequences were exons 1-2 of CREBBP (Table 1).
Both FusionMap and FusionFinder are among the most commonly used programs to detect fusion genes from RNA-Seq [19] and their sensitivity and specificity have been evaluated [19], [30], [31], [38]. We, too, have in previous studies analyzed RNA-Seq data using the FusionMap program finding that it identified fusion genes that corresponded well with the available cytogenetic information and that were biologically significant [23], [24], [25], [26], [27], [28]. The present study, on the other hand, shows that although the same approach detected hundreds (FusionMap) or tens (FusionFinder) of fusion genes, the programs failed to detect the biologically important KAT6A-CREBBP fusion gene although it was manually retrievable from the raw sequencing data.
The case illustrates that RNA-Seq with use of the FusionMap or FusionFinder programs may not be reliable as a stand-alone technique in the investigation of, at least, leukemias. Not only are there far too many false positives offered as fusion genes by this approach, but it may also fail to detect the truly important fusion gene; in the present case neither specificity nor sensitivity was satisfactory. Additional information about clinical, morphological, and cytogenetic features should be taken into account when searching for the crucial fusion genes in hematologic malignancies.
Supporting Information
Identified fusion genes using FusionMap on the raw sequencing data.
(XLSX)
Identified fusion genes using FusionFinder.
(XLSX)
Acknowledgments
The authors thank Jim Thorsen for help with bioinformatics and Anne Mette Eibak and Hege Brandt Gehrken for their technical help.
Funding Statement
This work was supported by grants from the Norwegian Cancer Society. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Schouten TJ, Hustinx TW, Scheres JM, Holland R, de Vaan GA (1983) Malignant histiocytosis. Clinical and cytogenetic studies in a newborn and a child. Cancer 52: 1229–1236. [DOI] [PubMed] [Google Scholar]
- 2. Bernstein R, Pinto MR, Spector I, Macdougall LG (1987) A unique 8;16 translocation in two infants with poorly differentiated monoblastic leukemia. Cancer Genet Cytogenet 24: 213–220. [DOI] [PubMed] [Google Scholar]
- 3. Heim S, Avanzi GC, Billstrom R, Kristoffersson U, Mandahl N, et al. (1987) A new specific chromosomal rearrangement, t(8;16) (p11;p13), in acute monocytic leukaemia. Br J Haematol 66: 323–326. [DOI] [PubMed] [Google Scholar]
- 4. Lai JL, Zandecki M, Jouet JP, Savary JB, Lambiliotte A, et al. (1987) Three cases of translocation (8;16)(p11;p13) observed in acute myelomonocytic leukemia: a new specific subgroup? Cancer Genet Cytogenet 27: 101–109. [DOI] [PubMed] [Google Scholar]
- 5. Haferlach T, Kohlmann A, Klein HU, Ruckert C, Dugas M, et al. (2009) AML with translocation t(8;16)(p11;p13) demonstrates unique cytomorphological, cytogenetic, molecular and prognostic features. Leukemia 23: 934–943. [DOI] [PubMed] [Google Scholar]
- 6. Hanslip JI, Swansbury GJ, Pinkerton R, Catovsky D (1992) The translocation t(8;16)(p11;p13) defines an AML subtype with distinct cytology and clinical features. Leuk Lymphoma 6: 479–486. [Google Scholar]
- 7. Brown T, Swansbury J, Taj MM (2012) Prognosis of patients with t(8;16)(p11;p13) acute myeloid leukemia. Leuk Lymphoma 53: 338–341. [DOI] [PubMed] [Google Scholar]
- 8. Borrow J, Stanton VP Jr, Andresen JM, Becher R, Behm FG, et al. (1996) The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet 14: 33–41. [DOI] [PubMed] [Google Scholar]
