Abstract
Pharmacogenetic research has the potential to explain the variation in treatment efficacy within patient populations. Understanding the interaction between genetic variation and medications may provide a method for matching patients to the most effective therapeutic options and improving overall patient outcomes. The OPRM1 gene has been a target of interest in a large number of pharmacogenetic studies due to its genetic and structural variation, as well as the role of opioid receptors in a variety of disorders. The mu-opioid receptor (MOR), encoded by OPRM1, naturally regulates the analgesic response to pain and also controls the rewarding effects of many drugs of abuse, including opioids, nicotine, and alcohol. Genetic variants in OPRM1, particularly the nonsynonymous polymorphism A118G, have been repeatedly associated with the efficacy of treatments for pain and various types of dependence. This review focuses on the current understanding of the pharmacogenetic impact of OPMR1, primarily in regards to the treatment of pain and addiction.
Keywords: OPRM1, Mu opioid receptor, Pharmacogenetics
Introduction
The efficacies of pharmacological treatments, from aspirin to chemotherapeutics, vary from patient to patient. These variations can be the difference between successful clinical outcomes and life-threatening failures. While decades of research have demonstrated that genetic polymorphisms affect the susceptibility to many diseases and disorders, it is only recently that a connection between patient genotype and pharmacological response has been shown. Pharmacogenetics, the study of the effects of patient genetics on medication responses (both therapeutic and adverse) treatment efficacy, arose with the goal of improving patient outcome by selecting treatments based on a patient's genetic background. Largely unstudied as recently as two decades ago, pharmacogenetics is now a rapidly expanding field due to both the realization of the potential benefits of these analyses and the prevalence of high throughput genotyping techniques. Although still in its infancy, the field of pharmacogenetics holds significant promise for improving treatment outcomes across a wide range of diseases.
The μ-Opioid Receptor
Opioid receptors are part of the Rhodopsin family of G-protein coupled receptors (GPCRs), which activate downstream signaling through interactions with heterotrimeric G proteins. The three most common types are the μ-opioid receptor (MOR), δ-opioid receptor (DOR), and κ-opioid receptor (KOR), encoded by the OPRM1, OPRD1, and OPRK1 genes, respectively. Each receptor type has seven transmembrane domains, three intracellular loops, three extracellular loops, an extracellular N-terminus, and an intracellular C-terminus. The three main receptor types are highly homologous within the transmembrane domains, which are arranged in a helical pattern, but have significantly less homology in the extracellular regions. Key residues within these domains create a ligand-binding pocket and binding of opioid agonists within the pocket results in activation of the opioid receptor and subsequent downstream signaling. Variation in the extracellular loops regulates ligandreceptor interaction and allows varying degrees of specificity between different endogenous peptides and opioid receptor types. MOR is activated by both endomorphins and β-endorphin, a cleavage product of the proopiomelanocortin precursor. Enkephalin and deltorphin have been shown to activate DOR, while the dynorphin class of peptides is specific for the KOR protein. Many of these peptides have some affinity for more than one receptor type.
Like the extracellular domains, differences in the intracellular regions allow for specificity of downstream signaling and account for the different pathways activated by the three receptor types. The intracellular domains interact with heterotrimeric Gi/G0 proteins, which are released as the α and βγ subunits following receptor activation. The release of the G protein subunits results in altered ion channel activity and decreased membrane potential, as well as activation of MAPK pathways leading to changes in gene expression (reviewed in [1]). Despite the similar mechanisms, the differences in the intracellular domains of MOR, DOR, and KOR result in different phenotypes when the receptors are activated. Activation of MOR or DOR results in rewarding effects and analgesia, while KOR is involved in aversion and dysphoria. Opioid receptor heterodimers also occur in vivo and have been shown to regulate unique phenotypes that differ from those regulated by the individual receptor types, adding further complexity to opioid receptor signaling (reviewed in [2]).
Whole genome sequencing of a wide range of ethnic groups has identified 3324 polymorphisms in the OPRM1 gene, which occupies a 200kb region on the long arm of chromosome 6 (http://www.1000genomes.org). Many of these polymorphisms occur at extremely low frequencies and have limited relevance at the population level. However, 1395 of the genetic variants have minor allele frequencies greater than 1% in the global population. These more common variants are more likely to be relevant in large scale genetic studies. There are 2 common nonsynonomous SNPs and an additional 3 synonymous coding variants. The small number of coding polymorphisms suggests selective pressure against genetic variation in the exonic regions of OPRM1.
The most common and most studied non-synonymous SNP is rs1799971, more often referred to as A118G, with a global minor allele frequency of 19%. The A118G variant causes an ASP>ASN residue change and occurs more frequently in non-African populations. A number of functional effects have been associated with the A118G polymorphism. The G allele of A118G creates a novel CpG-methylation site, preventing the upregulation of OPRM1 in response to chronic opioid use [3]. Copies of mRNA carrying the variant G allele were shown to be less abundant in human brain tissue than the A allele and studies of stably transfected cell lines have indicated that the A118G variant results in reduced expression of MOR at the cell surface [4, 5]. Decreased accumulation of the second-messenger cAMP transfected cells was observed in the presence of morphine, methadone, and DAMGO [5]. This reduced signaling following DAMGO activation has also been shown in human postmortem brain tissue [6]. In contrast, data suggest that β-endorphin has higher binding affinity and increased signaling at the variant receptor [7].
In addition to genetic variation, the OPRM1 gene also has substantial structural variation. Alternative splicing of 15 known exons produces at least 23 previously described splice variants, with 16 of these variants potentially translated into protein products (http://www.ensembl.org). Despite the large number of total exons, individual splice variants each contain only 3-5 exons. The 3′ UTR of OPRM1 is also known to vary in size, with some isoforms in both mice and humans known to have UTRs greater than10 kb in length [8, 9]. Given the known roles of 3′ UTRs in regulating transcript expression through miRNA binding and other mechanisms, the variable UTR length in OPRM1 may help regulate expression levels of the different isoforms [10].
Pain
Painful stimuli cause the release of endogenous opioids, activating MOR and causing analgesic responses. In this way MOR is responsible for mitigating the sensation of pain in the absence of opioid medication. There are a number of different classes of pain, which include nociceptive, neuropathic, inflammatory and pathological pain [11]. Various forms of painful stimuli result in different biochemical and physiological responses and it is, therefore, likely that there are differences in the effects of genetic variations on the thresholds and tolerance levels for different types of pain. Due to the involvement of MOR in analgesia, Fillingim et al. assessed the effects of the A118G polymorphism on pain from three different sources: pressure, heat, and ischemia [12]. Individuals carrying the G allele were found to have higher thresholds for pressure pain, while no differences were observed in ischemic pain [12]. For thermal pain, men with the G allele reported lower pain ratings, while women reported more pain [12]. Another study found the minor allele of the intronic variant rs9479757 to also be associated with a higher pressure pain threshold [13]. Although these experiments can provide valuable information about functional genetic variation, there is no guarantee that these effects are relevant to patient populations in less experimentally controlled settings. There is evidence, however, that OPRM1 variants do affect pain in some patients in a clinical setting. Women with the G allele of A118G report higher intensity pain from migraines, and have more pain and slower recovery from herniated discs [14, 15]. Fibromyalgia patients carrying the G allele also suffer from more pain, further suggesting that A118G is associated with pain sensitivity [16]. Patients with A/A genotypes have been found to have less pain from diabetic foot ulcers than G carriers [17]. Despite the evidence that the G allele of A118G is associated with increased pain from several different sources, a study of persistent widespread pain also found no association between pain ratings and genotype at A118G [18]. That study also found no association with rs563649 [18]. These findings suggest that OPRM1 polymorphisms effect pain sensations from some, but not all, stimuli.
Pharmacogenetics of Analgesia
Analgesics can be classified as either opioid or non-opioid based on the mechanism of action. The non-opioid class of analgesics primarily consists of non-steroidal anti-inflammatory drugs (NSAIDs), such as aspirin and ibuprofen, and paracetamol, also known as acetaminophen. Both NSAIDs and paracetamol act as cyclooxygenase inhibitors and are notably different than opioid analgesics in that they have low abuse potential and are available without a prescription. Both naturally-occuring and synthetic opioid receptor ligands can act as opioid analgesics. Some of the more common opioid analgesics include morphine, codeine, oxycodone, fentanyl, and hydrocodone. The majority of medications in this class act as agonists of MOR, although many also have some activity at either DOR or KOR. The relative level of analgesia provided by each drug is determined by the specific half-life of the drug and the affinity for individual opioid receptors types. Due to the activation of MOR, these opioid medications are potentially addictive and, therefore, their use as analgesics is highly regulated.
