Abstract
We recently reported that hairpin (or stem-loop) priming is better-suited than polyA tailing to generate cDNA for plant microRNA qPCR. One major limitation of this method is the need to perform individual cDNA synthesis reactions for the reference gene and test miRNAs. Here, we report a novel fusion primer that allows multiplexed hairpin cDNA synthesis of the most-commonly used reference gene, nucleolar small RNA U6, together with test miRNAs. We also propose the use of miR1515 as a house keeping control for tropical legumes. We show that multiplexed cDNA synthesis does not result in loss of sensitivity and reduces the amount of RNA required for miRNA gene expression assays.
Keywords: hairpin cDNA qPCR, U6, miR1515, multiplexing, miRNA
microRNAs (miRNAs) are short non-coding RNAs that regulate gene expression at the post-transcriptional level in both plants and animals (reviewed in refs. 1–4). In plants, their biogenesis starts with pri-miRNA transcripts which are capped and generally poly-adenylated and then cleaved by DICER-LIKE 1 to give rise to short duplex RNA molecules consisting of the mature miRNA (typically 21–22 nt in length) and miRNA* molecules. Both miRNA and miRNA* are then 2′ O-methylated at the 3′ end. Mature miRNAs are incorporated into RNA-induced silencing complexes and direct inhibition of translation or cleavage of complementary mRNAs (reviewed in refs. 5–7). Reliable quantification of individual mature miRNA levels is necessary to understand their biological roles. Hairpin/stem-loop priming8 and PolyA tailing9 are the two most commonly used cDNA synthesis methods for plant miRNA qPCR. We recently showed that hairpin priming method is better suited than polyA tailing method to synthesize cDNA for plant miRNA qPCR.10 One disadvantage of hairpin priming method is the use of specific RT primers for the cDNA synthesis and therefore individual cDNA synthesis reactions for each miRNA. This not only requires more input RNA, but also results in possible differences in cDNA synthesis efficiencies between test miRNAs and reference genes. It is crucial that the reference genes and test miRNAs undergo identical reaction conditions during cDNA synthesis and qPCR. This ensures equal efficiency of the reaction between the reference gene and the test miRNA(s) leading to accurate and reliable measurements.
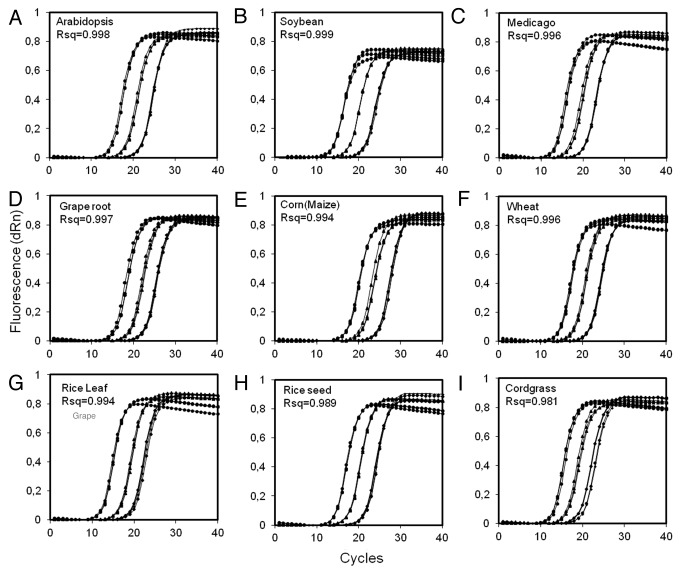
In general, other small endogenous noncoding RNAs such as U6, U24 or U26 are used to normalize the expression of both plant and animal miRNAs.11,12 In Northern hybridization and polyA technique, the small nucleolar RNA U6 has been successfully used as a normalization control for a wide range of experiments (e.g., see refs. 13 and 14). However, no suitable method exists for U6 cDNA synthesis coupled with hairpin priming. To resolve this issue, we designed cDNA synthesis and qPCR primers for U6 from a highly conserved region of the gene (Fig. S1A) For U6 cDNA synthesis primer design, we added the “universal reverse” primer used in hairpin cDNA qPCR to the 5′ end of U6-specific reverse primer sequence (Fig. S1B). When this modified U6 reverse primer is used for cDNA synthesis, U6 expression can thereafter be assayed using hairpin cDNA qPCR technique, using U6-specific forward primer and universal reverse primer. “Hairpin-primed” cDNAs were generated using U6-specific cDNA synthesis primer from RNA samples