- 9. Giles RH, Dauwerse JG, Higgins C, Petrij F, Wessels JW, et al. (1997) Detection of CBP rearrangements in acute myelogenous leukemia with t(8;16). Leukemia 11: 2087–2096. [DOI] [PubMed] [Google Scholar]
- 10. Chaffanet M, Mozziconacci MJ, Fernandez F, Sainty D, Lafage-Pochitaloff M, et al. (1999) A case of inv(8)(p11q24) associated with acute myeloid leukemia involves the MOZ and CBP genes in a masked t(8;16). Genes Chromosomes Cancer 26: 161–165. [PubMed] [Google Scholar]
- 11. Panagopoulos I, Isaksson M, Lindvall C, Bjorkholm M, Ahlgren T, et al. (2000) RT-PCR analysis of the MOZ-CBP and CBP-MOZ chimeric transcripts in acute myeloid leukemias with t(8;16)(p11;p13). Genes Chromosomes Cancer 28: 415–424. [DOI] [PubMed] [Google Scholar]
- 12. Rozman M, Camos M, Colomer D, Villamor N, Esteve J, et al. (2004) Type I MOZ/CBP (MYST3/CREBBP) is the most common chimeric transcript in acute myeloid leukemia with t(8;16)(p11;p13) translocation. Genes Chromosomes Cancer 40: 140–145. [DOI] [PubMed] [Google Scholar]
- 13. Schmidt HH, Strehl S, Thaler D, Strunk D, Sill H, et al. (2004) RT-PCR and FISH analysis of acute myeloid leukemia with t(8;16)(p11;p13) and chimeric MOZ and CBP transcripts: breakpoint cluster region and clinical implications. Leukemia 18: 1115–1121. [DOI] [PubMed] [Google Scholar]
- 14. Gervais C, Murati A, Helias C, Struski S, Eischen A, et al. (2008) Acute myeloid leukaemia with 8p11 (MYST3) rearrangement: an integrated cytologic, cytogenetic and molecular study by the groupe francophone de cytogenetique hematologique. Leukemia 22: 1567–1575. [DOI] [PubMed] [Google Scholar]
- 15. Panagopoulos I, Fioretos T, Isaksson M, Mitelman F, Johansson B, et al. (2002) RT-PCR analysis of acute myeloid leukemia with t(8;16)(p11;p13): identification of a novel MOZ/CBP transcript and absence of CBP/MOZ expression. Genes Chromosomes Cancer 35: 372–374. [DOI] [PubMed] [Google Scholar]
- 16. Terui K, Sato T, Sasaki S, Kudo K, Kamio T, et al. (2008) Two novel variants of MOZ-CBP fusion transcripts in spontaneously remitted infant leukemia with t(1;16;8)(p13;p13;p11), a new variant of t(8;16)(p11;p13). Haematologica 93: 1591–1593. [DOI] [PubMed] [Google Scholar]
- 17. Fujiki A, Imamura T, Furutani A, Hatano W, Asai D, et al. (2012) Quantitative RT-PCR analysis of the MOZ-CBP fusion transcript in therapy-related acute myeloid leukemia with t(8;16)(p11;p13). J Pediatr Hematol Oncol 34: 402–405. [DOI] [PubMed] [Google Scholar]
- 18. Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, et al. (2009) Transcriptome sequencing to detect gene fusions in cancer. Nature 458: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carrara M, Beccuti M, Lazzarato F, Cavallo F, Cordero F, et al. (2013) State-of-the-art fusion-finder algorithms sensitivity and specificity. Biomed Res Int 2013: 340620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Q, Xia J, Jia P, Pao W, Zhao Z (2012) Application of next generation sequencing to human gene fusion detection: computational tools, features and perspectives. Brief Bioinform 14(4): 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, et al. (2011) Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med 3: 98ra82. [DOI] [PubMed] [Google Scholar]
- 22. Lee CH, Ou WB, Marino-Enriquez A, Zhu M, Mayeda M, et al. (2012) 14-3-3 fusion oncogenes in high-grade endometrial stromal sarcoma. Proc Natl Acad Sci U S A 109: 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Panagopoulos I, Thorsen J, Gorunova L, Haugom L, Bjerkehagen B, et al. (2013) Fusion of the ZC3H7B and BCOR genes in endometrial stromal sarcomas carrying an X;22-translocation. Genes Chromosomes Cancer 52: 610–618. [DOI] [PubMed] [Google Scholar]