As one of the few non-synonymous SNPs in OPRM1, the vast majority of pharmacogenetic studies of pain management and OPRM1 have focused on the A118G variant (Table 1). Patients carrying the G allele have been found to require more medication to achieve analgesia when treated with fentanyl, morphine, or morphine-6-glucoronide (M6G) for a variety of pain sources, including surgery and chemotherapy [19-24]. In another trial, the A118G variant was associated with morphine analgesia only in the presence of a two minor alleles at the rs4680 locus located in the COMT gene [25]. Lö tsch et al. also found a trend towards significance towards decreased analgesia in G carriers treated with morphine in an outpatient setting [26]. The reduction in analgesia in carriers of the G allele has also been reported in trials of oxycodone, tramadol, and alfentanil [27-29]. A number of other studies have found the effect of the G allele to be recessive, with G/G individuals requiring more morphine or fentanyl to achieve analgesia [30-34]. The potentially contradictory findings of both dominant and recessive effects for A118G may be the result of patient ethnicity. A118G has a minor allele frequency of 38% in Asians, 16% in Europeans, and 3% in African-Americans (http://www.1000genomes.org). The lower frequency in Europeans and African-Americans results in only a small percentage of individuals carrying the G/G genotype, potentially making studies of these populations underpowered for an analysis of a recessive model. This issue would not be likely to arise in Asian populations, in which a recessive effect of the G allele has been repeatedly observed, due to the greater minor allele frequency [30, 32-34].
Table 1. Pharmacogenetic effects of OPRM1 variants in analgesia.
| SNP | Pain Source | Analgesic | Ethnicity | Subjects | Description | Ref. |
|---|---|---|---|---|---|---|
| A118G | Cancer pain | Morphine | Caucasian | 207 | G/G requires higher doses when patient is also Val/Val at Val158Met in COMT | [20] |
| A118G | Gynaecological surgery | Fentanyl | Han Chinese | 177 | G/G requires higher doses | [14] |
| A118G | Abdominal surgery | Fentanyl | Han Chinese | 189 | G carriers have more pain at same dose | [15] |
| A118G | Oral Surgery | Fentanyl | Japanese | 108 | G carriers require higher doses | [16] |
| A118G | Electric | Morphine-6-glucoronide | Caucasian | 16 | G carriers have more pain at same dose | [17] |
| A118G | Cold pressor | Fentanyl | Japanese | 280 | G carriers have more pain at same dose | [18] |
| rs93841 79 | Facial cosmetic surgery | Fentanyl | Japanese | 280 | G carriers require lower doses | [18] |
| A118G | Single electrical nerve stimulation | Oxycodone | Caucasian | 33 | G carriers have more pain at same dose and reduced ability to focus | [22] |
| A118G | Chemotherapy | Tramadol/Acetamin ophen | Chinese | 96 | G carriers have more pain at same dose | [23] |
| A118G | Electric and chemical | Alfentanil | Caucasian | 20 | G carriers have more electrical pain at same dose. G/G individuals have more chemical pain at same dose | [24] |
| A118G | Ceasarean section | Sufentanil | Caucasian | 41 | G carriers have less pain with same dose | [37] |
| A118G | Total knee arthroplasty | Morphine | Taiwanese | 120 | G/G requires higher doses | [25] |
| A118G | Cancer pain | Morphine | Caucasian | 99 | G/G requires higher doses | [26] |
| A118G | Abdominal surgery | Fentanyl/Morphine | Japanese | 138 | G/G requires higher doses | [27] |
| A118G | Abdominal surgery | Fentanyl | Han Chinese | 167 | G/G requires higher doses | [28] |
| A118G | Ceasarean section | Morphine | Various | 994 | G/G genotypes report higher pain scores and use higher doses | [29] |
| A118G | Labor | Fentanyl | Various | 224 | G carriers require lower doses | [36] |
| A118G | Cancer pain | Morphine | Caucasian | 137 | G carriers have more pain at same dose | [19] |
| rs634479 | Thoracotomy | Fentanyl/Oxycodone | Various | 90 | C carriers have more pain at same dose | [39] |
| rs499796 | Thoracotomy | Fentanyl/Oxycodone | Various | 90 | C carriers have more pain at same dose | [39] |
| rs548646 | Thoracotomy | Fentanyl/Oxycodone | Various | 90 | T carriers have more pain at same dose | [39] |
| rs679987 | Thoracotomy | Fentanyl/Oxycodone | Various | 90 | C carriers have more pain at same dose | [39] |
| A118G | Ceasarean section | Morphine | Various | 103 | G carriers have lower incidence of pruritus (itching) | [33] |
| A118G | Ceasarean section | Morphine | Taiwanese | 212 | G carriers have lower incidence of pruritus (itching) | [42] |
Despite the large amount of evidence supporting a role for A118G in the pharmacogenetics of opioid analgesics, other trials have failed to replicate the findings [35-40]. Two studies even found that women with the A/A genotype needed more fentanyl or sufentanil to achieve analgesia during labor [41, 42]. A meta-analysis of 8 previous studies found no association between the frequency of the G allele and analgesia, but did note a potentially significant effect of the G/G genotype [43]. Taken in aggregate these findings still suggest that A118G genotype may have a recessive effect on the doses of specific opioid medications required for analgesia from particular painful stimuli. Given the different types of painful stimuli and the wide range of available analgesics, however, the extent to which this effect is widely applicable is not clear. Other polymorphisms have also been associated with the efficacy of opioid analgesics. Patients with the G allele at rs9384179 require less fentanyl to achieve analgesia in the 24 hour period following surgery [23]. The analgesic effect of opioid medications after surgery was also associated with the genotypes at rs634479, rs499796, rs548646, and rs679987, which are all located in introns [44].
Both illicit and prescribed opioids have notable side effects including nausea, constipation, pruritus (itching), and respiratory depression. While pharmacogenetic studies often focus on treatment outcome, the effect of genetic variation on side effect severity is also relevant to appropriate treatment selection. Individuals with more severe side effects may be less likely to continue their prescribed treatment course, creating a link between the pharmacogenetics of side effects and treatment efficacy. These patients may also need additional treatment to manage the increased risk of side effects. Several studies have found no association between OPRM1 SNPs and opioid side effects. A118G did not have an effect on respiratory depression caused by morphine-6-glucuronide, a metabolite of morphine [22]. Nine SNPs in OPRM1 were found to not be associated with nausea and vomiting in cancer patients receiving opioids, including morphine, oxycodone, and fentanyl [45]. A similar lack of association was observed between A118G and nausea or vomiting due to fentanyl treatment in an additional study [33]. The intronic SNP rs2075572 was also only nominally associated with central side effects of morphine, such as nausea, drowsiness, and hallucinations [46]. In contrast to these negative findings, two independent studies observed decreased pruritus in women carrying the G allele of A118G after morphine treatment for post-caesarean section pain [38, 47]. A118G was also associated with reduced ability to focus when treated with oxycodone [27]. These studies suggest OPRM1 polymorphisms can alter the side effects of opioid medications. Substantially more research is required on this subject, however, since these pharmacogenetic effects may be specific to the particular genetic variant, opioid treatment, and side effect being studied.
Opioid Dependence
Heroin is the most common illicit opioid drug and is deacetylated in the brain to morphine, which directly activates MOR, as does heroin. Endorphin and enkephalin peptides are the main neurotransmitters that mediate reward processes in the brain. It is the activation of the MOR, and subsequent meso-limbic opioid and dopamine signaling, which lends opioid drugs their addictive properties [48, 49]. Positive reinforcement therefore, drives early use as opioids are consumed for their pleasurable effects. As opioid use progresses to abuse and addiction, attempts to discontinue opioid use result in withdrawal and dysphoria. Negative reinforcement now motivates drug use, and drugs are taken to avoid the negative effects of withdrawal [49]. This cycle of positive and negative reinforcement causes both heroin and morphine to be highly addictive. Since morphine is also a commonly used analgesic, it is not surprising that other opioid analgesics targeting MOR also have high potentials for addiction. Although millions of people worldwide are dependent on heroin, addiction to prescription opioids also poses a significant public health problem. Increased use of opioids for pain management has led to a rapidly growing number of people addicted to these analgesics.