obtained from different plant species. RNA preparations were obtained from rice (leaf and seed), wheat (leaf), Arabidopsis (seedlings), medicago (leaf), corn (leaf meristematic tissue), grape (root), soybean (root) and prairie cord grass (rhizomes). All cDNA synthesis reactions were performed on an ABI thermocycler (GeneAmp 9700). All qPCR assays were performed using a MX3000P thermocycler (Stratagene/Agilent Technologies) and SYBR Advantage qPCR premix (Product# 639676, Clontech). Three different dilutions (1/50, 1/500, 1/5,000) of cDNA from each plant species were examined for U6 expression by qPCR (Fig. 1). Suitability of these primers to synthesize cDNA and assay U6 expression were determined by examining the linearity of amplification, quality of the amplification curves and dissociation curves. We tested a range of monocotyledonous and dicotyledonous (both legumes and non-legumes) species. The primers we designed were able to successfully detect U6 expression with reliable amplification efficiencies and linearity in different plant species (Fig. 1).
Figure 1. Validation of the novel U6 fusion primer for cDNA synthesis on multiple plant species; (A) Arabidopsis, (B) soybean, (C) medicago, (D) grape root, (E) corn, (F) wheat, (G) rice leaf, (H) rice seed and (I) prairie cordgrass. Amplification plots of U6 in different plant species were obtained using primers designed in this study to adapt the use of U6 as normalization control in hairpin cDNA qPCR assays for miRNAs. Linearity (Rsq) values obtained by examining expression in different dilutions (circle: 1/50, triangle: 1/500 and diamond: 1/5,000) of cDNA are indicated in each panel.
One of the legume-specific miRNAs, miR1515 is uniformly expressed in a variety of tissues in soybean15 as well as other tropical legumes such as common bean and peanut.16 To utilize miR1515 as a reference gene for miRNA qPCR in soybean, we designed hairpin primers for this miRNA. Expression of miR1515 was assayed in different soybean tissues and in the roots during nodule development (Fig. 2). For expression analysis in different soybean organs, samples were collected from root, stem and young leaves of 12-d-old plants, 14 d post inoculation (dpi) mature nodules, flowers and young pods. For time course experiment with soybean, root tissues were harvested at 0, 1, 3, 6, 12 and 24 h and 3, 7 and 14 dpi from mock- (nitrogen free plant nutrient solution) and B. japonicum-inoculated plants. All root tissues harvested were first washed thoroughly, blot dried, immediately frozen in liquid N2 and stored at −80°C until RNA isolation. RTqPCR analysis showed the uniform expression of miR1515 in the different tissues (Fig. 2A) and along the course of nodulation (Fig. 2B), as previously identified using Northern analysis15 We propose the use of miR1515 as reference gene for miRNA expression assays in soybean and possibly other tropical legumes (e.g., common bean, cowpea, peanut). The use of a miRNA as reference gene for miRNA expression assays has some advantages over using protein coding genes such as actin, ubiquitin or EF1. Since miRNA biogenesis pathways are distinct from mRNAs, alterations in biosynthesis machinery (e.g., due to virus infection) might introduce artificial abundance difference between miRNAs and mRNA reference genes used as normalization controls. Using a miRNA as reference gene can help overcome such issues.