- 24. Nyquist KB, Panagopoulos I, Thorsen J, Haugom L, Gorunova L, et al. (2012) Whole-Transcriptome Sequencing Identifies Novel IRF2BP2-CDX1 Fusion Gene Brought about by Translocation t(1;5)(q42;q32) in Mesenchymal Chondrosarcoma. PLoS One 7: e49705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Panagopoulos I, Thorsen J, Gorunova L, Micci F, Haugom L, et al. (2013) RNA sequencing identifies fusion of the EWSR1 and YY1 genes in mesothelioma with t(14;22)(q32;q12). Genes Chromosomes Cancer 52: 733–740. [DOI] [PubMed] [Google Scholar]
- 26. Micci F, Thorsen J, Panagopoulos I, Nyquist KB, Zeller B, et al. (2013) High-throughput sequencing identifies an NFIA/CBFA2T3 fusion gene in acute erythroid leukemia with t(1;16)(p31;q24). Leukemia 27: 980–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Panagopoulos I, Micci F, Thorsen J, Haugom L, Buechner J, et al. (2013) Fusion of ZMYND8 and RELA genes in acute erythroid leukemia. PLoS One 8: e63663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Panagopoulos I, Gorunova L, Zeller B, Tierens A, Heim S (2013) Cryptic FUS-ERG fusion identified by RNA-sequencing in childhood acute myeloid leukemia. Oncol Rep 30: 2587–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaffer LG, Slovak ML, Campbell LJ (2009) ISCN 2009: an International System for Human Cytogenetic Nomenclature. Basel: Karger.
- 30. Ge H, Liu K, Juan T, Fang F, Newman M, et al. (2011) FusionMap: detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics 27: 1922–1928. [DOI] [PubMed] [Google Scholar]
- 31. Francis RW, Thompson-Wicking K, Carter KW, Anderson D, Kees UR, et al. (2012) FusionFinder: a software tool to identify expressed gene fusion candidates from RNA-Seq data. PLoS One 7: e39987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mercher T, Coniat MB, Monni R, Mauchauffe M, Nguyen Khac F, et al. (2001) Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc Natl Acad Sci U S A 98: 5776–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ma Z, Morris SW, Valentine V, Li M, Herbrick JA, et al. (2001) Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet 28: 220–221. [DOI] [PubMed] [Google Scholar]
- 34. Liu P, Tarle SA, Hajra A, Claxton DF, Marlton P, et al. (1993) Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science 261: 1041–1044. [DOI] [PubMed] [Google Scholar]
- 35. Gruber TA, Larson Gedman A, Zhang J, Koss CS, Marada S, et al. (2012) An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell 22: 683–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jackson CC, Medeiros LJ, Miranda RN (2010) 8p11 myeloproliferative syndrome: a review. Hum Pathol 41: 461–476. [DOI] [PubMed] [Google Scholar]
- 37. Macdonald D, Reiter A, Cross NC (2002) The 8p11 myeloproliferative syndrome: a distinct clinical entity caused by constitutive activation of FGFR1. Acta Haematol 107: 101–107. [DOI] [PubMed] [Google Scholar]
- 38. Carrara M, Beccuti M, Cavallo F, Donatelli S, Lazzarato F, et al. (2013) State of art fusion-finder algorithms are suitable to detect transcription-induced chimeras in normal tissues? BMC Bioinformatics 14 Suppl 7 S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identified fusion genes using FusionMap on the raw sequencing data.
(XLSX)
Identified fusion genes using FusionFinder.
(XLSX)