Despite the fact that the majority of opioid analgesics and illicit opioids directly target MOR, few studies have consistently found significant associations between OPRM1 polymorphisms and opioid dependence. The most well-replicated finding is the association between the A118G polymorphism and heroin dependence in the Indian population, which has been observed in four separate studies. In three of the studies, the minor G allele conferred increased risk of addiction [50-52]. The fourth study found decreased risk in individuals with the minor allele, although only 20 cases were analyzed [53]. Levran et al. observed a significant association between heroin dependence in European Americans and the combined genotypes of rs510769 in OPRM1 and rs2236861 in OPRD1, the gene encoding the δ-opoid receptor [54]. However, three studies of opioid addicts of European descent failed to find associations for five additional SNPs [55-57]. The negative findings in these analyses may be the result of analyzing only variants in OPRM1, and not OPRD1. The 118G allele was found to be protective against opioid dependence in a cohort of Malaysians, but this finding was not replicated in subsequent research [53, 58]. A118G and rs9479757 were also not significant in Han Chinese populations and no associations were found across 24 SNPs in two African American case-control analyses [57, 59-61]. In aggregate, these findings suggest that genetic variation in OPRM1 may affect susceptibility to opioid dependence in an ethnicity-dependent manner. In most ethnic groups, however, there is little evidence supporting an association between OPRM1 polymorphisms and opioid dependence. The statistical power of each study to detect relatively small effect sizes may also play a role in the conflicting results.
Pharmacogenetics of the Treatment of Opioid Dependence
There are two primary medications used to treat opioid dependence in the United States, methadone and buprenorphine, both of which interact with MOR. Methadone acts as a full MOR agonist, activating MOR downstream signaling in a similar manner to morphine. The activation of MOR enables methadone to reduce craving for illicit opioids and to decrease withdrawal symptoms. Buprenorphine is a partial MOR agonist, as well as an antagonist of KOR. Treatment with buprenorphine reduces craving much like methadone, but the reduced agonist activity somewhat reduces the addictive potential of the medication. Naltrexone, a MOR antagonist with high affinity for the MOR protein, is also efficacious for the treatment of opioid dependence. As a MOR antagonist, naltrexone causes rapid onset of withdrawal symptoms if given to an opioid dependent person. As a result, naltrexone is often used for rapid detoxification prior to standard buprenorphine or methadone treatment, rather than as a long term therapeutic option in most countries, Russia being a notable exception. Naloxone, another MOR antagonist, is often compounded with buprenorphine, blocking activation of MOR by buprenorphine if the medication is injected rather than taken orally [62, 63].
Few studies have analyzed the pharmacogenetic effects of OPRM1 variants on the standard treatments for opioid dependence and an even smaller number directly measure patient outcome or treatment efficacy (Table 2). A118G and the intronic SNP rs1074287 were both not associated with opioid positive urine drug screens during methadone maintenance therapy [64, 65]. A group of SNPs from multiple genes, including A118G, were associated with optimal methadone dose, although A118G was not significantly associated by itself [66]. While other research has not examined treatment outcome, several studies suggest differences in response to opioid dependence treatments based on OPRM1 genotype. Buprenorphine patients with the A/G genotype at the A118G locus have suppressed activation of the hypothalamic-pituitary-adrenal axis, which may indicate decreased craving, while those on methadone have higher plasma levels of the medication compared to homozygous A/A patients [67, 68]. Individuals with the A/G and G/G genotypes also have decreased pupillary constriction compared to A/A individuals in response to treatment with levomethadone, a methadone-like agonist [69]. This differential response is consistent with the hypothesis that the G allele of A118G may result in reduced analgesia after treatment with opioid medications. The minor alleles of an additional 12 SNPs are associated with insomnia and reduced libido in methadone maintenance patients [70]. Despite the logical connection between OPRM1 and opioid dependence, it is difficult to identify any firm pharmacogenetic connections between OPRM1 polymorphisms and treatment for opioid dependence with the limited number of studies available. Additional studies that examine treatment efficacy are clearly needed. Further pharmacogenetic analyses of specific ethnic groups may also be beneficial due to the potential ethnic differences in the association of the A118G polymorphism with opioid dependence.
Table 2. Pharmacogenetic effects of OPRM1 variants in addiction treatment.
| SNP | Addiction | Treatment | Ethnicity | Subjects | Description | Ref. |
|---|---|---|---|---|---|---|
| A118G | Opioids | Methadone | Han Chinese | 321 | Combined effects between G allele and variants in four other genes associated with maximum stabilized methadone dose | [61] |
| rs1074287 | Opioids | Methadone | Taiwanese | 336 | A/A patients have increased insomnia. A carriers have decreased libido | [65] |
| rs6912029 | Opioids | Methadone | Taiwanese | 336 | G/G patients have increased insomnia. G carriers have decreased libido | [65] |
| rs12209447 | Opioids | Methadone | Taiwanese | 336 | C/C patients have increased insomnia. C carriers have decreased libido | [65] |
| rs510769 | Opioids | Methadone | Taiwanese | 336 | G/G patients have increased insomnia. G carriers have decreased libido | [65] |
| rs3798676 | Opioids | Methadone | Taiwanese | 336 | C/C patients have increased insomnia. C carriers have decreased libido | [65] |
| rs7748401 | Opioids | Methadone | Taiwanese | 334 | T/T patients have increased insomnia. T carriers have decreased libido | [65] |
| rs495491 | Opioids | Methadone | Taiwanese | 335 | T/T patients have increased insomnia. T carriers have decreased libido | [65] |
| rs10457090 | Opioids | Methadone | Taiwanese | 336 | A/A patients have increased insomnia. A carriers have decreased libido | [65] |
| rs589046 | Opioids | Methadone | Taiwanese | 336 | G/G patients have increased insomnia. G carriers have decreased libido | [65] |
| rs3778152 | Opioids | Methadone | Taiwanese | 336 | A/A patients have increased insomnia. A carriers have decreased libido | [65] |
| rs563649 | Opioids | Methadone | Taiwanese | 336 | G/G patients have increased insomnia. G carriers have decreased libido | [65] |
| rs2075572 | Opioids | Methadone | Taiwanese | 336 | C/C patients have increased insomnia. C carriers have decreased libido | [65] |
| A118G | Nicotine | Transdermal patch | Caucasian | 320 | G carriers have higher rates of abstinence | [72] |
| A118G | Nicotine | Transdermal patch/nasal spray | Caucasian | 374 | G carriers have higher rates of abstinence | [73] |
| A118G | Nicotine | Transdermal patch | Caucasian | 710 | G carriers have lower rates of abstinence | [74] |
| A118G | Alcohol | Naltrexone | Various | 82 | G carriers have lower rates of relapse | [99] |
| A118G | Alcohol | Naltrexone | Korean | 32 | G carriers have lower rates of relapse | [102] |
| A118G | Alcohol | Naltrexone | Various | 604 | G carriers show decreased heavy drinking and increased abstinence | [103] |
| A118G | Alcohol | Naltrexone | Caucasian | 306 | G carriers show decreased heavy drinking | [104] |
| A118G | Alcohol | Naltrexone | Caucasian | 265 | A/A patients with at least one DAT 9 VNTR consume fewer drinks per day | [108] |
Nicotine Dependence
Tobacco use, including smoking and chewing, is highly addictive due to the effects of the nicotine present in tobacco. Smoking increases the risk of small-cell lung carcinoma and is responsible for millions of deaths each year due to cancer, heart disease, and stroke. Unlike the opioids involved in analgesia and addiction, nicotine does not directly target any of the opioid receptors. The primary targets for nicotine are instead the nicotinic acetylcholine receptors, a family of ligand-gated ion channels. Despite the differences in target receptors, however, nicotine still causes many downstream effects that are similar to those of opioid use and result in the onset of addiction. These effects, such as activation of reward pathways and increased dopamine release, are mediated by the release of endogenous opioids following nicotine use. With this important connection between the opioid system and nicotine, genetic variation in OPRM1 may affect susceptibility to nicotine dependence and the efficacy of smoking cessation treatments.