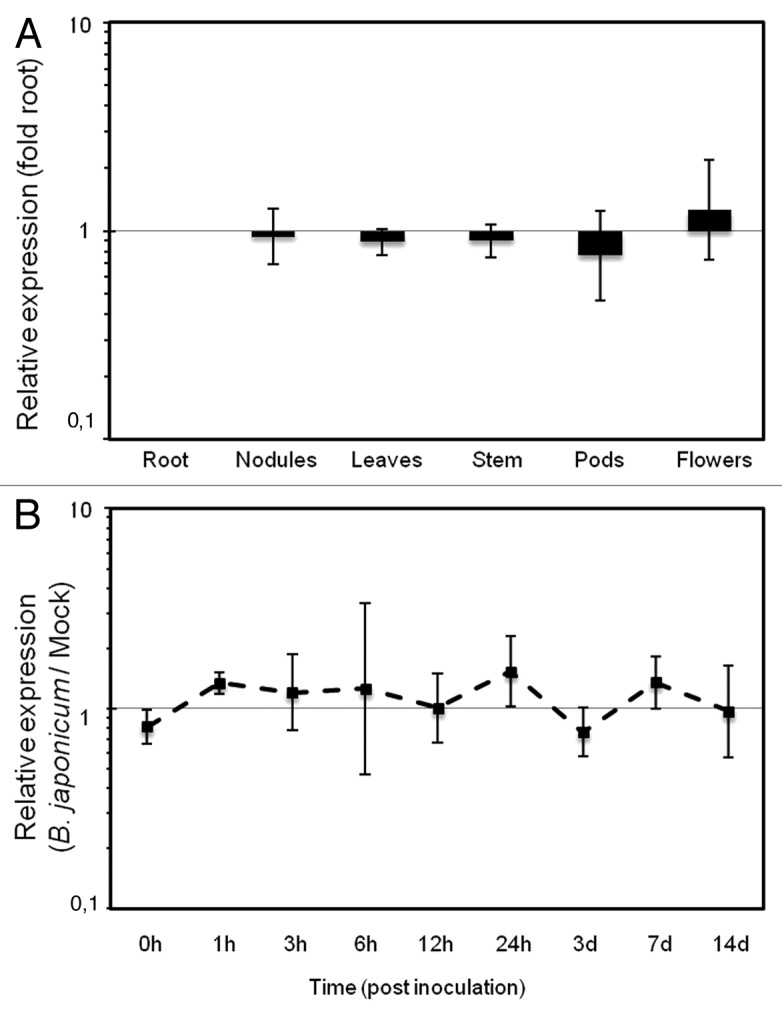
Figure 2. Expression of miR1515 in different soybean tissues (A) and along the course of nodulation (B) in soybean assayed by hairpin cDNA qPCR. (A) Relative expression compared with roots were calculated using average Ct values from four independent replicates (B) Relative expression at each time point post B. japonicum inoculation compared with mock inoculated roots. Data shown are average of four independent biological replicates. Error bars represent SD.
Finally, we examined if cDNA synthesis for U6 and miR1515 could be multiplexed along with test miRNAs without loss of efficiency. Hairpin primers for up to four different miRNAs (50 nM each final concentration) and the reference gene (U6 or miR1515) were added to a single cDNA synthesis reaction using soybean root RNA as template. Fold difference in miRNA expression between samples or between miRNAs and respective reference genes were calculated using the ddCt method.17 We compared Ct values between qPCR assays performed using multiplexed cDNA or independently synthesized cDNA (Fig. 3). There was no significant difference in efficiency or Ct values when miRNA abundance was assayed in single miRNA cDNA synthesis reactions compared with multiplexed cDNA synthesis reactions (ANCOVA; p > 0.05). Therefore, multiplexing during cDNA synthesis would not only enable the efficient use of minimal amounts of RNA for qPCR, but also enhance the accuracy of the assays by ensuring equal cDNA synthesis efficiency between test and reference genes.
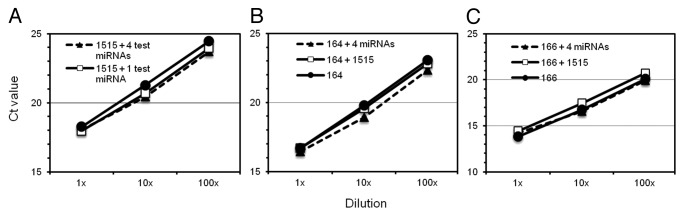
Figure 3. Multiplexing during hairpin cDNA synthesis did not affect assay efficiency. Ct values from hairpin cDNA qPCR assays to quantify (A) miR1515 (B) miR164 and (C) miR166 plotted using different dilutions of cDNAs obtained through independent (filled circles), duplex (open squares) or multiplex (filled triangles) synthesis reactions. Error bars indicate SD. In all cases, the slope of the curves or Ct values (at each dilution) did not differ significantly between different cDNAs.