A three SNP haplotype consisting of rs9479757, rs2075572, and rs10485057 was significantly associated with smoking initiation in a European-American cohort, while rs2075572 alone was also associated with nicotine dependence [71]. In the same study, A118G was not associated with nicotine dependence, a finding which was replicated in an independent case-control analysis [71, 72]. Although A118G was not found to be associated with nicotine dependence, other research has linked the variant to a variety of physical responses to nicotine. After smoking, A/A individuals have increased cerebral blood flow to areas of the brain associated with craving [73]. A/A patients also report increased reward from nicotine under negative mood induction and women with the A/A genotype find smoking to be for reinforcing than those with the A/G or G/G genotypes [74, 75]. In contrast to these findings, carriers of the G allele were found to experience more dopamine release in the striatum following smoking [76].
Pharmacogenetics of the Treatment of Nicotine Dependence
Medicinal treatments for nicotine dependence generally fall into three categories: nicotine replacement therapy (NRT), nicotine receptor agonists, and neurotransmitter regulators. In NRT, nicotine is administered to the patient through transdermal patches, gum, nasal sprays, or other sources. The goal is to progressively reduce the amount of nicotine provided to the patient until they are able to successfully discontinue nicotine use entirely. Several medications are also available for the treatment of nicotine dependence. Pharmacological options include varenicline and cytisine, partial agonists of nicotinic acetylcholine receptors, and buproprion, an antidepressant that regulates dopamine and norepinephrine release and reuptake. As with the MOR agonists used for treating opioid dependence, nicotine receptors agonists reduce withdrawal and craving experienced during smoking cessation and improve patient success rate.
The small number of pharmacogenetic studies analyzing the role of OPRM1 in nicotine dependence treatment have focused on the A118G polymorphism (Table 2). A clinical trial by Lerman et al. found that NRT patients carrying the G allele had better abstinence rates than A/A patients when treated with transdermal patches [77]. No effect of A118G was observed in patients receiving nasal spray treatments [77]. The effect of A118G on NRT efficacy was also observed in a multivariate analysis of another patient population [78]. However, a later trial in a British cohort found exactly the opposite association: patients with the A/A genotype were more likely to be abstinent when using transdermal patches [79]. A more recent trial found no effect of A118G genotype on NRT efficacy, although patients in the trial were treated with both transdermal patch and oral NRT, making a direct comparison difficult [80]. Since the original association was only found in transdermal patch efficacy, the addition of a second NRT type may be a confounding factor when directly comparing the findings to other studies. The contradictory findings of these four studies make it difficult to draw conclusions about the role of the A118G variant in smoking cessation. However, previous studies have still indicated several mechanisms by which A118G genotype may affect biochemical reactions to nicotine use and suggest that further pharmacogenetic studies are warranted.
Alcoholism
Alcohol is the second most commonly used drug of abuse worldwide, nicotine being the most common. More than 16 million people were abusing or dependent on alcohol in 2011 in the United States alone, creating a substantial societal burden (National Survey on Drug Use and Health, http://www.drugabuse.gov). Evidence suggests that the rewarding effects and positive reinforcement experienced after alcohol use are regulated by opioid receptors, with alcohol causing the release of endogenous opioids in the brain and indirectly activating the opioid signaling pathways [81, 82]. Like nicotine, the regulation of the effects of alcohol by the opioid system has led to substantial research on the role of OPRM1 polymorphisms in alcoholism treatment and risk.
Adolescents with the A/G or G/G genotypes at A118G are three times more likely to have an alcohol use disorder than those with the A/A genotype [83, 84]. Two studies have also found an association between A118G genotype and alcohol dependence in adult Caucasians; however, one found the G allele to be protective while the other found carriers of the G allele to be at increased risk [85, 86]. Other studies have failed to find any association between alcoholism and A118G, as well as other genetic variants including rs2075572, rs1799972, and a (CA)n repeat in intron 1 [56, 87-90]. A meta-analysis also found no effect of A118G genotype on risk of developing alcohol dependence [91]. In another study of European-Americans, three intronic SNPs (rs495491, rs6091485, and rs648893) were associated with alcohol dependence [92]. In African-Americans, A118G was not associated with dependence (although the 3% frequency of the G allele in this ethnic group substantially comprises power), while women with the T/T genotype at the rs1799972 polymorphism were found to consume more alcohol [89, 93]. Among Asian populations, the minor allele of A118G was associated with alcoholism in Indian and Japanese cohorts [51, 94]. This association was not observed in studies of Korean or Taiwanese patients, although the G allele did correlate with an increase in the number of drinking days over the course of the study [95-97]. A Meta-analysis of these Asian cohorts confirmed an effect of A118G genotype on susceptibility to alcohol dependence [91].
The association between A118G and alcohol dependence may be explained by different physiological responses to alcohol based on A118G genotype. People with the 118G variant have a stronger association between their desire to drink and subsequent drinking than people lacking the variant [98]. Two studies have also shown that heavy drinkers carrying the G allele have increased craving compared to individuals with the A/A genotype, although an additional study found conflicting results [99-101]. These findings included increases in cue-induced craving and G carriers are known to have greater response in the mesocorticolimbic regions of the brain following alcohol cues [99, 102]. Responses in these regions have been associated with increased alcohol craving, suggesting a potential mechanism behind the craving associated with A118G [103]. People carrying the 118G allele also experience more euphoria and subjective “highs” after drinking alcohol [101, 104]. Positron emission tomography scans of male social drinkers with the 118G variant showed a greater increase in dopamine release in the ventral striatum following alcohol consumption, compared to A/A participants, possibly explaining the increased feeling of euphoria [105].
Pharmacogenetics of the Treatment of Alcoholism
Treatment options for alcohol dependence have several mechanisms of action, including prevention of alcohol consumption, minimization of craving and withdrawal, and reduction of positive reinforcement. Naltrexone, a MOR antagonist, is also able to reduce relapse to heavy drinking and the frequency of alcohol consumption [106, 107]. Naltrexone prevents the activation of MOR by endogenous opioids, leading to decreases in the euphoria and subsequent attenuation of positive reinforcement associated with alcohol use. Over time patients experience reduced levels of craving for alcohol and gradually consume less alcohol (reviewed in [108]). Naltrexone did not increase rates of abstinence in nearly all the double-blind placebo-controlled trials.
Oslin et al. originally identified an association between A118G allele and naltrexone response (Table 2) [106]. Patients carrying the G allele had lower rates of relapse to heavy drinking on naltrexone than those with the A/A genotype; however, no difference in abstinence rates was observed between the genotypic groups [106]. Both the findings in relapse rates and abstinence rates were replicated in a Korean cohort [109]. Other studies have also demonstrated that naltrexone is more effective in reducing heavy drinking in individuals carrying the G allele [110, 111]. Two additional trials found no association at all between A118G and naltrexone efficacy, while a third trial found no association for either A118G or rs648893 [112-114]. Another study suggested that naltrexone reduced heavy drinking only in patients who had both the A/A genotype at A118G and the nine copy variant of the variable number tandem repeat (VNTR) in the dopamine transporter gene (DAT1) [115]. The inability of these studies to replicate the original association between A118G and naltrexone efficacy may be partially attributed to differences in patient demographics or outcome measurements. A recent meta-analysis concluded that individuals with the G/G or A/G genotypes did have reduced relapse rates compared to A/A individuals, but no difference in abstinence between genotypes was apparent [107]. This pharmacogenetic effect may be explained by differential responses to alcohol based on A118G genotype. Asian Americans carrying the G allele have decreased craving for alcohol when treated with naltrexone, while the medication increases the urge to drink in European Americans with the G allele [116, 117]. Naltrexone also reduces the euphoric effects of alcohol in European Americans with the A/G or G/G genotypes [101]. Confirmation of these findings, preferably taking into account potential ethnic differences, would help further explain the effect of A118G genotype on alcoholism treatment. While numerous pharmacogenetic studies have examined the role of A118G in naltrexone treatment, the amount of research on the interaction between OPRM1 genotype and other treatments is extremely limited. A single clinical trial showed no effect of two OPRM1 SNPs, including A118G, on the ability of nalmefene to reduce heavy drinking [118]. Future analyses of non-naltrexone treatment options are still required. In addition, there are no pharmacogenetic studies of naltrexone treatment for opioid addiction.
Conclusion
Opioid receptors are intrinsically linked to a variety of diseases and their respective treatments, either as direct drug targets or through downstream signaling activated by endogenous opioids. MOR naturally regulates the analgesic response to pain and also controls the rewarding effects of many drugs of abuse, including opioids, nicotine, and alcohol. Because of the opioid receptor's involvement, many analgesics are direct MOR agonists and treatments for addiction often act as either agonists, partial agonists, or antagonists of MOR. The connection between MOR and both addiction and pain makes OPRM1, the gene encoding MOR, an interesting target for pharmacogenetic studies (Figure 1). Genetic variants in OPRM1, particularly A118G, have been repeatedly associated with the efficacy of treatments for pain and alcohol dependence. In the two most well replicated findings, patients carrying the G allele had a reduced analgesic response to exogenous opioids and alcoholics with the G allele had reduced relapse rates when treated with naltrexone. Additional connections between OPRM1 and treatments for opioid and nicotine addiction are also promising, but require further study. Clear definitions of the phenotypes and ethnicities involved in these future analyses will be essential, as even minor variations in either factor could compromise the ability to replicate previous findings [119]. By confirming the pharmacogenetic effects of OPRM1 polymorphisms and using those findings to guide treatment decisions, patients can be prescribed the therapeutic options with the best efficacy and the greatest tolerability.
Figure 1.
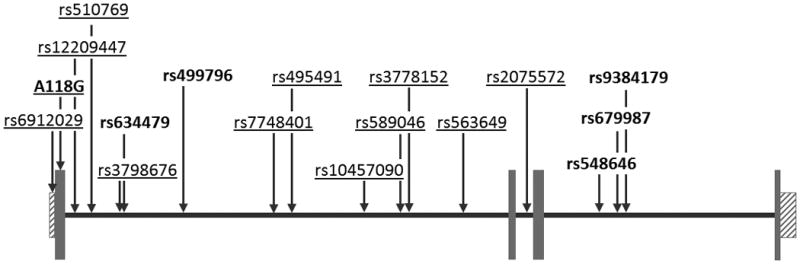
Diagram of the OPRM1 gene with the locations of genetic variants associated with pharmacogenetic effects in analgesia (bolded) or addiction treatment (underlined). Gray boxes indicate exons and boxes with diagonal lines indicate untranslated regions. SNP and exon locations taken from the February 2009 build of the human genome in UCSC Genome Browser (www.genome.ucsc.edu)
The vast majority of pharmacogenetic studies on OPRM1 have analyzed the effects of A118G. As one of the first genetic variants to be associated with pharmacological outcome and a relatively common non-synonymous SNP, the focus on A118G in pharmacogenetic trials is certainly understandable. However, intronic and synonymous coding variants in many genes have been shown to have important effects on transcription, mRNA stability, and splicing, thus affecting gene function despite not directly disrupting any specific residue. OPRM1 has numerous genetic and structural variations, all of which are potential relevant to the field of pharmacogenetics. The small number of studies analyzing OPRM1 polymorphisms other than A118G have revealed some effects on treatment efficacy [23, 44, 70]. By focusing on A118G and overlooking other variation in the gene, a significant portion of OPRM1's role in pharmacogenetics is likely being missed. As high throughput sequencing and other large scale genotyping methods become increasingly common, future studies can and must start to focus on all of the genetic variation present in the ORPM1 gene.
Highlights.
Pharmacogenetic studies of OPRM1 have focused on A118G, a non-synonymous variant
A118G and other polymorphisms are associated with the efficacy of opioid analgesics
A118G is also associated with treatment outcome in naltrexone patients
Future pharmacogenetic studies must expand beyond the current focus on A118G
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Al-Hasani R, Bruchas MR. Molecular mechanisms of opioid receptor-dependent signaling and behavior. Anesthesiology. 2011;115(6):1363–81. doi: 10.1097/ALN.0b013e318238bba6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stockton SD, Jr, Devi LA. Functional relevance of mu-delta opioid receptor heteromerization: a role in novel signaling and implications for the treatment of addiction disorders: from a symposium on new concepts in mu-opioid pharmacology. Drug Alcohol Depend. 2012;121(3):167–72. doi: 10.1016/j.drugalcdep.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oertel BG, et al. Genetic-epigenetic interaction modulates mu-opioid receptor regulation. Hum Mol Genet. 2012;21(21):4751–60. doi: 10.1093/hmg/dds314. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, et al. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem. 2005;280(38):32618–24. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]
- 5.Kroslak T, et al. The single nucleotide polymorphism A118G alters functional properties of the human mu opioid receptor. J Neurochem. 2007;103(1):77–87. doi: 10.1111/j.1471-4159.2007.04738.x. [DOI] [PubMed] [Google Scholar]
- 6.Oertel BG, et al. A common human micro-opioid receptor genetic variant diminishes the receptor signaling efficacy in brain regions processing the sensory information of pain. J Biol Chem. 2009;284(10):6530–5. doi: 10.1074/jbc.M807030200. [DOI] [PubMed] [Google Scholar]
- 7.Bond C, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95(16):9608–13. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikeda K, et al. The untranslated region of (mu)-opioid receptor mRNA contributes to reduced opioid sensitivity in CXBK mice. J Neurosci. 2001;21(4):1334–9. doi: 10.1523/JNEUROSCI.21-04-01334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ide S, et al. Characterization of the 3′ untranslated region of the human mu-opioid receptor (MOR-1) mRNA. Gene. 2005;364:139–45. doi: 10.1016/j.gene.2005.05.040. [DOI] [PubMed] [Google Scholar]
- 10.Wu Q, et al. Post-transcriptional regulation of mouse mu opioid receptor (MOR1) via its 3′ untranslated region: a role for microRNA23b. FASEB J. 2008;22(12):4085–95. doi: 10.1096/fj.08-108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merskey HBN, editor. Part III: Pain Terms, a Current List with Definitions and Notes on Usage. Second. IASP Press; Seattle: 1994. Classification of Chronic Pain; pp. 209–214. [Google Scholar]
- 12.Fillingim RB, et al. The A118G single nucleotide polymorphism of the mu-opioid receptor gene (OPRM1) is associated with pressure pain sensitivity in humans. J Pain. 2005;6(3):159–67. doi: 10.1016/j.jpain.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Huang CJ, et al. Association between human opioid receptor genes polymorphisms and pressure pain sensitivity in females*. Anaesthesia. 2008;63(12):1288–95. doi: 10.1111/j.1365-2044.2008.05760.x. [DOI] [PubMed] [Google Scholar]
- 14.Menon S, et al. The human mu-opioid receptor gene polymorphism (A118G) is associated with head pain severity in a clinical cohort of female migraine with aura patients. J Headache Pain. 2012;13(7):513–9. doi: 10.1007/s10194-012-0468-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olsen MB, et al. Pain intensity the first year after lumbar disc herniation is associated with the A118G polymorphism in the opioid receptor mu 1 gene: evidence of a sex and genotype interaction. J Neurosci. 2012;32(29):9831–4. doi: 10.1523/JNEUROSCI.1742-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finan PH, et al. Genetic influences on the dynamics of pain and affect in fibromyalgia. Health Psychol. 2010;29(2):134–42. doi: 10.1037/a0018647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng KI, et al. Association of the functional A118G polymorphism of OPRM1 in diabetic patients with foot ulcer pain. J Diabetes Complications. 2010;24(2):102–8. doi: 10.1016/j.jdiacomp.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Holliday KL, et al. Do genetic predictors of pain sensitivity associate with persistent widespread pain? Mol Pain. 2009;5:56. doi: 10.1186/1744-8069-5-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, et al. Association of human micro-opioid receptor gene polymorphism A118G with fentanyl analgesia consumption in Chinese gynaecological patients. Anaesthesia. 2010;65(2):130–5. doi: 10.1111/j.1365-2044.2009.06193.x. [DOI] [PubMed] [Google Scholar]