There are other situations where multiplexing in hairpin cDNA qPCR might be handy. The ability of hairpin cDNA qPCR to distinguish family members or mature sequence variants can sometimes be a limitation, e.g., when attempting to assay miRNA abundance of multiple/all family members. Nevertheless, this could be addressed by multiplexing cDNA synthesis for multiple family members/variants in a single reaction using specific hairpin primers for each variant/family member. This limitation can also be addressed by the use of a recently developed method (miQPCR, reviewed in ref. 18) where an RNA adaptor is ligated to the 3′ end of mature miRNAs and the ligated molecules are reverse transcribed using a primer complementary to the adaptor sequence. However, this approach might as well suffer from the inability to distinguish methylated vs non-methylated RNA molecules, especially in RNA preparations from plants.
In this study, we have developed tools to enhance hairpin cDNA qPCR method to assay plant miRNA gene expression. First, we developed a cDNA synthesis primer for U6 that allows multiplexed cDNA synthesis and validated it using multiple plant species. Next, we developed miR1515 as a reference gene to normalize miRNA expression in soybean and possibly other tropical legumes. Finally, we successfully multiplexed cDNA synthesis of multiple miRNAs with reference genes without loss of efficiency.
Supplementary Material
Acknowledgments
We thank Drs Jerome Verdier and Bikram Pant (Noble Foundation, Ardmore, OK) for comments on the manuscript and Drs Sharon Clay, Anne Fennell, Jose Gonzalez, Xing-You Gu, Jai Rohila and Yajun Wu (South Dakota State University, Brookings, SD) for plant materials or RNA preparations to test U6 primers. Data for gene expression assays were obtained using instruments available at the South Dakota State University’s Functional Genomics Core Facility supported in part by the National Science Foundation/EPSCoR Grant No. 0091948 and by the State of South Dakota. Research in the authors’ laboratory is supported by funds from USDA-AFRI, DOE Sun Grant Initiative, South Dakota Soybean Research and Promotion Council and South Dakota State University and Agricultural Experiment station.
Glossary
Abbreviations:
- RNA
ribonucleic acid
- DNA
deoxyribonucleic acid
- miRNA
microRNA
- cDNA
complementary DNA
- RTqPCR
reverse transcriptase quantitative polymerase chain reaction
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/24918
References
- 1.Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sunkar R, Li YF, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Alonso CR. A complex ‘mRNA degradation code’ controls gene expression during animal development. Trends Genet. 2012;28:78–88. doi: 10.1016/j.tig.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–87. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–87. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 8.Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–25. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- 10.Adhikari S, Turner M, Subramanian S. Hairpin priming is better suited than in vitro polyadenylation to generate cDNA for plant miRNA qPCR. Mol Plant. 2013;6:229–31. doi: 10.1093/mp/sss106. [DOI] [PubMed] [Google Scholar]
- 11.D’haene B, Mestdagh P, Hellemans J, Vandesompele J. miRNA expression profiling: from reference genes to global mean normalization. Methods Mol Biol. 2012;822:261–72. doi: 10.1007/978-1-61779-427-8_18. [DOI] [PubMed] [Google Scholar]
- 12.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–52. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lelandais-Brière C, Naya L, Sallet E, Calenge F, Frugier F, Hartmann C, et al. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell. 2009;21:2780–96. doi: 10.1105/tpc.109.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Zheng Y, Jagadeeswaran G, Li Y, Gowdu K, Sunkar R. Identification and temporal expression analysis of conserved and novel microRNAs in Sorghum. Genomics. 2011;98:460–8. doi: 10.1016/j.ygeno.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Deng Y, Wu T, Subramanian S, Yu O. Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol. 2010;153:1759–70. doi: 10.1104/pp.110.156950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai J, Jeong D-H, De Paoli E, Park S, Rosen BD, Li Y, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–53. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010;50:244–9. doi: 10.1016/j.ymeth.2010.01.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.