- 20.Wu WD, et al. Polymorphism of the micro-opioid receptor gene (OPRM1 118A>G) affects fentanyl-induced analgesia during anesthesia and recovery. Mol Diagn Ther. 2009;13(5):331–7. doi: 10.1007/BF03256337. [DOI] [PubMed] [Google Scholar]
- 21.Fukuda K, et al. Diversity of opioid requirements for postoperative pain control following oral surgery--is it affected by polymorphism of the mu-opioid receptor? Anesth Prog. 2010;57(4):145–9. doi: 10.2344/0003-3006-57.4.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romberg RR, et al. Polymorphism of mu-opioid receptor gene (OPRM1:c.118A>G) does not protect against opioid-induced respiratory depression despite reduced analgesic response. Anesthesiology. 2005;102(3):522–30. doi: 10.1097/00000542-200503000-00008. [DOI] [PubMed] [Google Scholar]
- 23.Fukuda K, et al. Association between OPRM1 gene polymorphisms and fentanyl sensitivity in patients undergoing painful cosmetic surgery. Pain. 2009;147(1-3):194–201. doi: 10.1016/j.pain.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Campa D, et al. Association of ABCB1/MDR1 and OPRM1 gene polymorphisms with morphine pain relief. Clin Pharmacol Ther. 2008;83(4):559–66. doi: 10.1038/sj.clpt.6100385. [DOI] [PubMed] [Google Scholar]
- 25.Reyes-Gibby CC, et al. Exploring joint effects of genes and the clinical efficacy of morphine for cancer pain: OPRM1 and COMT gene. Pain. 2007;130(1-2):25–30. doi: 10.1016/j.pain.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lotsch J, et al. Cross-sectional analysis of the influence of currently known pharmacogenetic modulators on opioid therapy in outpatient pain centers. Pharmacogenet Genomics. 2009;19(6):429–36. doi: 10.1097/fpc.0b013e32832b89da. [DOI] [PubMed] [Google Scholar]
- 27.Zwisler ST, et al. The antinociceptive effect and adverse drug reactions of oxycodone in human experimental pain in relation to genetic variations in the OPRM1 and ABCB1 genes. Fundam Clin Pharmacol. 2010;24(4):517–24. doi: 10.1111/j.1472-8206.2009.00781.x. [DOI] [PubMed] [Google Scholar]
- 28.Liu YC, Wang WS. Human mu-opioid receptor gene A118G polymorphism predicts the efficacy of tramadol/acetaminophen combination tablets (ultracet) in oxaliplatin-induced painful neuropathy. Cancer. 2012;118(6):1718–25. doi: 10.1002/cncr.26430. [DOI] [PubMed] [Google Scholar]
- 29.Oertel BG, et al. The mu-opioid receptor gene polymorphism 118A>G depletes alfentanil-induced analgesia and protects against respiratory depression in homozygous carriers. Pharmacogenet Genomics. 2006;16(9):625–36. doi: 10.1097/01.fpc.0000220566.90466.a2. [DOI] [PubMed] [Google Scholar]
- 30.Chou WY, et al. Association of mu-opioid receptor gene polymorphism (A118G) with variations in morphine consumption for analgesia after total knee arthroplasty. Acta Anaesthesiol Scand. 2006;50(7):787–92. doi: 10.1111/j.1399-6576.2006.01058.x. [DOI] [PubMed] [Google Scholar]
- 31.Klepstad P, et al. The 118 A > G polymorphism in the human mu-opioid receptor gene may increase morphine requirements in patients with pain caused by malignant disease. Acta Anaesthesiol Scand. 2004;48(10):1232–9. doi: 10.1111/j.1399-6576.2004.00517.x. [DOI] [PubMed] [Google Scholar]
- 32.Hayashida M, et al. Analgesic requirements after major abdominal surgery are associated with OPRM1 gene polymorphism genotype and haplotype. Pharmacogenomics. 2008;9(11):1605–16. doi: 10.2217/14622416.9.11.1605. [DOI] [PubMed] [Google Scholar]
- 33.Zhang W, et al. Study of the OPRM1 A118G genetic polymorphism associated with postoperative nausea and vomiting induced by fentanyl intravenous analgesia. Minerva Anestesiol. 2011;77(1):33–9. [PubMed] [Google Scholar]
- 34.Tan EC, et al. Ethnicity and OPRM variant independently predict pain perception and patient-controlled analgesia usage for post-operative pain. Mol Pain. 2009;5:32. doi: 10.1186/1744-8069-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zwisler ST, et al. Lack of Association of OPRM1 and ABCB1 Single-Nucleotide Polymorphisms to Oxycodone Response in Postoperative Pain. J Clin Pharmacol. 2011 doi: 10.1177/0091270010397729. [DOI] [PubMed] [Google Scholar]
- 36.Huehne K, et al. High post surgical opioid requirements in Crohn 's disease are not due to a general change in pain sensitivity. Eur J Pain. 2009;13(10):1036–42. doi: 10.1016/j.ejpain.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Coulbault L, et al. Environmental and genetic factors associated with morphine response in the postoperative period. Clin Pharmacol Ther. 2006;79(4):316–24. doi: 10.1016/j.clpt.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Wong CA, et al. Observational study of the effect of mu-opioid receptor genetic polymorphism on intrathecal opioid labor analgesia and post-cesarean delivery analgesia. Int J Obstet Anesth. 2010;19(3):246–53. doi: 10.1016/j.ijoa.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Klepstad P, et al. Influence from genetic variability on opioid use for cancer pain: a European genetic association study of 2294 cancer pain patients. Pain. 2011;152(5):1139–45. doi: 10.1016/j.pain.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 40.Pettersson FD, et al. The A118G single-nucleotide polymorphism of human mu-opioid receptor gene and use of labor analgesia. Reprod Sci. 2012;19(9):962–7. doi: 10.1177/1933719112438970. [DOI] [PubMed] [Google Scholar]
- 41.Landau R, et al. Genetic variability of the mu-opioid receptor influences intrathecal fentanyl analgesia requirements in laboring women. Pain. 2008;139(1):5–14. doi: 10.1016/j.pain.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Capraris A, et al. Micro opioid receptor A118G polymorphism and post-operative pain: opioids' effects on heterozygous patients. Int J Immunopathol Pharmacol. 2011;24(4):993–1004. doi: 10.1177/039463201102400417. [DOI] [PubMed] [Google Scholar]
- 43.Walter C, Lotsch J. Meta-analysis of the relevance of the OPRM1 118A>G genetic variant for pain treatment. Pain. 2009;146(3):270–5. doi: 10.1016/j.pain.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Ochroch EA, et al. Natural variation in the mu-opioid gene OPRM1 predicts increased pain on third day after thoracotomy. Clin J Pain. 2012;28(9):747–54. doi: 10.1097/AJP.0b013e3182442b1c. [DOI] [PubMed] [Google Scholar]
- 45.Laugsand EA, et al. Clinical and genetic factors associated with nausea and vomiting in cancer patients receiving opioids. Eur J Cancer. 2011;47(11):1682–91. doi: 10.1016/j.ejca.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 46.Droney JM, et al. Analgesia and central side-effects: two separate dimensions of morphine response. Br J Clin Pharmacol. 2012 doi: 10.1111/bcp.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai FF, et al. Human opioid mu-receptor A118G polymorphism may protect against central pruritus by epidural morphine for post-cesarean analgesia. Acta Anaesthesiol Scand. 2010;54(10):1265–9. doi: 10.1111/j.1399-6576.2010.02310.x. [DOI] [PubMed] [Google Scholar]
- 48.Volkow ND, et al. Neurochemical mechanisms underlying responses to psychostimulants. NIDA Res Monogr. 1996;159:322–48. [PubMed] [Google Scholar]
- 49.Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35(1):217–38. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar D, Chakraborty J, Das S. Epistatic effects between variants of kappa-opioid receptor gene and A118G of mu-opioid receptor gene increase susceptibility to addiction in Indian population. Prog Neuropsychopharmacol Biol Psychiatry. 2012;36(2):225–30. doi: 10.1016/j.pnpbp.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 51.Deb I, et al. Single-nucleotide polymorphism (A118G) in exon 1 of OPRM1 gene causes alteration in downstream signaling by mu-opioid receptor and may contribute to the genetic risk for addiction. J Neurochem. 2010;112(2):486–96. doi: 10.1111/j.1471-4159.2009.06472.x. [DOI] [PubMed] [Google Scholar]
- 52.Kapur S, et al. A118g polymorphism in mu opioid receptor gene (oprm1): association with opiate addiction in subjects of Indian origin. J Integr Neurosci. 2007;6(4):511–22. doi: 10.1142/s0219635207001635. [DOI] [PubMed] [Google Scholar]
- 53.Tan EC, et al. Mu opioid receptor gene polymorphisms and heroin dependence in Asian populations. Neuroreport. 2003;14(4):569–72. doi: 10.1097/00001756-200303240-00008. [DOI] [PubMed] [Google Scholar]
- 54.Levran O, et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav. 2008;7(7):720–9. doi: 10.1111/j.1601-183X.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nikolov MA, et al. No evidence of association between 118A>G OPRM1 polymorphism and heroin dependence in a large Bulgarian case-control sample. Drug Alcohol Depend. 2011;117(1):62–5. doi: 10.1016/j.drugalcdep.2010.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franke P, et al. Nonreplication of association between mu-opioid-receptor gene (OPRM1) A118G polymorphism and substance dependence. Am J Med Genet. 2001;105(1):114–9. [PubMed] [Google Scholar]
- 57.Crowley JJ, et al. A genetic association study of the mu opioid receptor and severe opioid dependence. Psychiatr Genet. 2003;13(3):169–73. doi: 10.1097/00041444-200309000-00006. [DOI] [PubMed] [Google Scholar]
- 58.Nagaya D, et al. A118G mu opioid receptor polymorphism among drug addicts in Malaysia. J Integr Neurosci. 2012;11(1):117–22. doi: 10.1142/S0219635212500082. [DOI] [PubMed] [Google Scholar]
- 59.Glatt SJ, et al. Evaluation of OPRM1 variants in heroin dependence by family-based association testing and meta-analysis. Drug Alcohol Depend. 2007;90(2-3):159–65. doi: 10.1016/j.drugalcdep.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi J, et al. Sequence variations in the mu-opioid receptor gene (OPRM1) associated with human addiction to heroin. Hum Mutat. 2002;19(4):459–60. doi: 10.1002/humu.9026. [DOI] [PubMed] [Google Scholar]
- 61.Levran O, et al. Heroin addiction in African Americans: a hypothesis-driven association study. Genes Brain Behav. 2009;8(5):531–40. doi: 10.1111/j.1601-183X.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fudala PJ, et al. Effects of buprenorphine and naloxone in morphine-stabilized opioid addicts. Drug Alcohol Depend. 1998;50(1):1–8. doi: 10.1016/s0376-8716(98)00008-8. [DOI] [PubMed] [Google Scholar]
- 63.Mendelson J, et al. Buprenorphine and naloxone combinations: the effects of three dose ratios in morphine-stabilized, opiate-dependent volunteers. Psychopharmacology (Berl) 1999;141(1):37–46. doi: 10.1007/s002130050804. [DOI] [PubMed] [Google Scholar]
- 64.Fonseca F, et al. Response to methadone maintenance treatment is associated with the MYOCD and GRM6 genes. Mol Diagn Ther. 2010;14(3):171–8. doi: 10.1007/BF03256370. [DOI] [PubMed] [Google Scholar]
- 65.Crettol S, et al. Association of dopamine and opioid receptor genetic polymorphisms with response to methadone maintenance treatment. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32(7):1722–7. doi: 10.1016/j.pnpbp.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 66.Hung CC, et al. Impact of genetic polymorphisms in ABCB1, CYP2B6, OPRM1, ANKK1 and DRD2 genes on methadone therapy in Han Chinese patients. Pharmacogenomics. 2011;12(11):1525–33. doi: 10.2217/pgs.11.96. [DOI] [PubMed] [Google Scholar]
- 67.Barratt DT, et al. ABCB1 haplotype and OPRM1 118A > G genotype interaction in methadone maintenance treatment pharmacogenetics. Pharmgenomics Pers Med. 2012;5:53–62. doi: 10.2147/PGPM.S29272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kakko J, et al. Mood and neuroendocrine response to a chemical stressor, metyrapone, in buprenorphine-maintained heroin dependence. Biol Psychiatry. 2008;63(2):172–7. doi: 10.1016/j.biopsych.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 69.Lotsch J, et al. Modulation of the central nervous effects of levomethadone by genetic polymorphisms potentially affecting its metabolism, distribution, and drug action. Clin Pharmacol Ther. 2006;79(1):72–89. doi: 10.1016/j.clpt.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 70.Wang SC, et al. Genetic polymorphisms in the opioid receptor mu1 gene are associated with changes in libido and insomnia in methadone maintenance patients. Eur Neuropsychopharmacol. 2012;22(10):695–703. doi: 10.1016/j.euroneuro.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 71.Zhang L, Kendler KS, Chen X. The mu-opioid receptor gene and smoking initiation and nicotine dependence. Behav Brain Funct. 2006;2:28. doi: 10.1186/1744-9081-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Verde Z, et al. 'Smoking genes': a genetic association study. PLoS One. 2011;6(10):e26668. doi: 10.1371/journal.pone.0026668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z, et al. Nicotine abstinence-induced cerebral blood flow changes by genotype. Neurosci Lett. 2008;438(3):275–80. doi: 10.1016/j.neulet.2008.04.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perkins KA, et al. Dopamine and opioid gene variants are associated with increased smoking reward and reinforcement owing to negative mood. Behav Pharmacol. 2008;19(5-6):641–9. doi: 10.1097/FBP.0b013e32830c367c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ray R, et al. Association of OPRM1 A118G variant with the relative reinforcing value of nicotine. Psychopharmacology (Berl) 2006;188(3):355–63. doi: 10.1007/s00213-006-0504-2. [DOI] [PubMed] [Google Scholar]
- 76.Domino EF, et al. Tobacco smoking produces greater striatal dopamine release in G-allele carriers with mu opioid receptor A118G polymorphism. Prog Neuropsychopharmacol Biol Psychiatry. 2012;38(2):236–40. doi: 10.1016/j.pnpbp.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 77.Lerman C, et al. The functional mu opioid receptor (OPRM1) Asn40Asp variant predicts short-term response to nicotine replacement therapy in a clinical trial. Pharmacogenomics J. 2004;4(3):184–92. doi: 10.1038/sj.tpj.6500238. [DOI] [PubMed] [Google Scholar]
- 78.Ray R, et al. Genetic variation in mu-opioid-receptor-interacting proteins and smoking cessation in a nicotine replacement therapy trial. Nicotine Tob Res. 2007;9(11):1237–41. doi: 10.1080/14622200701648367. [DOI] [PubMed] [Google Scholar]
- 79.Munafo MR, et al. Association of the mu-opioid receptor gene with smoking cessation. Pharmacogenomics J. 2007;7(5):353–61. doi: 10.1038/sj.tpj.6500432. [DOI] [PubMed] [Google Scholar]
- 80.Munafo MR, et al. Lack of Association of OPRM1 Genotype and Smoking Cessation. Nicotine Tob Res. 2012 doi: 10.1093/ntr/nts174. [DOI] [PubMed] [Google Scholar]
- 81.Volpicelli JR, et al. Naltrexone in the treatment of alcohol dependence. Arch Gen Psychiatry. 1992;49(11):876–80. doi: 10.1001/archpsyc.1992.01820110040006. [DOI] [PubMed] [Google Scholar]
- 82.O'Malley SS, et al. Naltrexone decreases craving and alcohol self-administration in alcohol-dependent subjects and activates the hypothalamo-pituitary-adrenocortical axis. Psychopharmacology (Berl) 2002;160(1):19–29. doi: 10.1007/s002130100919. [DOI] [PubMed] [Google Scholar]
- 83.Miranda R, et al. Initial evidence of an association between OPRM1 and adolescent alcohol misuse. Alcohol Clin Exp Res. 2010;34(1):112–22. doi: 10.1111/j.1530-0277.2009.01073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miranda R, Jr, et al. Preliminary Evidence for a Gene-Environment Interaction in Predicting Alcohol Use Disorders in Adolescents. Alcohol Clin Exp Res. 2012 doi: 10.1111/j.1530-0277.2012.01897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koller G, et al. Possible association between OPRM1 genetic variance at the 118 locus and alcohol dependence in a large treatment sample: relationship to alcohol dependence symptoms. Alcohol Clin Exp Res. 2012;36(7):1230–6. doi: 10.1111/j.1530-0277.2011.01714.x. [DOI] [PubMed] [Google Scholar]
- 86.Bart G, et al. Increased attributable risk related to a functional mu-opioid receptor gene polymorphism in association with alcohol dependence in central Sweden. Neuropsychopharmacology. 2005;30(2):417–22. doi: 10.1038/sj.npp.1300598. [DOI] [PubMed] [Google Scholar]
- 87.Bergen AW, et al. Mu opioid receptor gene variants: lack of association with alcohol dependence. Mol Psychiatry. 1997;2(6):490–4. doi: 10.1038/sj.mp.4000331. [DOI] [PubMed] [Google Scholar]
- 88.Kranzler HR, et al. Association of alcohol or other drug dependence with alleles of the mu opioid receptor gene (OPRM1) Alcohol Clin Exp Res. 1998;22(6):1359–62. [PubMed] [Google Scholar]
- 89.Olfson E, Bierut LJ. Convergence of genome-wide association and candidate gene studies for alcoholism. Alcohol Clin Exp Res. 2012;36(12):2086–94. doi: 10.1111/j.1530-0277.2012.01843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xuei X, et al. The opioid system in alcohol and drug dependence: family-based association study. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(7):877–84. doi: 10.1002/ajmg.b.30531. [DOI] [PubMed] [Google Scholar]
- 91.Chen D, et al. Ethnic-specific meta-analyses of association between the OPRM1 A118G polymorphism and alcohol dependence among Asians and Caucasians. Drug Alcohol Depend. 2012;123(1-3):1–6. doi: 10.1016/j.drugalcdep.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 92.Zhang H, et al. Association between two mu-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet. 2006;15(6):807–19. doi: 10.1093/hmg/ddl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Crystal HA, et al. A C17T polymorphism in the mu opiate receptor is associated with quantitative measures of drug use in African American women. Addict Biol. 2012;17(1):181–91. doi: 10.1111/j.1369-1600.2010.00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nishizawa D, et al. Association of mu-opioid receptor gene polymorphism A118G with alcohol dependence in a Japanese population. Neuropsychobiology. 2006;53(3):137–41. doi: 10.1159/000093099. [DOI] [PubMed] [Google Scholar]
- 95.Kim SG, et al. Association of functional opioid receptor genotypes with alcohol dependence in Koreans. Alcohol Clin Exp Res. 2004;28(7):986–90. doi: 10.1097/01.alc.0000130803.62768.ab. [DOI] [PubMed] [Google Scholar]
- 96.Kim SA, et al. Association of polymorphisms in nicotinic acetylcholine receptor alpha 4 subunit gene (CHRNA4), mu-opioid receptor gene (OPRM1), and ethanol-metabolizing enzyme genes with alcoholism in Korean patients. Alcohol. 2004;34(2-3):115–20. doi: 10.1016/j.alcohol.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 97.Loh el W, et al. Endogenous opioid receptor genes and alcohol dependence among Taiwanese Han. Alcohol Clin Exp Res. 2004;28(1):15–9. doi: 10.1097/01.ALC.0000106303.41755.B8. [DOI] [PubMed] [Google Scholar]
- 98.Kranzler HR, et al. Variation in OPRM1 moderates the effect of desire to drink on subsequent drinking and its attenuation by naltrexone treatment. Addict Biol. 2012 doi: 10.1111/j.1369-1600.2012.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ray LA. Stress-induced and cue-induced craving for alcohol in heavy drinkers: Preliminary evidence of genetic moderation by the OPRM1 and CRH-BP genes. Alcohol Clin Exp Res. 2011;35(1):166–74. doi: 10.1111/j.1530-0277.2010.01333.x. [DOI] [PubMed] [Google Scholar]
- 100.van den Wildenberg E, et al. A functional polymorphism of the mu-opioid receptor gene (OPRM1) influences cue-induced craving for alcohol in male heavy drinkers. Alcohol Clin Exp Res. 2007;31(1):1–10. doi: 10.1111/j.1530-0277.2006.00258.x. [DOI] [PubMed] [Google Scholar]
- 101.Ray LA, Hutchison KE. Effects of naltrexone on alcohol sensitivity and genetic moderators of medication response: a double-blind placebo-controlled study. Arch Gen Psychiatry. 2007;64(9):1069–77. doi: 10.1001/archpsyc.64.9.1069. [DOI] [PubMed] [Google Scholar]
- 102.Filbey FM, et al. Differential neural response to alcohol priming and alcohol taste cues is associated with DRD4 VNTR and OPRM1 genotypes. Alcohol Clin Exp Res. 2008;32(7):1113–23. doi: 10.1111/j.1530-0277.2008.00692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Filbey FM, et al. Exposure to the taste of alcohol elicits activation of the mesocorticolimbic neurocircuitry. Neuropsychopharmacology. 2008;33(6):1391–401. doi: 10.1038/sj.npp.1301513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ray LA, Hutchison KE. A polymorphism of the mu-opioid receptor gene (OPRM1) and sensitivity to the effects of alcohol in humans. Alcohol Clin Exp Res. 2004;28(12):1789–95. doi: 10.1097/01.alc.0000148114.34000.b9. [DOI] [PubMed] [Google Scholar]
- 105.Ramchandani VA, et al. A genetic determinant of the striatal dopamine response to alcohol in men. Mol Psychiatry. 2011;16(8):809–17. doi: 10.1038/mp.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oslin DW, et al. A functional polymorphism of the mu-opioid receptor gene is associated with naltrexone response in alcohol-dependent patients. Neuropsychopharmacology. 2003;28(8):1546–52. doi: 10.1038/sj.npp.1300219. [DOI] [PubMed] [Google Scholar]
- 107.Chamorro AJ, et al. Association of micro-opioid receptor (OPRM1) gene polymorphism with response to naltrexone in alcohol dependence: a systematic review and meta-analysis. Addict Biol. 2012;17(3):505–12. doi: 10.1111/j.1369-1600.2012.00442.x. [DOI] [PubMed] [Google Scholar]
- 108.Johnson BA. Update on neuropharmacological treatments for alcoholism: scientific basis and clinical findings. Biochem Pharmacol. 2008;75(1):34–56. doi: 10.1016/j.bcp.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim SG, et al. A micro opioid receptor gene polymorphism (A118G) and naltrexone treatment response in adherent Korean alcohol-dependent patients. Psychopharmacology (Berl) 2009;201(4):611–8. doi: 10.1007/s00213-008-1330-5. [DOI] [PubMed] [Google Scholar]
- 110.Anton RF, et al. An evaluation of mu-opioid receptor (OPRM1) as a predictor of naltrexone response in the treatment of alcohol dependence: results from the Combined Pharmacotherapies and Behavioral Interventions for Alcohol Dependence (COMBINE) study. Arch Gen Psychiatry. 2008;65(2):135–44. doi: 10.1001/archpsyc.65.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Oroszi G, et al. OPRM1 Asn40Asp predicts response to naltrexone treatment: a haplotype-based approach. Alcohol Clin Exp Res. 2009;33(3):383–93. doi: 10.1111/j.1530-0277.2008.00846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Coller JK, et al. OPRM1 A118G genotype fails to predict the effectiveness of naltrexone treatment for alcohol dependence. Pharmacogenet Genomics. 2011;21(12):902–5. doi: 10.1097/FPC.0b013e32834c5445. [DOI] [PubMed] [Google Scholar]
- 113.Gelernter J, et al. Opioid receptor gene (OPRM1, OPRK1, and OPRD1) variants and response to naltrexone treatment for alcohol dependence: results from the VA Cooperative Study. Alcohol Clin Exp Res. 2007;31(4):555–63. doi: 10.1111/j.1530-0277.2007.00339.x. [DOI] [PubMed] [Google Scholar]
- 114.Tidey JW, et al. Moderators of naltrexone's effects on drinking, urge, and alcohol effects in non-treatment-seeking heavy drinkers in the natural environment. Alcohol Clin Exp Res. 2008;32(1):58–66. doi: 10.1111/j.1530-0277.2007.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Anton RF, et al. Naltrexone Modification of Drinking Effects in a Subacute Treatment and Bar-Lab Paradigm: Influence of OPRM1 and Dopamine Transporter (SLC6A3) Genes. Alcohol Clin Exp Res. 2012;36(11):2000–7. doi: 10.1111/j.1530-0277.2012.01807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ray LA, et al. Pharmacogenetics of naltrexone in asian americans: a randomized placebo-controlled laboratory study. Neuropsychopharmacology. 2012;37(2):445–55. doi: 10.1038/npp.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McGeary JE, et al. Genetic moderators of naltrexone 's effects on alcohol cue reactivity. Alcohol Clin Exp Res. 2006;30(8):1288–96. doi: 10.1111/j.1530-0277.2006.00156.x. [DOI] [PubMed] [Google Scholar]
- 118.Arias AJ, et al. Effects of opioid receptor gene variation on targeted nalmefene treatment in heavy drinkers. Alcohol Clin Exp Res. 2008;32(7):1159–66. doi: 10.1111/j.1530-0277.2008.00735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nielsen DA, Kreek MJ. Common and specific liability to addiction: approaches to association studies of opioid addiction. Drug Alcohol Depend. 2012;123(Suppl 1):S33–41. doi: 10.1016/j.drugalcdep.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]


